Transcriptional analyses of Acinetobacter baumannii ATCC 17978 showed that the expression of A1S_2091 was enhanced in cells cultured in darkness at 24°C through a process that depended on the BlsA photoreceptor. Disruption of A1S_2091, a component of the A1S_2088-A1S_2091 polycistronic operon predicted to code for a type I chaperone/usher pilus assembly system, abolished surface motility and pellicle formation but significantly enhanced biofilm formation on plastic by bacteria cultured in darkness.
KEYWORDS: Acinetobacter baumannii, BlsA, temperature, G. mellonella, A549 cells
ABSTRACT
Transcriptional analyses of Acinetobacter baumannii ATCC 17978 showed that the expression of A1S_2091 was enhanced in cells cultured in darkness at 24°C through a process that depended on the BlsA photoreceptor. Disruption of A1S_2091, a component of the A1S_2088-A1S_2091 polycistronic operon predicted to code for a type I chaperone/usher pilus assembly system, abolished surface motility and pellicle formation but significantly enhanced biofilm formation on plastic by bacteria cultured in darkness. Based on these observations, the A1S_2088-A1S_2091 operon was named the photoregulated pilus ABCD (prpABCD) operon, with A1S_2091 coding for the PrpA pilin subunit. Unexpectedly, comparative analyses of ATCC 17978 and prpA isogenic mutant cells cultured at 37°C showed the expression of light-regulated biofilm biogenesis and motility functions under a temperature condition that drastically affects BlsA production and its light-sensing activity. These assays also suggest that ATCC 17978 cells produce alternative light-regulated adhesins and/or pilus systems that enhance bacterial adhesion and biofilm formation at both 24°C and 37°C on plastic as well as on the surface of polarized A549 alveolar epithelial cells, where the formation of bacterial filaments and cell chains was significantly enhanced. The inactivation of prpA also resulted in a significant reduction in virulence when tested by using the Galleria mellonella virulence model. All these observations provide strong evidence showing the capacity of A. baumannii to sense light and interact with biotic and abiotic surfaces using undetermined alternative sensing and regulatory systems as well as alternative adherence and motility cellular functions that allow this pathogen to persist in different ecological niches.
INTRODUCTION
Acinetobacter bacteria are Gram-negative, aerobic, non-spore-forming bacteria that do not require complex media for growth (1). Members of this genus, which comprises more than 30 species (2), are present in the environment (3) and frequently found in different food sources, even under refrigerated conditions or after irradiation (4). They are also normal inhabitants of the human skin and are widely distributed in hospital environments (5, 6). Not surprisingly, these microorganisms have been implicated in a variety of food spoilage processes and human infections (7, 8). These diseases involve a group of isolates represented by members of the “A. calcoaceticus-A. baumannii (Acb) complex,” which are relevant to human health since they are associated with intra- and extranosocomial infections worldwide (9). A. baumannii is the most medically relevant member of the Acb complex since it has been associated with hospital infections as well as community-acquired and injury infections, particularly those caused after natural disasters or wound infections in military personnel deployed to Iraq and Afghanistan (10). Furthermore, A. baumannii has been isolated from a wide range of environmental sources and samples, including water and aquaculture environments (11), soil (12), different food sources (13), animals (14, 15), and insects (16), all of which could be reservoirs for this bacterium outside the hospital environment (10).
As a facultative pathogen that lives in different ecological niches, A. baumannii uses a wide range of extracellular signals to adapt to either an environmental or a host lifestyle, with each style playing a critical role in the less understood life cycle and ecology of this bacterium. Accordingly, our work and that of others showed that A. baumannii senses and responds to extracellular signals, including iron limitation, changes in salt concentrations, desiccation stress, and the presence of antibiotics and disinfectants (17). These signals affect A. baumannii's ability to interact with the host, form biofilms on abiotic surfaces, and display motility on semisolid media (18–21). However, these are only some of the numerous extracellular signals that modulate the physiology of A. baumannii and its interaction with environments in which this microorganism is normally found. Thus, while we were studying the effects of factors such as temperature and medium composition on the expression of some of the above-mentioned phenotypes, we unexpectedly observed that A. baumannii senses and responds to light by differentially affecting biofilm formation, motility, and the interaction with Candida albicans, a response that is widespread among different members of the Acinetobacter genus (22–24). The ability of A. baumannii to sense and respond to light when cultured at 24°C depends on the expression of blsA, a gene that codes for a “short” photoreceptor that harbors an N-terminal blue-light sensing using flavin (BLUF) domain, which binds the chromophore flavin adenine dinucleotide (FAD) but lacks identifiable regulatory output motifs (24, 25). Albeit limited, there is some understanding of the molecular interactions that play a role in the photosensing functions exerted by BlsA (26). In contrast, almost nothing is known regarding the mechanisms by which this protein works as a regulator and the cellular targets that it affects in response to illumination.
In this report, we show that the light-regulated motility and biofilm and pellicle formation responses displayed by A. baumannii ATCC 17978 (17978) depend on the active expression of the photoregulated pilus assembly system PrpABCD in a BlsA-mediated process when bacteria are cultured at 24°C. PrpABCD also proved to be critical for the interaction of polarized A549 human alveolar epithelial cells with 17978 and the virulence of this strain to Galleria mellonella when tested at 37°C, a temperature at which BlsA-mediated light regulation is lost because of a drastic reduction in blsA expression and an alteration of the BlsA reversible photocycle at temperatures higher than 30°C (24, 27). However, the ability of a 17978 prpA mutant to display light-regulated surface motility and biofilm responses at 37°C indicates that undetermined additional 17978 factors involved in these responses are controlled by light in a BlsA-independent regulatory process that remains to be identified and characterized.
RESULTS
Identification of a light-regulated locus coding for a predicted type I pilus.
Our initial analysis of transcriptome sequencing (RNA-Seq) data collected from 17978 cells cultured in swimming broth (SB) in the presence or absence of light (B. A. Arivett, S. E. Fiester, C. C. R. Wood, and L. A. Actis, unpublished data) led to the identification of the A1S_2088-A1S_2091 4-gene locus (Fig. 1A), in which the expression of all of its components was significantly repressed when bacteria were cultured at 24°C under illumination (see Table S1 in the supplemental material). The initial annotation of the 17978 genome describes A1S_2089 and A1S_2090 as genes coding for putative fimbrial usher and P pilus assembly proteins, respectively (28). Our in silico analysis using several bioinformatics tools, including Phyre2, predicted that (i) A1S_2091 is a 147-amino-acid protein significantly related to type I fimbrial proteins, (ii) A1S_2090 is a 234-amino-acid protein significantly related to fimbrial chaperone proteins, (iii) A1S_2089 is an 819-amino-acid protein significantly related to type I fimbrial usher proteins, and (iv) A1S_2088 is a 339-amino-acid protein significantly related to fimbrial tip adhesins. Thus, we named this 4-gene cluster the photoregulated pilus ABCD (prpABCD) operon, which has the potential for assembling a type I pilus in A. baumannii differentially produced in response to illumination. Interestingly, BLAST searches proved that the prpABCD operon is present in the genome of a large number of A. baumannii strains, and analysis of strain MAR002 showed that this biofilm-hyperproducing isolate harbors a prpABCD homolog, which was annotated as the LH9211085-LH9211080-LH9211075-LH9211070 operon (29). The prpABCD was also detected in the genomes of different members of the Acinetobacter genus, including the clinical species A. gyllenbergii, A. pittii, and A. nosocomialis and the environmental species A. calcoaceticus, A. oleivorans, and A. nectaris.
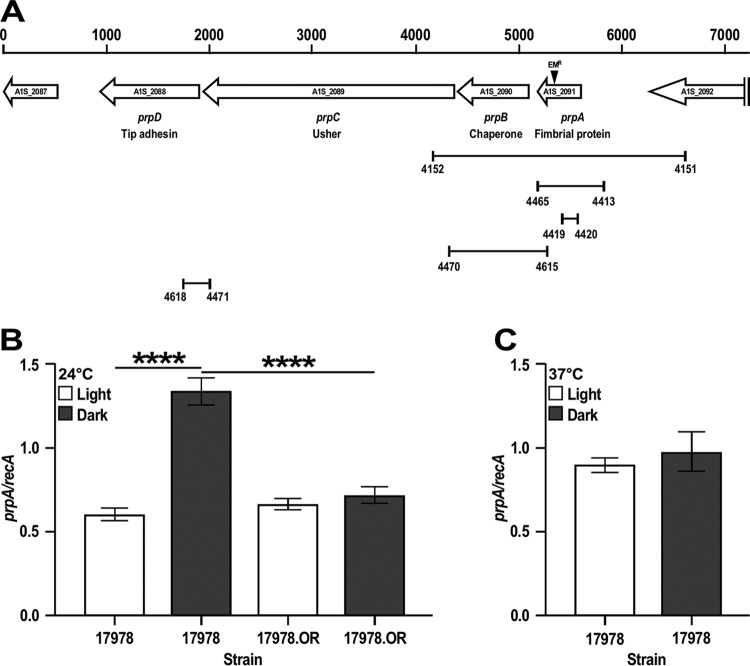
FIG 1.
Genetic organization and expression analysis of the prpABCD operon. (A) In silico analysis of the A1S_2089-A1S_2091 coding region. The horizontal arrows represent predicted coding regions and their direction of transcription. Coding regions are identified by their original genomic annotation, the names assigned in this work, and their predicted protein products. Numbers on the top of the vertical bars represent size in base pairs. The location of the insertion of the Emr DNA cassette within prpA is indicated by the black triangle. The connected vertical bars indicate primer locations and the lengths of cognate amplicons used to clone and mutagenize prpA, construct the complementing vector pMU1269 (Table 1), test the expression of prpA by qRT-PCR, and determine the polycistronic nature of the prpABCD operon. Numbers underneath the vertical bars identify primers listed in Table S2 in the supplemental material. (B) qRT-PCR of prpA using RNA isolated from 17978 and 17978.OR cells cultured in SB at 24°C under darkness or illumination. (C) qRT-PCR of prpA using RNA isolated from 17978 cells cultured in SB at 37°C under darkness or illumination. recA was used as a constitutively expressed control gene. Horizontal bars identify statistically different values (****, P ≤ 0.0001), and error bars represent the standard errors of each data set.
Taken together, these observations, which are in agreement with our reports describing light-regulated responses in A. baumannii and different members of the Acinetobacter genus (23, 24), support the potential importance of PrpABCD in the biology and physiology of a bacterium that has a remarkable ability to persist under different environmental conditions.
Expression analysis of the prp locus.
The prp locus is preceded by a 638-nucletide intergenic region that separates prpA from A1S_2092, which is predicted to code for a PepN homolog, and is followed by a 367-nucleotide (nt) intergenic region that separates prpD from A1S_2087, a gene encoding a putative glutathione S-transferase (Fig. 1A). This genomic structure together with the fact that prpA-prpB, prpB-prpC, and prpC-prpD are separated by 72-, 9-, and 5-nucleotide intergenic regions, respectively, suggest that prpABCD is a polycistronic operon. This possibility was tested by reverse transcription-PCR (RT-PCR) of total RNA isolated from 17978 wild-type bacteria cultured in darkness at 24°C. Figure S1A in the supplemental material shows that the amplification of 17978 cDNA with primers 4470 and 4615 resulted in the predicted 975-nucleotide amplicon encompassing the 3′ end of prpA and the 5′ end of prpC. The size of this amplicon matches the size of the amplicon obtained by using total DNA as a template. Similarly, PCR amplification of 17978 cDNA using primers 4471 and 4618 produced a 288-nucleotide amplicon linking the 3′ end of prpC with the 5′ end of prpD, which matches the amplicon obtained with total DNA (Fig. S1C). In contrast, no amplicons were produced when the same primer sets were used with total RNA samples that were not reverse transcribed. The same results were obtained when total RNA isolated from bacteria cultured in the presence of light was used as a template (data not shown).
The initial RNA-Seq data showing that the prpABCD locus is differentially expressed in response to light was further confirmed by RT-quantitative PCR (qRT-PCR) using recA as a constitutively expressed internal control. Our RNA-Seq data show that there are not significant differences in the transcription of recA in 17978 cells cultured under darkness or illumination (Arivett et al., unpublished). This approach showed that the transcription of prpA is increased about 2.2-fold in cells cultured in darkness compared to transcript levels detected in bacteria cultured under illumination at 24°C (Fig. 1B). This observation together with the polycistronic nature of this pilus-encoding operon are in agreement with our RNA-Seq data showing a significant reduction in the transcription of all four prp coding regions in cells cultured in the presence of light compared to data collected from cells cultured in darkness (Table S1). Since the BlsA photoreceptor protein plays a critical role in 17978 light responses (24), the transcription of prpA was further measured in the 17978 parental strain and the 17978.OR blsA::aph isogenic derivative. Figure 1B shows that the insertion inactivation of blsA results in comparable prpA transcriptional levels in bacteria cultured at 24°C in the presence and absence of light.
Taken together, these results indicate that prpABCD is a polycistronic operon that is differentially transcribed in response to light via a BlsA-mediated regulatory process when bacteria are cultured at 24°C by a mechanism that remains to be determined.
Functional analysis of the prp locus in bacteria cultured at 24°C.
The biological role of the prpABCD locus was tested by using the isogenic 17978.prpA mutant (Table 1), which harbors the insertion of a DNA cassette coding for erythromycin (Em) resistance (Emr) within a 61-bp prpA intragenic deletion (Fig. 1A) that resulted in the predicted 800-nucleotide size increase compared with the 2.4-kbp parental amplicon (see Fig. S2A in the supplemental material). As reported previously (24), the 17978 parental strain displayed surface motility when inoculated onto the surface of a swimming agar (SA) plate incubated at 24°C in darkness (Fig. 2A). In contrast, only growth at the inoculation site without detectable surface motility was observed when the 17978.prpA derivative was tested under the same experimental conditions (Fig. 2A). The effect of the prpA mutation was further examined by transmission electron microscopy (TEM) of cells lifted from SA plates incubated in darkness at 24°C. Figure 3A shows the presence of long and thin pili associated with bacteria located at the edge of the motility zone. These pili, some of which formed bundles (Fig. 3B, white arrow), were clearly detected in the area located immediately in front of the motility zone. In contrast, none of the pilus structures produced by the parental strain could be detected in 17978.prpA samples collected and analyzed under the same experimental conditions (Fig. 3C).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| A. baumannii | ||
| ATCC 17978 | Clinical isolate | ATCC |
| 17978.OR | blsA::aph derivative of ATCC 17978; Kmr | 24 |
| 17978.prpA | prpA::ermAM derivative of ATCC 17978; Emr | This work |
| 17978.prpA.EV | prpA mutant harboring pWH1266; Emr Tetr | This work |
| 17978.prpA.CV | prpA mutant harboring pMU1269; Ampr Emr Tets | This work |
| E. coli | ||
| DH5α | DNA recombinant methods | Gibco-BRL |
| Top10 | DNA recombinant methods | Life Technologies |
| Lactococcus lactis subsp. cremoris | Strain MG1363 harboring pIL252; Emr | H. Smidt and T. Kruse |
| Plasmids | ||
| pCR8-TOPO | Gateway cloning vector; Spr | Life Technologies |
| pEX100T | Suicide vector for allelic exchange; Ampr | ATCC |
| pIL252 | Cloning vector; source of ermAM encoding Emr | H. Smidt and T. Kruse |
| pWH1266 | A. baumannii-E. coli shuttle vector; Ampr Tetr | 42 |
| pMU1135 | pCR8-TOPO harboring prpA; Spr | This work |
| pMU1142 | pCR8-TOPO harboring prpA::ermAM; Spr Emr | This work |
| pMU1143 | pEX100T harboring prpA::ermAM; Ampr Emr | This work |
| pMU1269 | pWH1266 harboring prpA; Ampr Tets | This work |
Ampr, ampicillin resistance; Emr, erythromycin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance; Tetr, tetracycline resistance; Tets, tetracycline sensitivity.
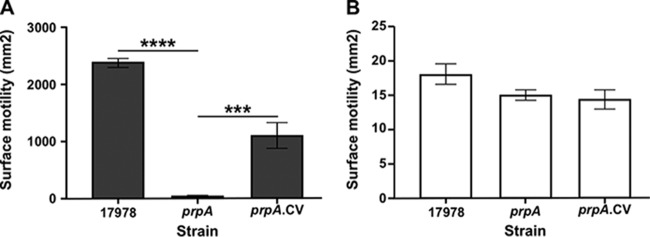
FIG 2.
Role of PrpA in surface motility at 24°C. Cells of the parental strain (17978) and the prpA insertion mutant (prpA) and its complemented derivative (prpA.CV) were surface inoculated onto SA plates and incubated at 24°C under darkness (A) or illumination (B). Horizontal bars in panel A represent statistically different values (****, P ≤ 0.0001; ***, P ≤ 0.001) between cognate samples, and error bars represent the standard errors of each data set.
FIG 3.
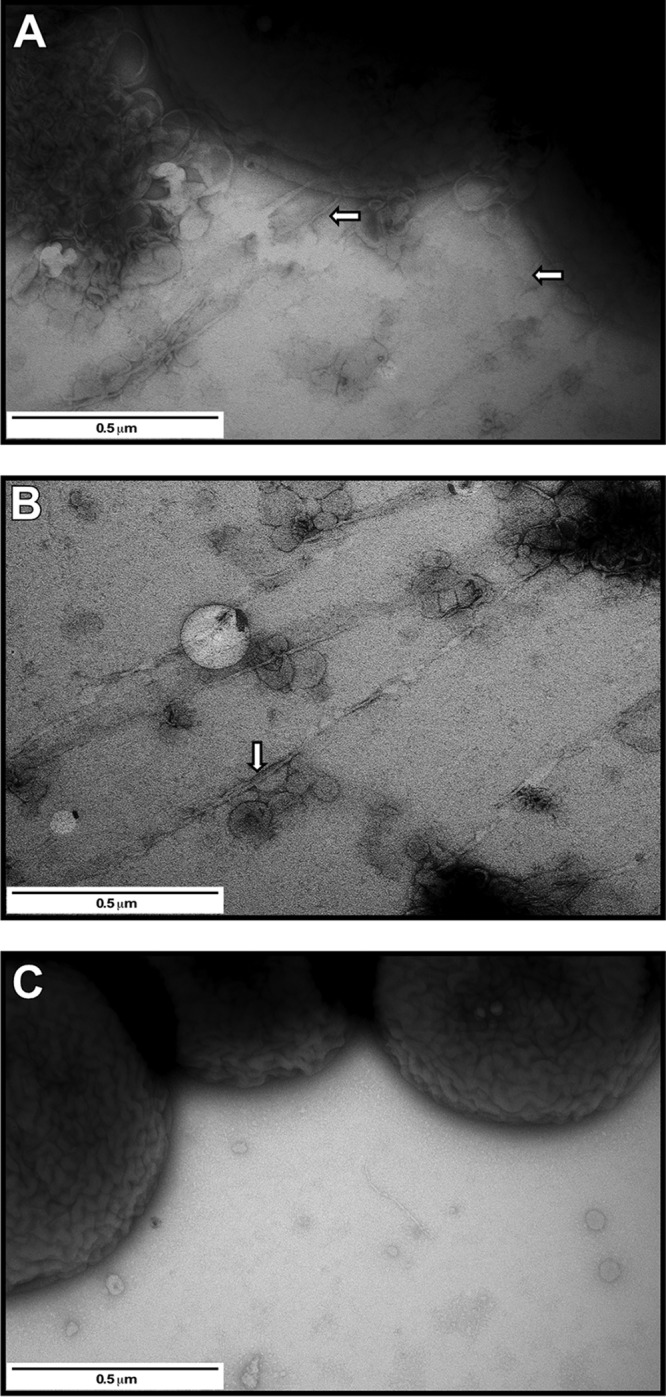
TEM analysis of samples collected from the surface of SA plates. (A and B) TEM grids were placed substrate-side down on 17978 cells at the edge of the motility zone (A) or immediately in front of this zone, where cells were not visible (B). (C) Micrograph of cells lifted from the edge of the colony formed by the nonmotile 17978.prpA mutant. The white arrows in panels A and B identify pili attached to bacterial cells as well as pili present in front of the motility zone. Plates were incubated overnight at 24°C under darkness. Images are representative of at least three fields of view for each sample at a ×50,000 magnification.
Electroporation of the complementing vector pMU1269, a derivative of the pWH1266 shuttle cloning vector that harbors the prpA parental allele expressed from its native promoter (Table 1), into 17978.prpA mutant cells (17978.prpA.CV) significantly increased surface motility (P ≤ 0.05) albeit not to wild-type levels (Fig. 2A). This response by 17978.prpA.CV cells could be due to a potential polar effect on the expression of downstream coding regions because of the insertion of the Emr cassette within prpA. RT-PCR of RNA isolated from 17978.prpA cells with primer pairs 4470-4615 and 4471-4618 (Fig. 1A), which were used to determine the polycistronic nature of the prpABCD operon (Fig. S1A and S1C), produced prpA-prpC and prpC-prpD intergenic amplicons of the same sizes as those detected by using 17978 RNA as a template (compare Fig. S1A and S1B and Fig. S1C and S1D, respectively). These results demonstrate that the insertion of the Emr DNA cassette within prpA did not cause polar effects on the prpBCD downstream coding regions. Alternatively, the reduced motility response displayed by 17978.prpA.CV cells could be due to a gene dosage effect because of the genetic complementation of this mutant with a plasmid copy of the prpA parental allele. The qRT-PCR data shown in Fig. S1E prove that the electroporation of pMU1269 indeed results in a 2.2-fold increase (P = 0.008) in prpA transcript levels compared with the parental 17978 strain. Thus, the higher copy number of prpA in the complemented strain most likely affects the stoichiometry of the PrpABCD protein components needed for proper motility functions. As expected from our previous observations (24), 17978 cells displayed no motility when SA plates were incubated at 24°C under illumination, a response that was also observed with 17978.prpA and 17978.prpA.CV bacteria (Fig. 2B). The motility phenotype of the 17978.prpA.EV strain, which was electroporated with the empty vector pWH1266, was comparable to that displayed by the 17978.prpA mutant when tested under the same experimental conditions (data not shown). Taken together, these results show that the PrpABCD pilus assembly system is required for 17978 bacterial cells to display motility on a semisolid surface in the absence of light. Growth analysis of the 17978 parental strain and the prpA derivative proved that the insertion inactivation of this coding region did not result in significant growth defects when the strains were cultured in SB (Fig. S2B) or Luria-Bertani (LB) broth (data not shown) without selective pressure.
Biofilm assays showed that 17978 cells were able to form readily detectable pellicles on the surface of SB cultures statically incubated at 24°C in darkness (Fig. 4A). In contrast, 17978.prpA cells produced no detectable pellicles when incubated under the same experimental conditions. As expected from our previous observations (24), illumination resulted in no detectable pellicle formation by cells of the 17978 parental strain as well as by cells of the 17978.prpA mutant when the SB cultures were statically incubated in the presence of light (data not shown).
FIG 4.
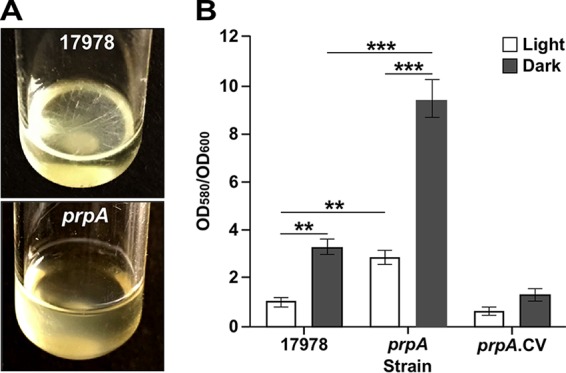
Role of PrpA in pellicle and biofilm formation in response to illumination at 24°C. (A) Pellicle formation by the parental strain (17978) and the prpA insertion mutant (prpA) on the surface of SB cultures statically incubated at 24°C in darkness. (B) Biofilm formation by the same two strains and the complemented prpA derivative (prpA.CV) on microtiter plate wells statically incubated at 24°C under darkness or illumination. Horizontal bars identify statistically different values (**, P ≤ 0.01; ***, P ≤ 0.001), and error bars represent the standard errors of each data set.
The biofilm assays showed that the inactivation of prpA significantly affected biofilm biogenesis on plastic, with 2.5-fold and 3.0-fold increases under illumination and darkness, respectively, when the 17978 and 17978.prpA strains were compared side by side under the same experimental conditions (Fig. 4B). Genetic complementation of the 17978.prpA derivative with a plasmid copy of the prpA parental allele reduced biofilm formation to levels comparable to those produced by 17978 under illumination but lower than those produced by the parental strain under darkness. The latter response could also be due to a prpA gene dosage effect that affects the proper stoichiometry and function of the PrpABCD pilus assembly system.
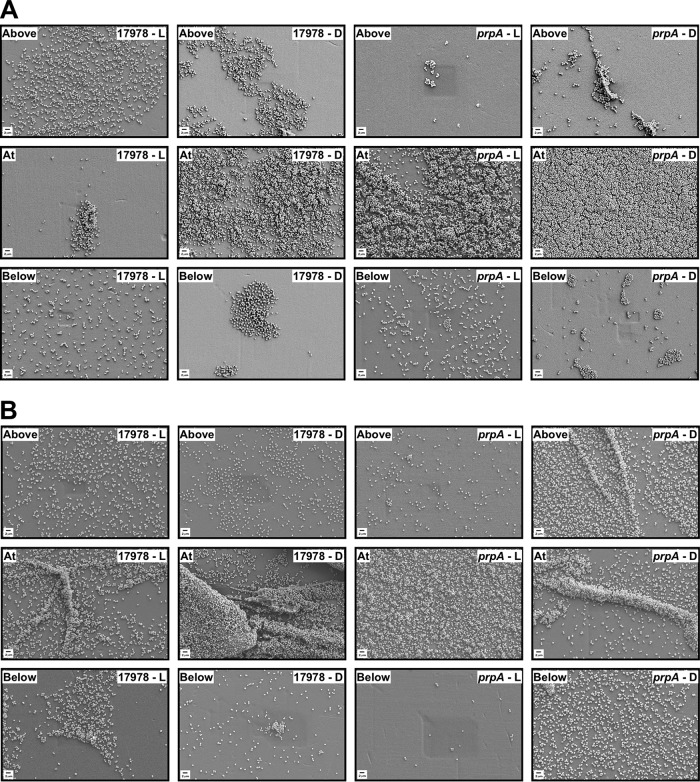
Scanning electron microscopy (SEM) analysis of plastic coverslips semisubmerged in SB inoculated with 17978 bacteria showed that this strain produced denser and more-structured biofilms under darkness than under illuminated conditions, particularly at the liquid-air interface (Fig. 5A). A similar outcome was detected with the 17978.prp strain, which covered the surface of the coverslips with a more uniform and denser layer of cells at the liquid-air interface when incubated in darkness (Fig. 5A). This analysis also showed that overall, the 17978.prp insertion derivative produced more biofilm than the 17978 strain under both darkness and illumination, an observation that matches the biofilm data collected by using standard crystal violet assays, as shown in Fig. 4B.
FIG 5.
SEM analysis of biofilms formed on plastic. Plastic coverslips were semisubmerged in SB inoculated with 17978 or prpA bacteria and statically incubated at 24°C (A) or 37°C (B) under illumination (L) or darkness (D) for 96 h or 48 h, respectively. Samples were fixed, gold coated, and examined by SEM. Images taken above, at, and below the air-liquid interface are representative of at least three fields of view for each sample at a ×5,000 magnification.
Taken together, these observations underscore the positive roles that the prpABCD locus and the products that it codes for play in the capacity of 17978 cells to differentially display surface motility and produce pellicles at the surface of static broth cultures incubated at 24°C. On the other hand, the biofilm results indicate that cellular products other than those coded for by prpABCD could be responsible for the light-mediated responses shown in Fig. 4B.
Functional analysis of the prp locus in bacteria incubated at 37°C.
Considering that the interaction of A. baumannii with the human host occurs mostly at 37°C, we determined whether the prpABCD operon plays a role when bacterial cells are incubated at this temperature. Surface motility assays showed that 17978 cells displayed comparable high-motility responses on SA plates incubated 37°C in the presence and absence of light (Fig. 6A). These responses are in accordance with the finding that the transcriptional levels of prpA were not significantly different in cells cultured in darkness and those cultured under illumination (Fig. 1C). Altogether, these results reinforce the correlation between the light-regulated expression of the prpABCD operon and the consequent differential display of surface motility on a semisolid surface, which was apparent when bacteria were cultured at 24°C but not at 37°C.
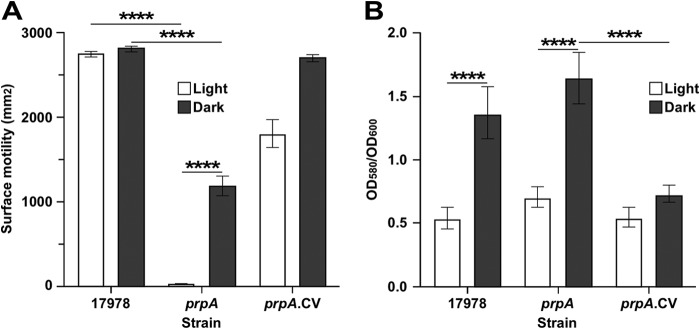
FIG 6.
Role of PrpA in surface motility and biofilm formation in response to illumination at 37°C. (A) Cells of the parental strain (17978) and the prpA insertion mutant (prpA) and its complemented derivative (prpA.CV) were surface inoculated onto SA plates and incubated at 37°C in the presence or absence of light. (B) Biofilm formation by the same three strains on microtiter plate wells statically incubated at 37°C under darkness or illumination. Horizontal bars identify statistically different values (****, P ≤ 0.001), and error bars represent the standard errors of each data set.
In contrast to the parental strain, 17978.prpA cells showed no motility and about half of the motility displayed by 17978 parental cells when the SA plates were incubated in the presence and absence of light, respectively (Fig. 6A). Electroporation of 17978.prpA cells with pMU1269, a pWH1266 derivative harboring a copy of the prpA parental allele, fully restored motility in the absence of light and significantly increased the motility response to illumination compared to the prpA mutant although not to the levels displayed by the 17978 parental cells (Fig. 6A).
Crystal violet assays of biofilms formed on plastic surfaces using cells cultured in SB at 37°C showed that the 17978 parental strain and the prpA mutant formed more biofilm in darkness than under illuminated conditions, although the amounts of biofilm produced by these two strains under illumination and darkness were not significantly different from each other (Fig. 6B). Genetically complemented 17978.prpA.CV cells displayed the same response as 17978 parental and 17978.prpA cells under illumination and significantly reduced biofilm formation when cultured in darkness (Fig. 6B). Further analysis of biofilms using SEM supports the data collected with crystal violet assays. The 17978 strain formed biofilms with more complex structures under darkness than in the presence of light, mainly at the liquid-air interface (Fig. 5B). Interestingly, 17978.prpA cells formed more biofilms under darkness not only at the liquid-air interface but also above and below this interface than in samples of this strain cultured under illumination. Overall, the SEM results support the data collected with standard crystal violet assays shown in Fig. 6B. Although not shown, incubation of the 17978 parental strain and the 17978.prpA derivative in SB at 37°C resulted in nondetectable formation of pellicles on the surface of SB in either the presence or absence of light.
Taken together, these results not only reveal the importance of the PrpABCD pilus system in surface motility and biofilm biogenesis but also suggest the expression of alternative light-regulated motility and biofilm mechanisms when 17978.prpA cells are cultured at 37°C.
Contribution of the prp locus to virulence.
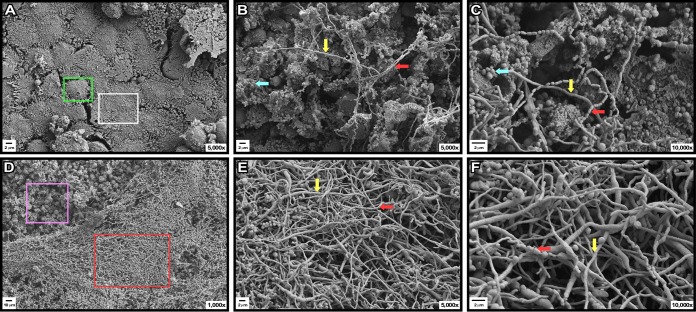
The virulence role of the 17978 PrpABCD pilus assembly system was tested by using the ex vivo polarized A549 human alveolar epithelial cell system and the in vivo G. mellonella experimental infection model that we used previously to examine the role of A. baumannii factors and products in its pathophysiology (18, 29). Infection of polarized A549 cells with 17978 bacteria resulted in remarkable damage to the cells and the disappearance of the surfactant layer that covers their surfaces, which was readily detectable on polarized cells cultured under sterile conditions (compare Fig. 7A with B to D). Furthermore, 17978 bacteria formed clusters, filaments, and cell chains attached to the surface of damaged A549 cells (Fig 7B and C, aqua, yellow, and red arrows, respectively). Examination at higher magnification showed that some of the bacterial filaments were formed by incomplete or a lack of cell division, while chains were formed by bacterial cells attached by their ends (Fig. 7C, yellow and red arrows, respectively). Infection with the 17978.prpA insertion derivative resulted in apparent A549 cell damage, with bacterial clusters attached to damaged cells, as well as the formation of areas covered by a dense layer of bacterial filaments (Fig. 7D, pink and red squares, respectively). Examination at higher magnification showed that, as shown for the 17978 strain, cell chains were formed by bacteria attached by their ends, while filaments were formed by incomplete or a lack of cell division, with the latter being the predominant structures (Fig. 7E and F, red and yellow arrows, respectively).
FIG 7.
Role of PrpA in A549 cell-bacterium interactions. Shown are SEM micrographs of uninfected (sterile) polarized A549 human alveolar epithelial cells (A) or polarized cells infected with bacteria of the 17978 strain (B and C) or the prpA insertion derivative (D to F) and incubated for 48 h at 37°C in the presence of 5% CO2. The green and white boxes in panel A identify the surface of a healthy A549 cell and the mucin layer covering the cells when cultured under sterile conditions, respectively. The red and pink boxes in panel D identify areas covered by a layer of bacterial filaments and areas where the mucin layer was degraded upon bacterial infection, respectively. The aqua arrows in panels B and C identify bacterial clusters attached to damaged epithelial cells. The yellow and red arrows in panels B, C, E, and F identify bacterial filaments and bacterial chains attached to A549 cell surfaces, respectively.
Infection of G. mellonella larvae, which were incubated for 5 days at 37°C after injection with cells of the 17978 strain, resulted in a survival rate of 47.5%. This rate is significantly lower (P < 0.05) than the rates displayed by noninjected larvae or larvae injected with the same volume of sterile phosphate-buffered saline (PBS) (Fig. 8). Figure 8 also shows that infection of caterpillars with 17978.prpA bacteria resulted in an 82.5% survival rate, which is significantly higher (P ≤ 0.05) than the rates scored with larvae infected with cells of the 17978 parental strain but comparable to the survival rates displayed by the negative controls.
FIG 8.
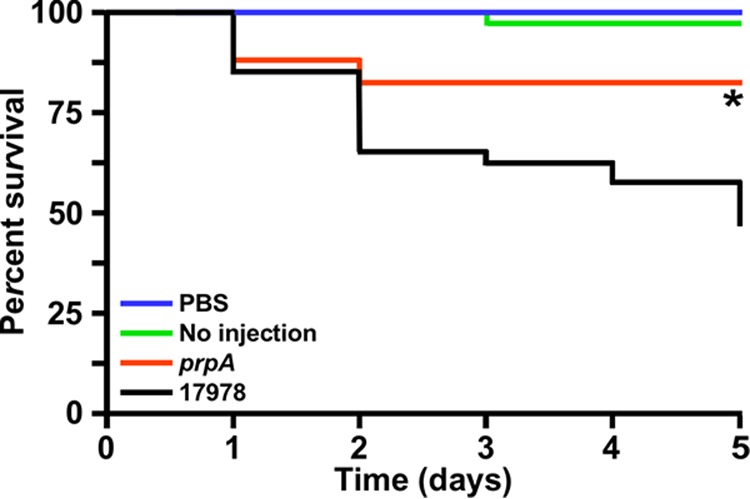
Role of PrpA in virulence. G. mellonella larvae (n = 40 per experimental condition) were injected with 105 bacterial cells of the 17978 parental strain or the prpA isogenic insertion derivative. Larvae injected with sterile PBS and noninjected larvae were used as negative controls. Larvae were incubated at 37°C and assessed for death at 24-h intervals over 5 days, with the removal of dead larvae at times of inspection. The asterisk below the prpA line data indicates a P value of ≤0.05 compared with the data representing the virulence response of 17978.
Taken together, these observations indicate that the PrpABCD pilus assembly system plays a role in the interaction of host cells with 17978 and its virulence when tested with two experimental models already used to study A. baumannii virulence (17).
DISCUSSION
Our original report describing the unexpected ability of A. baumannii ATCC 17978 to respond to light showed that this ubiquitous environmental signal controls surface motility, biofilm biogenesis, and virulence through the function of BlsA, a “short” BLUF-containing photoreceptor (24). The fact that the molecular and cellular factors involved in these light-regulated responses are mostly unknown prompted us to identify and characterize some of these factors and associated responses in more detail using RNA-Seq data as a guide. This approach (see Table S1 in the supplemental material) together with qRT-PCR analyses (Fig. 1B) resulted in the identification of prpABCD, a polycistronic operon coding for a predicted type I pilus assembly system, the expression of which was significantly increased under darkness by an uncharacterized regulatory mechanism that depended on the production of active BlsA when cells were cultured at 24°C in SB (Fig. 1C). Such a response strongly indicates that the effect of light on surface motility could be associated with the expression of this operon when cells are incubated under the above-mentioned conditions. This hypothesis is fully supported by the failure of the 17978.prpA derivative, which harbors a deletion-insertion that removed an internal region of this gene, to display surface motility on SA plates incubated at 24°C in darkness (Fig. 2). Furthermore, the comparative analysis of 17978 and 17978.prpA cells lifted from SA plates incubated under the above-mentioned conditions using TEM showed that prpA inactivation abolishes the production of long and thin pilus structures detected only in the 17978 samples (Fig. 3). Interestingly, the prpA mutation also resulted in a significant reduction in the presence of vesicle-like structures that were readily apparent in the 17978 samples. Currently, it is not known whether these structures play a role in surface motility and/or any aspect of A. baumannii pathophysiology. The lack of motility of the parental strain, the 17978.prpA mutant, and the 17978.prpA.CV derivative when cultured at 24°C under illumination is not surprising considering our initial observation that light drastically reduces this cellular response (24). Altogether, these results not only demonstrate the effect of light on the differential expression of the prpABCD operon but also provide novel insights into the cellular factors and the mechanism by which a microorganism is able to move on the surface of a semisolid medium in spite of a genus name that derives from the Greek term “akineto,” which denotes a lack of motility.
The pellicle and biofilm biogenesis responses suggest different relationships between these two cellular responses and motility (Fig. 4). In the case of pellicle formation, it is apparent that this relationship is direct, since the inactivation of prpA drastically reduced this cellular response, a behavior that most likely depends on the ability of bacterial cells to migrate toward the liquid-air interface by a mechanism that so far has not been described for any member of the Acinetobacter genus. The role of PrpA, and predictably the entire PrpABCD pilus assembly system, in pellicle formation is further supported by the identification of PrpA as a component of the proteins embedded in the matrix of pellicles formed by 17978 cells statically cultured at 25°C in Mueller-Hinton broth or T broth (30). Taken together, our surface motility and pellicle formation results provide strong support for the potential involvement of the PrpABCD pili in A. baumannii motility functions, which remain to be understood at the functional and cellular levels.
In contrast to pellicle formation at 24°C, the relationship between motility and biofilm formation on an abiotic surface is inverse; nonmotile 17978.prpA bacteria formed more biofilms on plastic, under both illumination and darkness, when tested by using crystal violet assays (Fig. 4B). This observation is supported by data from SEM analyses, which showed that 17978.prpA bacteria form more-uniform and denser biofilms at the liquid-air interface under illumination and darkness, respectively, than 17978 (Fig. 5A). This outcome could be due to a decrease in motility and the consequent increased sessility that results in enhanced bacterial adherence and biofilm development, as reported previously for other bacteria (31–33). Alternatively, the absence of prpABCD-mediated pili could facilitate or trigger the interaction of 17978 bacteria with solid substrates through additional or alternative light-regulated adhesins. The annotation of additional 17978 putative pilus assembly systems, including the A1S_0690-A1S_0695 and A1S_1507-A1S_1510 operons, and predicted pilus proteins, such as A1S_3167 (PilY1) and A1S_3177 (fimbrial protein), lends support to this possibility, which remains to be tested experimentally.
The studies conducted at 37°C resulted in particularly interesting and novel information, since they showed that light still controls cellular functions under conditions that have a negative impact on BlsA's production and function. Our original report describing light-regulated responses in A. baumannii ATCC 17978 showed that the incubation of bacteria at 37°C resulted in the constitutive expression of surface motility and biofilm biogenesis on plastic (24). This response could be attributed to the significantly reduced constitutive transcription of blsA in cells cultured at this temperature (24). The negative effect of a temperature of 37°C on blsA transcription was recently confirmed by work reported by Abatedaga et al. (27), which also showed the predicted reduction in BlsA production as well as the observation that temperatures higher than 30°C significantly impair the photocycle of purified BlsA protein. Thus, the ability of the 17978 parental strain incubated at 37°C to produce comparable surface motility responses in the presence and absence of light under the conditions described in this report (Fig. 6A) is in accordance with all the above-mentioned observations regarding the effect of temperature on BlsA production and activity and the consequent constitutive expression of the PrpABCD pilus assembly system (Fig. 1C). Conversely, the ability of 17978.prpA cells to display light-regulated motility at 37°C was an unexpected response that has relevant functional and regulatory implications. First, the expression of light-regulated motility at 37°C by 17978.prpA (Fig. 6A) strongly indicates that this cellular response involves additional motility functions that are not operational at 24°C, since this mutant is nonmotile at this temperature under darkness or light (Fig. 2A). Second, considering the observation that the amounts of biofilm produced by the 17978 parental strain under darkness and illumination are similar to those produced by the 17978.prpA mutant, it is possible to speculate that the same cellular component and/or process is responsible for the biofilm responses shown in Fig. 6B. Third, the detection of light-regulated responses by the 17978 parental strain and the isogenic 17978.prpA strain under temperature conditions that negatively affect the production of BlsA and its photoreceptor activity (Fig. 6) strongly indicates that an alternative and uncharacterized photodetection and regulatory system must be operational when A. baumannii cells persist and/or grow at 37°C. Taking into account all these observations, it is tempting to speculate that the BlsA-mediated system is one of the mechanisms that A. baumannii uses to persist outside the host in nosocomial environments at temperatures lower than 37°C, while at least one different light-dependent regulatory system, which remains to be identified and characterized, is responsible for the capacity of this pathogen to adapt to and persist under higher-temperature conditions imposed by the human host.
Our data also suggest that A. baumannii is capable of producing different adhesins and pilus assembly systems in response to changes in environmental conditions. This possibility is supported by our previous reports showing that an A. baumannii ATCC 19606T derivative deficient in the expression of the CsuAB-A-B-C-D-E chaperone-usher pilus assembly system does not produce pili and does not to attach to and develop biofilms on plastic when cultured in LB broth at 37°C (21, 34). However, the inactivation of this system enhanced rather than abolished the adherence of the ATCC 19606T Csu-deficient mutant to A549 monolayers and sheep erythrocytes compared to the parental strain (35). Furthermore, the Csu mutant produced an increased amount of thin and short pili compared with the ATCC 19606T parental strain, whose surface contained thin and short as well as long pilus-like structures (35). Thus, several uncharacterized sensory and regulatory functions, in addition to those mediated by BlsA, should be involved in these adaptive responses, which allow facultative pathogens, such as A. baumannii, to respond to different extracellular cues that they encounter throughout their life cycles.
The A. baumannii MAR002 biofilm hyperproducer clinical isolate is among the strains that harbor a homolog of the prpABCD operon, which was annotated as the LH92_11070-LH92_11085 4-gene cluster (29). That report (29) showed that LH92_11085 and its 17978 prpA homolog were significantly overexpressed in biofilm cells compared to planktonic bacteria of the cognate strains cultured at 37°C. That observation indicates that the expression of the prpABCD operon is controlled not only by light but also by the bacterial lifestyle. It is interesting to note that the analysis of the MAR002 Δ11085 derivative, which harbors a partial LH92_11085 deletion, resulted in functional outcomes that were similar to as well as different from those that we observed with the 17978.prpA derivative. The MAR002 Δ11085 derivative produced less rather than more biofilms on plastic as well as on the surface of polarized A549 cells than the parental strain, without detectable formation of filamentous bacteria as we observed with the 17978 strain and particularly the 17978.prpA derivative (Fig. 7). In contrast, the polarized A549 cell culture model showed that the inactivation of LH92_11085 reduced the virulence of MAR002 against host cells (29), a response that parallels our data collected with the G. mellonella experimental infection model. This virulence model showed that the rates of survival of caterpillars infected with the 17978.prpA mutant were not only significantly higher than those displayed by larvae infected with the 17978 parental strain but also comparable to those of the negative controls (Fig. 8). Overall, these observations indicate that the 17978 PrpABCD system and its MAR002 LH92_11070-LH92_11085 homolog play a role in the physiology and virulence of A. baumannii. However, some of the biological roles associated with these pilus assembly systems, such as adherence and biofilm biogenesis, seem to be strain specific, a phenomenon that has been described for other A. baumannii traits, including the different effects of mutations in genes coding for the same antimicrobial resistance functions in different clinical isolates (36–38). Furthermore, it is important to note that A. baumannii MAR002 was classified as an ST271 genotype isolate that is not related to any of the high-biofilm producers reported and does not map to any international clonal lineage (29). Whether these taxonomical differences or the potential ability to differentially express distinct adhesins and biofilm functions under different conditions is responsible for the distinct MAR002 phenotypes compared to those of 17978 is a possibility that remains to be explored.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All bacterial strains and plasmids used in this work are listed in Table 1. Bacterial strains were routinely maintained and cultured on Luria-Bertani (LB) agar or broth (39), unless otherwise indicated, and supplemented with the appropriate antibiotics. All cultures were grown at 37°C for 16 to 18 h (overnight) or at 24°C under dark or illuminated conditions as described previously (24).
General DNA procedures.
A commercial kit (Qiagen) and a phenol-based method adapted from the one described previously by Barcak et al. (40) were used to isolate plasmid and total DNA, respectively. DNA was amplified with DNA polymerases and digested with restriction enzymes as indicated by the supplier (New England BioLabs). Automated DNA sequencing using BigDye-based chemistry (Life Technologies) was performed prior to subcloning reactions and electroporation using primers supplied with cloning vector kits (Life Technologies) or custom-designed primers (Integrated DNA Technologies) listed in Table S2 in the supplemental material.
Construction and complementation of a 17978 prpA::ermAM isogenic insertion derivative.
A 2.4-kb genomic fragment including the prpA coding region and 1-kb up- and downstream flanking regions were PCR amplified by using Taq DNA polymerase and primers 4151 and 4152 (Fig. 1A; see also Table S2 in the supplemental material). This amplicon was ligated into pCR8-TOPO, generating pMU1135 (Table 1), which was further confirmed by automated DNA sequencing. Inverse PCR of pMU1135 with Phusion DNA polymerase and primers 4153 and 4154 (Table S2), which anneal within the prpA coding region and result in a 61-bp intragenic deletion, were used to generate an amplicon that was ligated into the Lactococcus lactis pIL252 ermAM cassette, which codes for erythromycin resistance (Emr) (Table 1). Phusion DNA polymerase and primers 4100 and 4101 (Table S2) were used to PCR amplify the ermAM cassette from pIL252. The resulting pMU1142 derivative harboring the prpA::ermAM fragment (Table 1) was PCR amplified by using primers 4151 and 4152 (Fig. 1A and Table S2). The amplicon was ligated into pEX100T, which was previously digested with SmaI, generating pMU1143 (Table 1). This plasmid was then electroporated into electrocompetent 17978 cells as described previously (41). Gene disruption mutants were selected based on their erythromycin and sucrose resistance phenotypes. PCR using primers 4151 and 4152 (Fig. 1A and Table S2) was performed to verify the mutation by allelic exchange in the 17978.prpA isogenic derivative (Fig. 1A and Table 1). The nature of this mutant was further confirmed by automated DNA sequencing.
For complementation studies, a 662-nt chromosomal region encompassing the prpA wild-type allele and its promoter region was PCR amplified from 17978 total genomic DNA with Q5 high-fidelity DNA polymerase (New England BioLabs) using primers 4413 and 4465 (Fig. 1A and Table S2), each flanked with BamHI restriction sites, and cloned into the cognate site of the A. baumannii-Escherichia coli shuttle vector pWH1266 (42) to generate the pMU1269 complementing plasmid (Table 1). This plasmid, which was checked by automated DNA sequencing, was electroporated into 17978.prpA electrocompetent cells, and the resulting 17978.prpA.CV transformants (Table 1) were selected on LB agar plates containing 500 μg/ml ampicillin (Amp) and 20 μg/ml Em. The 17978.prpA.EV derivative harboring the empty pWH1266 shuttle vector (Table 1) was used as a negative control.
RNA isolation and transcriptional analysis of prpABCD.
Three independent cultures of the 17978 parental strain and the blsA (17978.OR) mutant were grown in 25 ml of swimming broth (SB) (10 g/liter tryptone, 5 g/liter NaCl) at 24°C or 37°C under illumination or darkness until an optical density at 600 nm (OD600) of 0.8 was reached. Bacterial cells were collected by centrifugation, and total RNA was isolated by using the Direct-Zol Miniprep Plus kit (Zymo Research) according to the manufacturer's protocol. Following treatment with DNase I (Invitrogen), RNA quality was assessed by using the Bioanalyzer 2100 system and the RNA Pico 6000 kit (Agilent Technologies). cDNA used for RT-PCR and qRT-PCR assays was synthesized by using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol. Briefly, 100 ng of DNA-free total RNA isolated from 17978 cells cultured as described above was used as the template with random hexamer primers and the following cycling conditions: priming for 5 min at 25°C, reverse transcription for 20 min at 46°C, and inactivation for 1 min at 95°C.
RT-PCR was performed by using the cDNA samples described above as the template and primer pairs 4470-4615 and 4471-4618, which link the prpA-prpC and prpC-prpD coding regions, respectively (Fig. 1 and Table S2), to determine the predicted polycistronic nature of the prp operon and the potential polar effect of the insertion of the ermAM cassette under the following conditions: initial denaturation at 95°C for 3 min, denaturation at 95°C for 30 s, annealing for 30 s, and extension for 1 min at 72°C. The denaturation, annealing, and extension steps were repeated for 29 more cycles, followed by a final extension step for 5 min at 72°C. The level of production of the predicted amplicons was determined by ethidium bromide agarose gel electrophoresis (39). Amplification of total RNA without reverse transcription was used as a negative control.
The differential transcription of prpA was measured by qRT-PCR using cDNA synthesized from DNA-free total RNA isolated from cells of the 17978 parental strain, the 7978.OR blsA mutant, and the 17978.prpA.CV derivative cultured under illumination or darkness as described above and primers 4419 and 4420 (Fig. 1A and Table S2) under experimental conditions described previously (43). Primers 4488 and 4489 (Table S2) were used to detect the transcription of an internal fragment of the recA housekeeping gene, which was used as a constitutively expressed gene. Triplicates of three independent biological samples were used to statistically validate the data collected for each tested strain.
Bacterial growth analysis.
Fresh bacterial cultures of 17978 and the prpA insertion derivative were passaged from LB agar plates to LB broth and grown overnight at 37°C in a shaking incubator set at 200 rpm. Bacterial cultures were then diluted 100 times into fresh SB and transferred to a 96-well microtiter plate. OD600 measurements were recorded hourly for 24 h at 37°C in a shaking plate reader (BioTek Instruments, Inc.), using two independent biological samples in six technical replicates.
Motility assays.
Fresh bacterial cultures grown on LB agar plates supplemented with antibiotics when necessary were used to inoculate the surface of swimming agar (SA) (SB containing 0.3% agarose) plates and incubated overnight at 24°C or for 16 to 18 h at 37°C as described previously (24). Motility assays were performed in triplicate using three independent biological samples each time. The surface motility area of each replicate was measured by using the ImageJ image-processing program (National Institutes of Health).
Transmission electron microscopy.
Bacterial cells were inoculated onto the surfaces of SA plates that were incubated overnight at 24°C in darkness. Freshly prepared carbon-coated, nitrocellulose-substrated TEM grids were placed substrate side down at the edge or immediately in front of the motility zone to analyze the 17978 strain. Grids were placed at the edge of the colony formed by the nonmotile 179878.prpA mutant. The grids were carefully removed and negative stained with 5 μl of 1.5% (wt/vol) ammonium molybdate for 5 min. The grids were then wicked dry with filter paper and allowed to air dry. Images were captured at 120 kV with a JEOL 1200-EX II TEM instrument.
Biofilm and pellicle assays.
Fresh bacterial cultures of 17978 and the prpA mutant were grown in SB overnight with shaking at 37°C. To assess pellicle formation, cultures grown overnight were diluted 1:100 into fresh SB, and 1 ml of each diluted culture was inoculated into glass tubes, which were incubated statically at 24°C or 37°C for 96 h or 48 h, respectively, as reported previously (24). The formation of pellicles on the surface of liquid cultures was assessed visually. To assess biofilm formation, bacterial cultures were prepared as described above, and 200 μl of each strain was inoculated into 96-well microtiter plates. Plates were incubated at 24°C for 96 h or at 37°C for 48 h without shaking. Each incubation was performed in light or darkness, for a total of four different conditions for each culture. Biofilms were quantified by crystal violet staining as described previously (21). Biofilm experiments were performed in duplicate, using fresh biological samples for each experiment.
To examine biofilm structures formed on plastic surfaces, three independent fresh SB cultures of 17978 or the isogenic prpA derivative grown overnight at 37°C were diluted 1:100, and 5 ml of each diluted culture was inoculated into 50-ml conical tubes. A sterile plastic coverslip was placed semisubmerged into each tube, as described previously (44). All cultures were incubated statically at either 24°C for 96 h or 37°C for 48 h in light or darkness. Coverslips were recovered from each 50-ml conical tube, washed, processed, and examined by scanning electron microscopy (SEM) on a Zeiss Supra 35 VP microscope as described previously (44).
Infection of polarized A549 human alveolar epithelial cells.
A549 cells were polarized and infected with bacteria cultured in LB broth as described previously (45), using 106 bacteria/ml, and incubated for 48 h at 37°C in the presence of 5% CO2. All polarized cell samples were fixed after 48 h of infection and analyzed by SEM as described previously (44). Polarized cell assays were performed in duplicate under each condition, using different biological samples each time.
Galleria mellonella virulence assays.
Fresh cultures of 17978 or the prpA mutant were passaged from LB agar plates and grown overnight in LB broth without antibiotics. Cells were harvested by centrifugation, washed twice in a sterile phosphate-buffered saline (PBS) solution, and then resuspended in 1 ml PBS. Bacterial cells were standardized to an OD600 of 0.5, which corresponded to 108 bacterial cells/ml. Healthy G. mellonella larvae weighing between 250 and 300 mg, which were previously sorted into groups of 10 based on yellow coloration and high activity, were injected with 5 μl of either the 17978 parental strain or the prpA mutant. Control groups included larvae inoculated with 5 μl sterile PBS to account for physical trauma caused by the injection process or larvae that were not injected to confirm larva viability throughout the experiment. Infected and control larvae were stored for 5 days at 37°C in a humidified chamber and assessed for death at 24-h intervals. Death events were based on a lack of movement and melanization, as described previously (18). If more than two deaths occurred in the control groups, the experiment was discarded and repeated. Kaplan-Meier survival curves were plotted by using GraphPad Prism 7, and log rank tests were performed by using the statistical analysis tools included with GraphPad Prism software. A total of four replicates per experimental condition (n = 40) were used to validate the experimental data. P values of ≤0.05 were considered statistically significant for the log rank test of survival curves (SAS Institute, Inc., Cary, NC).
Statistics.
The GraphPad 7 InStat software package (GraphPad Software, Inc.) was used to analyze the statistical significance of data sets by Student's t test or analysis of variance (ANOVA) as appropriate for each experiment. P values of ≤0.05 were considered statistically significant for all above-mentioned assays. Error bars represent the standard errors for each data set shown in the figures.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by National Institutes of Health Public Health grant R15GM117478-01 as well as by Miami University research funds.
We thank Hauke Smidt and Thomas Kruse (Wageningen University, The Netherlands) for providing the Lactococcus lactis subsp. cremoris strain that was used as a source of plasmid pIL252.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00442-18.
REFERENCES
- 1.Towner KJ, Bergogne-Berezin E, Fewson CA. 1991. Acinetobacter: portrait of a genus, p 1–24. In Towner KJ, Bergogne-Berezin E, Fewson CA (ed), The biology of Acinetobacter. Plenum Press, New York, NY. [Google Scholar]
- 2.Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L. 2012. The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS One 7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann P. 1968. Isolation of Acinetobacter from soil and water. J Bacteriol 96:39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firstenberg-Eden R, Rowley DB, Shattuck GE. 1980. Factors affecting inactivation of Moraxella-Acinetobacter cells in an irradiation process. Appl Environ Microbiol 40:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Berezin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villegas MV, Hartstein AI. 2003. Acinetobacter outbreaks, 1977-2000. Infect Control Hosp Epidemiol 24:284–295. doi: 10.1086/502205. [DOI] [PubMed] [Google Scholar]
- 7.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 8.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemec A, Krizova L, Maixnerova M, Sedo O, Brisse S, Higgins PG. 2015. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int J Syst Evol Microbiol 65:934–942. doi: 10.1099/ijs.0.000043. [DOI] [PubMed] [Google Scholar]
- 10.Eveillard M, Kempf M, Belmonte O, Pailhories H, Joly-Guillou ML. 2013. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int J Infect Dis 17:e802–e805. doi: 10.1016/j.ijid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Huys G, Bartie K, Cnockaert M, Hoang Oanh DT, Phuong NT, Somsiri T, Chinabut S, Yusoff FM, Shariff M, Giacomini M, Teale A, Swings J. 2007. Biodiversity of chloramphenicol-resistant mesophilic heterotrophs from Southeast Asian aquaculture environments. Res Microbiol 158:228–235. doi: 10.1016/j.resmic.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Vangnai AS, Petchkroh W. 2007. Biodegradation of 4-chloroaniline by bacteria enriched from soil. FEMS Microbiol Lett 268:209–216. doi: 10.1111/j.1574-6968.2006.00579.x. [DOI] [PubMed] [Google Scholar]
- 13.Rafei R, Hamze M, Pailhories H, Eveillard M, Marsollier L, Joly-Guillou ML, Dabboussi F, Kempf M. 2015. Extrahuman epidemiology of Acinetobacter baumannii in Lebanon. Appl Environ Microbiol 81:2359–2367. doi: 10.1128/AEM.03824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endimiani A, Hujer KM, Hujer AM, Bertschy I, Rossano A, Koch C, Gerber V, Francey T, Bonomo RA, Perreten V. 2011. Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data. J Antimicrob Chemother 66:2248–2254. doi: 10.1093/jac/dkr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zordan S, Prenger-Berninghoff E, Weiss R, van der Reijden T, van den Broek P, Baljer G, Dijkshoorn L. 2011. Multidrug-resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerg Infect Dis 17:1751–1754. doi: 10.3201/eid1709.101931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kempf M, Rolain JM, Diatta G, Azza S, Samb B, Mediannikov O, Gassama Sow A, Diene SM, Fenollar F, Raoult D. 2012. Carbapenem resistance and Acinetobacter baumannii in Senegal: the paradigm of a common phenomenon in natural reservoirs. PLoS One 7:e39495. doi: 10.1371/journal.pone.0039495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.McConnell MJ, Actis L, Pachón J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaddy JA, Arivett BA, McConnell MJ, Lopez-Rojas R, Pachon J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McQueary CN, Kirkup BC, Si Y, Barlow M, Actis LA, Craft DW, Zurawski DV. 2012. Extracellular stress and lipopolysaccharide modulate Acinetobacter baumannii surface-associated motility. J Microbiol 50:434–443. doi: 10.1007/s12275-012-1555-1. [DOI] [PubMed] [Google Scholar]
- 20.Nwugo CC, Arivett BA, Zimbler DL, Gaddy JA, Richards AM, Actis LA. 2012. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS One 7:e51936. doi: 10.1371/journal.pone.0051936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 22.Bitrian M, Gonzalez RH, Paris G, Hellingwerf KJ, Nudel CB. 2013. Blue-light-dependent inhibition of twitching motility in Acinetobacter baylyi ADP1: additive involvement of three BLUF-domain-containing proteins. Microbiology 159:1828–1841. doi: 10.1099/mic.0.069153-0. [DOI] [PubMed] [Google Scholar]
- 23.Golic A, Vaneechoutte M, Nemec A, Viale AM, Actis LA, Mussi MA. 2013. Staring at the cold sun: blue light regulation is distributed within the genus Acinetobacter. PLoS One 8:e55059. doi: 10.1371/journal.pone.0055059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192:6336–6345. doi: 10.1128/JB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Losi A, Gartner W. 2008. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci 7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- 26.Brust R, Haigney A, Lukacs A, Gil A, Hossain S, Addison K, Lai CT, Towrie M, Greetham GM, Clark IP, Illarionov B, Bacher A, Kim RR, Fischer M, Simmerling C, Meech SR, Tonge PJ. 2014. Ultrafast structural dynamics of BlsA, a photoreceptor from the pathogenic bacterium Acinetobacter baumannii. J Phys Chem Lett 5:220–224. doi: 10.1021/jz4023738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abatedaga I, Valle L, Golic AE, Muller GL, Cabruja M, Moran Vieyra FE, Jaime PC, Mussi MA, Borsarelli CD. 2017. Integration of temperature and blue-light sensing in Acinetobacter baumannii through the BlsA sensor. Photochem Photobiol 93:805–814. doi: 10.1111/php.12760. [DOI] [PubMed] [Google Scholar]
- 28.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Fraga L, Perez A, Rumbo-Feal S, Merino M, Vallejo JA, Ohneck EJ, Edelmann RE, Beceiro A, Vazquez-Ucha JC, Valle J, Actis LA, Bou G, Poza M. 2016. Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7:443–455. doi: 10.1080/21505594.2016.1145335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nait Chabane Y, Marti S, Rihouey C, Alexandre S, Hardouin J, Lesouhaitier O, Vila J, Kaplan JB, Jouenne T, De E. 2014. Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS One 9:e111660. doi: 10.1371/journal.pone.0111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol 16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 35.de Breij A, Gaddy J, van der Meer J, Koning R, Koster A, van den Broek P, Actis L, Nibbering P, Dijkshoorn L. 2009. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC 19606T to human airway epithelial cells and their inflammatory response. Res Microbiol 160:213–218. doi: 10.1016/j.resmic.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Kröger C, Kary SC, Schauer K, Cameron AD. 2016. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes 8:e12. doi: 10.3390/genes8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montana S, Vilacoba E, Traglia GM, Almuzara M, Pennini M, Fernandez A, Sucari A, Centron D, Ramirez MS. 2015. Genetic variability of AdeRS two-component system associated with tigecycline resistance in XDR-Acinetobacter baumannii isolates. Curr Microbiol 71:76–82. doi: 10.1007/s00284-015-0829-3. [DOI] [PubMed] [Google Scholar]
- 38.Wright MS, Iovleva A, Jacobs MR, Bonomo RA, Adams MD. 2016. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med 8:26. doi: 10.1186/s13073-016-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol 204:321–342. doi: 10.1016/0076-6879(91)04016-H. [DOI] [PubMed] [Google Scholar]
- 41.Dorsey CW, Tomaras AP, Actis LA. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl Environ Microbiol 68:6353–6360. doi: 10.1128/AEM.68.12.6353-6360.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. doi: 10.1016/0378-1119(90)90494-C. [DOI] [PubMed] [Google Scholar]
- 43.Fiester SE, Arivett BA, Schmidt RE, Beckett AC, Ticak T, Carrier MV, Ghosh R, Ohneck EJ, Metz ML, Sellin Jeffries MK, Actis LA. 2016. Iron-regulated phospholipase C activity contributes to the cytolytic activity and virulence of Acinetobacter baumannii. PLoS One 11:e0167068. doi: 10.1371/journal.pone.0167068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohneck EJ, Arivett BA, Fiester SE, Wood CR, Metz ML, Simeone GM, Actis LA. 2018. Mucin acts as a nutrient source and a signal for the differential expression of genes coding for cellular processes and virulence factors in Acinetobacter baumannii. PLoS One 13:e0190599. doi: 10.1371/journal.pone.0190599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.