Abstract
Fungal endophytes as a source of bioactive metabolites have led to the development of pharmaceutical products finding new applications. In a survey of endophytic fungal biodiversity, an antimicrobial endophytic strain CLB32 was isolated from the leaf of Combretum latifolium Blume (Combretaceae) from the Western Ghats of Southern India. CLB32 was then identified as Gliomastix polychroma (KR704576) by morphological and phylogenetic analysis based on internal transcribed spacer (ITS) nuclear rDNA and intervening 5.8S rRNA gene. CLB32 here constituted the first report on incidence of endophytic fungi from C. latifolium Blume. Ethyl acetate fraction of strain CLB32 was evaluated for antimicrobial activity by disc diffusion assay. Secondary metabolites produced effectively inhibited methicillin-resistant Staphylococcus aureus (18.33 ± 0.33 mm), Pseudomonas aeruginosa (14.66 ± 0.33 mm) and Candida albicans (14.00 ± 0.57 mm). Biosynthesis of these antimicrobial compounds was detected by analytical TLC-bioautography method as depicted by zone of inhibition on intensive the band. These findings suggest that G. polychroma CLB32, as a producer of natural antimicrobial drugs, could help to combat against multidrug-resistant infections and also provide baseline information for industrial applications.
Keywords: endophytic fungi, multidrug resistance, antimicrobial metabolites, TLC-bioautography
1. Introduction
Emergence of multidrug-resistant (MDR) microorganism infections has generated considerable attention in recent decades (Boucher et al. 2008; Cars et al. 2008). Antibiotic resistance is one of the greatest challenges facing modern medicine (Spellberg et al. 2008). Antibiotics that lose their effectiveness for treating human disease through antibiotic resistance must be replaced with new drugs (Hamad 2010). The massive increases in trade and human mobility brought about by globalization have enabled the rapid spread of infectious agents, including those that are drug-resistant. World Health Organization (WHO 2014) report on global surveillance of antimicrobial resistance reveals that antibiotic resistance is no longer a prediction for the future, it is happening right now, across the world and is putting the treatment of common infections in the community and hospitals at risk.
Biodiscovery from microbial resources helps in the exploration of microbial metabolic products to detect, identify and evaluate their potential for medicinal, agricultural and biotechnological operations (Krutboke 2010; Baker et al. 2014). Plants lack immune response to certain pathogens, but the endophytes that reside inside the plant tissue enhance the immune response of the plants to fight against invading pathogens (Melotto et al. 2008). Fungal endophytes are polyphyletic group of highly diverse fungi that are defined functionally by their occurrence within tissues of plants without causing any immediate overt effects (Rodriguez et al. 2009). This endophyte–plant interaction induces the production of novel antimicrobial agents and endophyte–endophyte interactions within plants also have the potential to produce novel antimicrobial agents (Bandara et al. 2006; Nutzmann et al. 2011). In recent decades, endophytic fungi from plants have been widely accepted as major sources of drugs. A large number of bioactive compounds with new structures are continuously being isolated from endophytes (Strobel et al. 2004). They are strongly considered as largely unexploited metabolic resources. Therefore, new antimicrobial metabolites continue to be identified from fungal endophyte source.
Combretum latifolium Blume (Combretaceae) is a large climbing shrub that has great ethnomedicinal values (Shrisha et al. 2011). The stem and bark of this shrub are used as insecticides (Suthari et al. 2014). Leaf juice is used in the treatment for dysentery and goitre (Debnath et al. 2014). In view of this, C. latifolium Blume is selected to explore hidden potential endophytic fungi that can inhibit the growth of methicillin-resistant Staphylococcus aureus (MRSA) and other human pathogens.
2. Materials and methods
2.1. Study site location and source of endophytic fungi
Pushpagiri Sanctuary (12°35′N 75°40′E, elevation 1748 m) is located in the Western Ghats of Kodagu (Coorg), Karnataka, which is a part of Southern India. It is covered with thick evergreen forests and shoal grassland habitat. It is one of the eight ‘hottest hotspots’ of biological diversity in the world (Myres et al. 2000). C. latifolium Blume from this region was selected for the study.
2.2. Collection of samples
Specimens of C. latifolium Blume were carefully collected. In order to secure the endophytic nature of the isolates, the cut ends of bark and root specimens were sealed with wax. All the specimen samples were brought to the laboratory in an icebox and stored at 4°C. Each sample was used for the isolation of endophytic fungi within 24 h of collection.
2.3. Isolation of endophytic fungi
Isolation of endophytic fungi was carried out according the method described by Wang et al. (2012) with some modifications. Each sample tissue was washed under running water for 15 min and dried at room temperature prior to surface-sterilization. All the slight, visibly damaged material was excluded. To remove the epiphytic microorganisms, samples were rinsed with 70% ethanol for 2 min, surface-sterilized by sodium hypochlorite (4%) for 5 min and rinsed with 70% ethanol for 30 s. Then the sample tissues were rinsed with sterile double distilled water and kept for surface drying in sterile conditions. To confirm the success of the surface disinfection process, aliquots of the sterile distilled water from the final rinse were inoculated on the isolation media plates. The plant tissue samples were cut into small segments (5 mm size) and placed on water agar plates (distilled water, 1.5% agar) amended with chloramphenicol (250 ppm) and incubated at 30°C for 3–4 days to few weeks, until the growth initiated. The hyphal tips that emerged from the plant tissues were picked and maintained on PDA plates for further studies.
2.4. Molecular profiling of endophytic strain CLB32
2.4.1. Genomic DNA extraction
G. polychroma was cultured in PDB for 7 days at 30°C, and the mycelium was harvested by vacuum filtration. The chilled mycelia were ground with pestle and mortar under liquid nitrogen, transferred into the micro centrifuge tube with 1 ml of 2× CTAB extraction buffer and incubated at 65°C for 30 min with gentle swirling. After centrifugation, the aqueous phase of the mixture containing total DNA was extracted with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1). Residual phenol was removed with the addition of chloroform:isoamyl alcohol (24:1) twice. Two volumes of ethanol and 0.1 volume of 3 M sodium acetate were added to the aqueous phase of DNA for precipitation and then incubated at −20°C overnight (Kim et al. 2010). The DNA pellet was washed with 70% ethanol twice, and suspended in 15 µl of TE buffer. Polymerase chain reaction (PCR) amplification was carried according to the protocol of Bhagat et al. (2012) using ITS1 (5ʹ TCCGTAGGTGAACCTGCGG 3ʹ) and ITS4 (5ʹ TCCTCCGCTTATTGATATGC 3ʹ) universal primers (White et al. 1990).
2.4.2. Taxon sampling and phylogenetic affiliation
Internal transcribed spacer (ITS) sequence data of strain G. polychroma CLB32 obtained was annotated using Geneious 6.1.6 (Biomatters, 2013, Auckland, New Zealand) software and submitted to the National Centre for Biotechnology Information (NCBI) GenBank. ITS rDNA sequences with maximum identity to that of strain CLB32 were retrieved from NCBI nucleotide database using Basic Local Alignment Search Tool (BLAST). ITS sequences were filter-searched, and the closest sequences were selected for the phylogenetic analysis. The multiple sequence alignments were performed using CLUSTALW software utilizing default settings, and dendrograms were generated by MEGA 4.0 software with a bootstrap consensus of 1000 replicates (Padhi and Tayung 2013).
2.5. Evaluation of antimicrobial activity
Antimicrobial susceptibility testing of ethyl acetate extract was carried out by the disc diffusion assay (Sadrati et al. 2013). Sterile discs (6 mm dia) impregnated with 20 μl of ethyl acetate extract were dried in laminar hood and placed on the surface of the media seeded with test pathogens in Petri plates. One negative control disc impregnated with only 20 μl of ethyl acetate was also placed for each test organism with Gentamicin as a positive control. The plates were incubated at 37 ± 2°C and diameter of the zone of inhibition was recorded.
2.6. Thin-layer chromatography-bioautography
Ten microlitres of ethyl acetate extract was spotted on TLC silica gel plates (TLC, Alugram® SIL G/UV254; Machereye-Nagel, Duren, Germany) in petroleum ether/ethyl acetate (1:2) optimized solvent system. The developed TLC plates were observed under visible light and UV light at 254 and 365 nm, respectively. TLC plates were then air-dried for complete removal of the solvent traces and UV sterilized for 30 min. TLC plates were then encased in sterile Petri plates overlaid with Brain heart infusion medium containing 0.65% agar incorporated with 1 mg ml−1 concentration of 2,3,5-triphenyl tetrazolium chloride (TTC) (Sigma-Aldrich) inoculated with 1% standardized MRSA inocula. After 8 h of diffusion at 8°C, the plates were incubated for 24 h at 37°C. The areas of inhibition zones on the active spot were recorded (Valgas et al. 2007).
3. Results and discussion
3.1. Isolation of endophytic fungi
It has becoming evidence that all higher plants host has one or more untapped endophytic microorganisms (Strobel 2003). The results obtained in the present investigation attribute considerable interest towards endophytic research. In this study, an endophytic fungus, G. polychroma CLB32, was isolated from the asymptomatic bark tissue of C. latifolium Blume. In addition, media Petri plates spread with the final rinse of sterile water showed no microbial growth even after 10 days of incubation at 28°C. This indicates that the surface sterilization method was effective in killing the epiphytic microorganisms. Thus, subsequent isolates can be considered as true endophytic fungi (Barnett and Hunter 1956). Earlier studies also define the successful surface sterilization process for the isolation of endophytes with no contamination of epiphytes (Kusari et al. 2009; Lin et al. 2010). The isolated endophytic fungus was then identified as Gliomastix sp. on the basis of morphological characteristics of growth pattern, colour of colony, surface texture, hyphae, margin and characteristics of the spores (Figure 1). Further to identify the strain at species level molecular analysis has been carried out. This study constitutes the first report on incidence of endophytic fungus inhabiting C. latifolium Blume.
Figure 1.
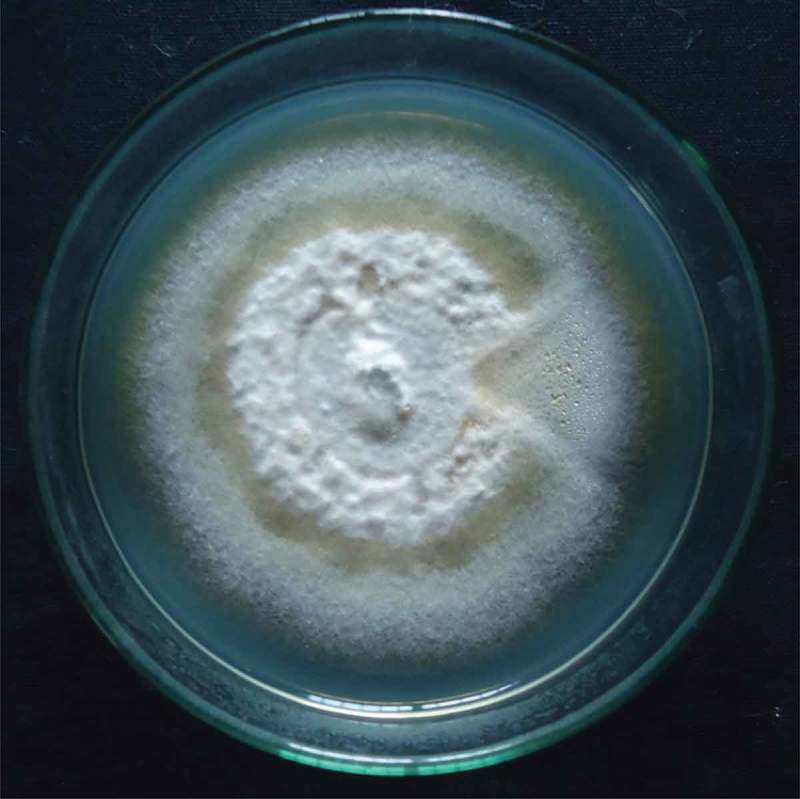
Colony morphology of Gliomastix polychroma CLB32 on potato dextrose agar after 10 days.
3.2. Taxon sampling and phylogenetic affiliation
Strain CLB32 was identified using both morphological characters and phylogenetic data for precise identification. Phylogenetic affiliation was carried out based on ITS region of rDNA and intervening 5.8S rRNA gene (Figure 2). The amplified ITS region of rDNA was sequenced and aligned with ITS sequences of the different strains retrieved from NCBI databases using CLUSTALW (Thompson et al. 1994). Alignment with those from NCBI database resulted in several closely related sequences. Corresponding neighbour joining (NJ) tree showed clearly that strain CLB32 fell into the group of Gliomastix polychroma with strong support (Figure 3). ITS rDNA sequence profiling is one of the cost-effective tools that enable the researchers to identify the complex microbial communities in species level at exceptional depth and resolution (Rastogi and Sani 2011). The number of previously phylogenetically unstudied fungi is large (Summerbell et al. 2011); however, morphological identification has long been the basis and tradition, which is, beyond doubt, of great importance (Wang et al. 2014). Thus combined with morphology and phylogenetic data, strain CLB32 was defined as Gliomastix polychroma ultimately. The ITS sequence data of this fungus is deposited in GenBank under the accession no. KR704576.
Figure 2.
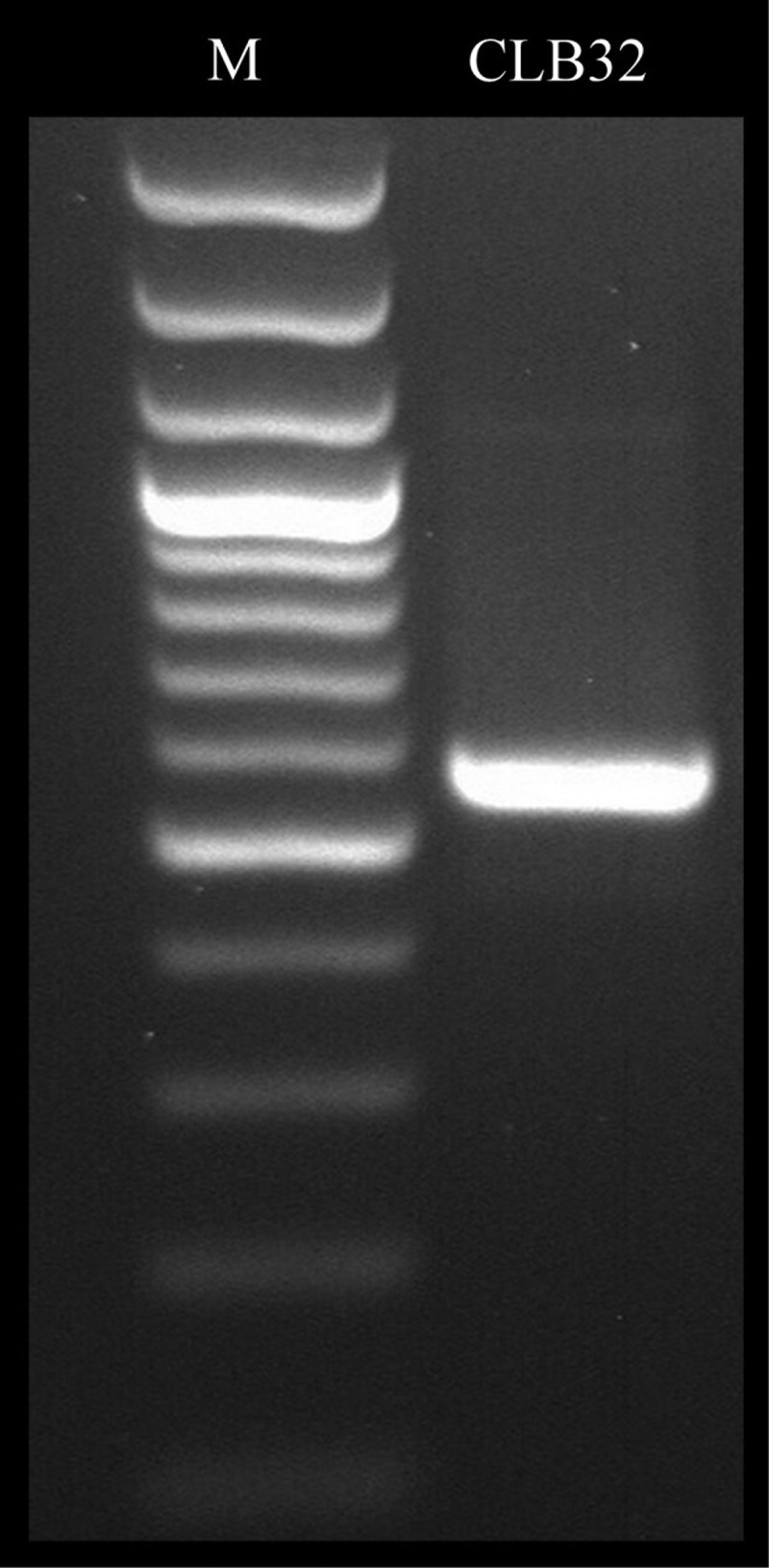
Polymerase chain reaction (PCR) amplification of Gliomastix polychroma CLB32 rDNA by internal transcribed spacer (ITS1 and ITS4) universal primers.
Note: Lane: M = 100 bp DNA ladder, CLB32 = ~580 bp amplicon representing ITS region of rDNA.
Figure 3.
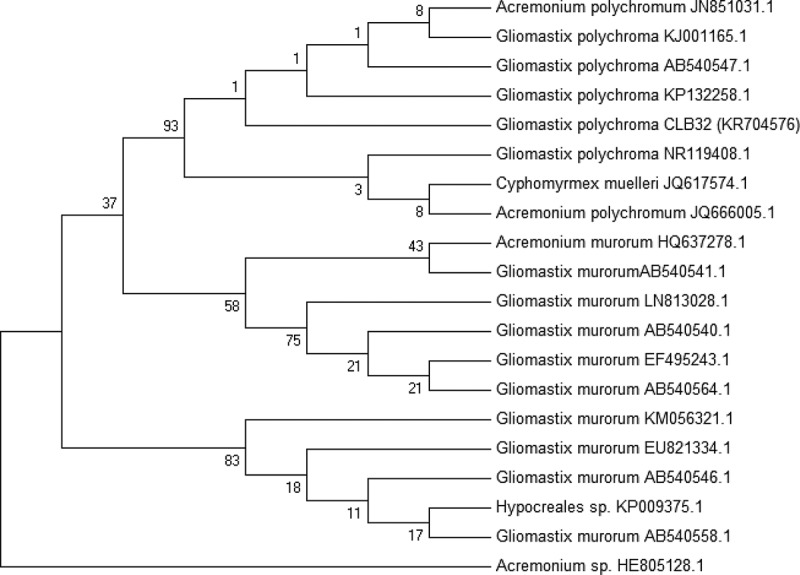
Phylogenetic tree derived from NJ analysis showing the evolutionary relationship of Gliomastrix polychroma CLB32 with its closest BLAST hits. Bootstrap values (1000 replications) based on multiple sequence alignment using the MEGA-5 software.
3.3. Antimicrobial susceptibility testing
Antimicrobial potential of ethyl acetate extract of CBL32 was analysed by disc diffusion assay (Figure 4). Ethyl acetate has been used for the extraction of secondary metabolites as most of secondary metabolites are miscible with ethyl acetate with strong literature support (Jumpathong et al. 2011; Rao and Satish 2015). The results obtained were validated with standard antibiotics, namely gentamicin for antibacterial activity and nystatin for antifungal activity. During antimicrobial susceptibility test, strain CBL32 displayed strong antimicrobial activity, where significant activity was observed against MRSA (18.33 ± 0.33 mm), followed by E. coli (17.66 ± 0.33 mm) and Pseudomonas aeruginosa (16.33 ± 0.66 mm). The strain also showed antifungal activity against Candida albicans and Microsporum canis (Table 1). The ability to suppress the growth of both bacterial and fungal pathogens implies that metabolites have a broad spectrum of antimicrobial activity (Rao et al. 2015). This fungus, as an endophyte, may be involved in protecting the host against invading pathogens, and this could have led to the ability of this endophyte to biosynthesize some phytochemicals originally associated with the host (Stierle et al. 1993, Wang et al. 2012). Streptomyces longisporoflavus as endophyte was isolated from L. ciliata Benth collected from Western Ghats, India, which exhibited anti-diabetic activity (Akshatha et al. 2014). Antimicrobial activity of endophytic fungi inhabiting eight medicinal plant species sampled in different locations from the Western Ghats of India was recorded (Raviraja et al. 2006). Therefore, the present work expands our understanding of potential hidden endophytic fungi underexplored area are the promising source for novel antimicrobial agents. Biodiscovery of antimicrobial producing endophytic fungi associated with C. latifolium Blume is valuable for industrial interest and for basic research (see supplemental data).
Figure 4.
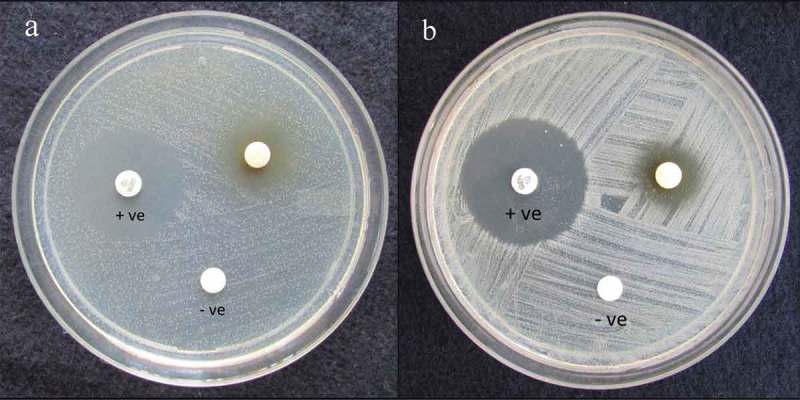
Antimicrobial activity of ethyl acetate extract of endophytic Gliomastix polychroma CLB32 against (a) MRSA, (b) Salmonella typhi.
Note: +ve = Gentamicin as positive control; –ve = Ethyl acetate as negative control.
Table 1.
Determination of antimicrobial activity of ethyl acetate extract of Gliomastix polychroma CLB32 (100 μg/disc) by disc diffusion assay.
| Test pathogens | Zone of inhibition (mm) | Standard (mm) |
|---|---|---|
| MRSA | 18.33 ± 0.33a | 24.00 ± 0.00b (G) |
| Staphylococcus epidermidis | 14.66 ± 0.33c | 24.66 ± 0.33b (G) |
| Salmonella typhi | 15.33 ± 0.33bc | 26.33 ± 0.00a (G) |
| Pseudomonas aeruginosa | 16.33 ± 0.66abc | 24.66 ± 0.33b (G) |
| Escherichia coli | 17.66 ± 0.33ab | 26.00 ± 0.00a (G) |
| Candida albicans | 14.00 ± 0.57c | 19.66 ± 0.33c (N) |
| Microsporum canis | 14.33 ± 0.66c | 19.00 ± 0.00c (N) |
Note: Values represent the diameter of the zone of inhibition. Data are means from three replicates ± SE and those with the same superscript letters in the appropriate columns are significantly different (ANOVA, Tukey’s HSD at p < 0.05).
G = Gentamicin (100 μg/disc), N = Nystatin (100 μg/disc).
3.4. Thin layer chromatography-bioautography
Chromatographic profile of ethyl acetate extract of CLB32 signalled the presence of an intense band under 254 and 365 nm, which might be due to the increased production of secondary metabolites. In the bioautography assay, a clear zone of inhibition was observed on the intense band against the red background where the medium was preinoculated with MRSA and TTC agent. This confirmed that the presence of antimicrobial compound in the ethyl acetate extract, which was active against MRSA strain. Detection of antimicrobial compound by TLC-bioautography is one of the economical, simplest and reproducible methods for drug discovery from natural products (Hota 2010; Patra et al. 2012). The antimicrobial activity of ethyl acetate extract of Phomopsis sp. FPSP-25 was determined using the same approach (Rakshith et al. 2013). Further chemical investigation is needed to characterize the metabolite to combat against human pathogens and MDR microorganisms for antibacterial activity.
4. Conclusion
This work suggests that the screening of potential endophytic actinomycetes from an underexplored area can give a new facelift for antimicrobial research and drug development. Effective inhibition of MDR microorganisms by this fungus implies that this strain could be a reliable source for industrially and pharmaceutically important bioactive compounds. This work is the first report on the incidence of endophytic fungi inhabiting C. latifolium Blume.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding Statement
This work was supported by a PhD fellowship awarded to the first author by the University Grants Commission (UGC) and University of Mysore, India [DV9/192/NON-NETFS/2013-2014, dated 11 November 2013]. This work was also supported by Institution of Excellence, University of Mysore, India.
Acknowledgements
Authors thank Institution of Excellence and Department of Studies in Microbiology for providing instrumentation facilities.
References
- Akshatha VJ, Nalini MS, D’Souza C, Prakash HS.. 2014. Streptomycete endophytes from anti-diabetic medicinal plants of the Western Ghats inhibit alpha-amylase and promote glucose uptake. Lett Appl Microbiol. 58:433–439. [DOI] [PubMed] [Google Scholar]
- Baker S, Kumar KM, Santosh P, Rakshith D, Satish S. 2014. Extracellular synthesis of silver nanoparticles by novel Pseudomonas veronii AS41G inhabiting Annona squamosa L. and their bactericidal activity. Spectrochim Acta Mol Biomol Spectros. doi: 10.1016/j.saa.2014.10.033 [DOI] [PubMed] [Google Scholar]
- Bandara WMMS, Seneviratne G, Kulasooriya SA. 2006. Interactions among endophytic bacteria and fungi: effects and potentials. J Biosci. 31:645–650. [DOI] [PubMed] [Google Scholar]
- Barnett HL, Hunter BB. 1956. Illustrated genera of imperfect fungi. 2nd ed. Minneapolis: Burgess publishing; p. 218. [Google Scholar]
- Bhagat J, Kaur A, Sharma M, Saxena AK, Chadha BS. 2012. Molecular and functional characterization of endophytic fungi from traditional medicinal plants. World J Microbiol Biotechnol. 28:963–971. [DOI] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2008. Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin Infect Dis. 48:1–12. [DOI] [PubMed] [Google Scholar]
- Cars O, Hogberg LD, Murray M, Nordberg O, Sivaraman S, Lundborg CS, So AD, Tomson G. 2008. Meeting the challenge of antibiotic resistance. BMJ. 337:a1438. [DOI] [PubMed] [Google Scholar]
- Debnath B, Debnath A, Shilsharma A, Paul C. 2014. Ethnomedicinal knowledge of Mog and Reang communities of south district of Tripura, India. Indian J Adv Plant Res. 1:49–54. [Google Scholar]
- Hamad B. 2010. The antibiotics market. Nat Rev Drug Discov. 9:675–676. [DOI] [PubMed] [Google Scholar]
- Hota D. 2010. Evalution of plant extracts In: Bioactive medicinal plants. New Delhi: Gene-Tech Books; p. 86–87. [Google Scholar]
- Jumpathong J, Seshime Y, Fujii I, Peberdy J, Lumyong S. 2011. Genome screening for reducing type I polyketide synthase genes in tropical fungi associated with medicinal plants. World J Microb Biot. 27:1989–1995. [Google Scholar]
- Kim J-S, Seo S-G, Jun B-K, Kim J-W, Kim S-H. 2010. Simple and reliable DNA extraction method for the dark pigmented fungus, Cercospora sojina. J Plant Pathol. 6:289–292. [Google Scholar]
- Krutboke I. 2010. Biodiscovery from microbial resources: actinomycetes leading the way. Focus Microbiol Aust. 53–57. [Google Scholar]
- Kusari S, Zuhlke S, Spiteller M. 2009. An endophytic fungus from Camptotheca acuminate that produces camptothecin and analogues. J Nat Prod. 72:2–7. [DOI] [PubMed] [Google Scholar]
- Lin X, Jian Y, Zhong J, Zheng H, Su WJ, Qian XM, et al. 2010. Endophytes from the pharmaceutical plant, Annona squamosa: isolation, bioactivity, identification and diversity of its polyketide synthase gene. Fungal Divers. 41:41–51. [Google Scholar]
- Melotto M, Underwood W, He SY. 2008. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol. 46:101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myres N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature. 403:853–858. [DOI] [PubMed] [Google Scholar]
- Nutzmann H-W, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schumann J, Hertweck C, Strauss J, Brakhage AA. 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc Natl Acad Sci. 108:14282–14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi S, Tayung K. 2013. Antimicrobial activity and molecular characterization of an endophytic fungus, Quambalaria sp. isolated from Ipomoea carnea. Ann Microbiol. 63:793–800. [Google Scholar]
- Patra JK, Gouda S, Sahoo SK, Thatoi HN. 2012. Chromatography separation, 1H NMR analysis and bioautography screening of methanol extract of Excoecaria agallocha L. from Bhitarkanika, Orissa, India. Asian Pac J Trop Biomed. 2:S50–S56. [Google Scholar]
- Rakshith D, Santosh P, Satish S. 2013. Isolation and characterization of antimicrobial metabolite producing endophytic Phomopsis sp. from Ficus pumila Linn. (Moraceae). Inter J Chem Anal Sci. 4:156–160. [Google Scholar]
- Rao HCY, Santosh P, Rakshith D, Satish S. 2015. Molecular characterization of an endophytic Phomopsis liquidambaris CBR-15 from Cryptolepis buchanani Roem. and impact of culture media on biosynthesis of antimicrobial metabolites. 3Biotech. 5:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao HCY, Satish S. 2015. Genomic and chromatographic approach for the discovery of polyketide antimicrobial metabolites from an endophytic Phomopsis liquidambaris CBR-18. Front Life Sci. 8:200–207. [Google Scholar]
- Rastogi G, Sani RK. 2011. Molecular techniques to assess microbial community structure, function, and dynamics in the environment In: Ahmad I, et al., editors. Microbes and microbial technology: agricultural and environmental applications. New York (NY): Springer. doi: 10.1007/978-1-4419-7931-5_2 [DOI] [Google Scholar]
- Raviraja NS, Maria GL, Sridhar KR. 2006. Antimicrobial evaluation of endophytic fungi inhabiting medicinal plants of the Western Ghats of India. Eng Life Sci. 6:515–520. [Google Scholar]
- Rodriguez RJ, White JF, Arnold AE, Redman RS. 2009. Fungal endophytes: diversity and functional roles. New Phytol. 182:314–330. [DOI] [PubMed] [Google Scholar]
- Sadrati N, Daoud H, Zerroug A, Dahamna S, Bouharati S. 2013. Screening of antimicrobial and antioxidant secondary metabolites from endophytic fungi isolated from wheat (Triticum durum). J Plant Prot Res. 53:128–136. [Google Scholar]
- Shrisha DL, Raveesha KA, Nagabhushan S. 2011. Bioprospecting of selected medicinal plants for antibacterial activity against some pathogenic bacteria. J Med Plant Res. 5:4087–4093. [Google Scholar]
- Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the infectious diseases society of America. Clin Infec Dis. 46:155–164. [DOI] [PubMed] [Google Scholar]
- Stierle A, Strobel G, Stierle D. 1993. Taxol and taxane produc-tion by Taxomyces andreanae, an endophytic fungus of pacific yew. Science. 260:214–216. [DOI] [PubMed] [Google Scholar]
- Strobel G, Daisy B, Castillo U, Harper J. 2004. Natural products from endophytic microorganisms. J Nat Prod. 67:257–268. [DOI] [PubMed] [Google Scholar]
- Strobel GA. 2003. Endophytes as sources of bioactive products. Microbes Infect. 5:535–544. [DOI] [PubMed] [Google Scholar]
- Summerbell RC, Gueidan C, Schroers H-J, Hong GS, Starink M, Rosete Y, Guarro J, Scott JA. 2011. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol. 68:139–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthari S, Sreeramalu N, Omkar K, Raju VS. 2014. The climbing plants of northern telangana in india and their ethnomedicinal and economic uses. Indian J Plant Sci. 3:86–100. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 11:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgas C, De Souza SM, Smania EFA, Smania AJ. 2007. Screening methods to determine antibacterial activity of natural products. Braz J Microbiol. 38:369–380. [Google Scholar]
- Wang J, Wang G, Zhang Y, Zheng B, Zhang C, Wang L. 2014. Isolation and identification of an endophytic fungus Pezicula sp. in Forsythia viridissima and its secondary metabolites. World J Microbiol Biotechnol. doi: 10.1007/s11274-014-1686-0 [DOI] [PubMed] [Google Scholar]
- Wang LW, Xu G, Wang JY, Su ZZ, Lin FC, Zhang CL, Kubicek CP. 2012. Bioactive metabolites from Phoma species, an endophytic fungus from the Chinese medicinal plant Arisaema erubescens. Appl Microbiol Biotechnol. 93:1231–1239. [DOI] [PubMed]
- White TJ, Bruns T, Lee S, Taylor JW. 1990. PCR protocols: a guide to methods and applications amplification and direct sequencing of fungal genes for phylogenetics In: Innis M, Gelfand DH, Sninsky JJ, White TJ, editors. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. San Diego: Academic Press; p. 315–322. [Google Scholar]
- World Health Organization 2014. Antimicrobial resistance. Global report on surveillance-2014. Geneva: World Health Organization. [Google Scholar]


