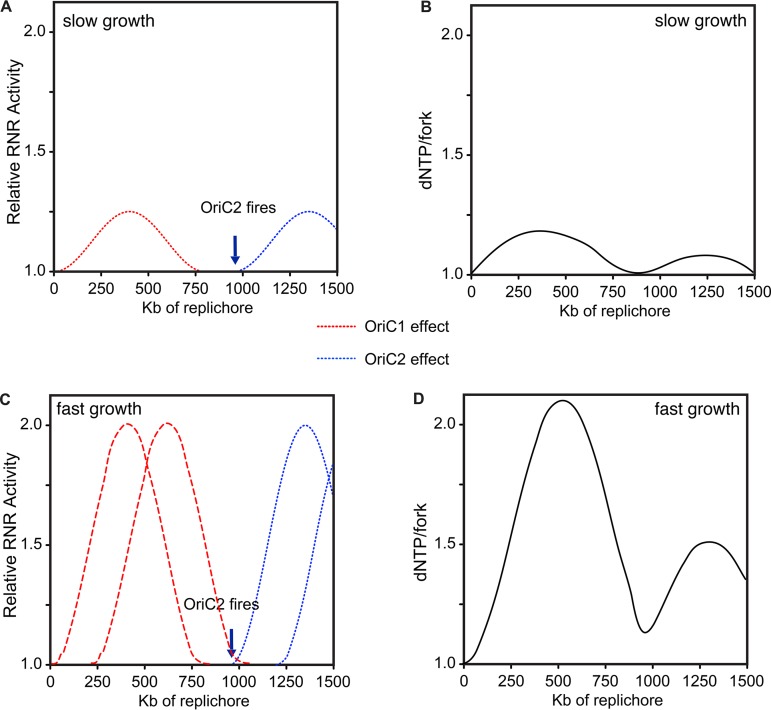
FIG 6 .
Hypothesized model of the relationship between replication timing, ribonucleotide reductase (RNR) activity, and the resulting availability of dNTPs per active replication fork. The model is fit to the V. cholerae genome with two chromosomes (chr1 and -2): one of 3.0 Mb and one of 1.1 Mb. RNR activity follows a wave that rises after the firing of the origin of chr1 and then steadily declines until additional origins fire. The chr2 origin should fire after ~950 kb of replication on each replichore of chr1 to ensure termination synchrony between chromosomes, stimulating a second wave of RNR activity. The right axis uses arbitrary relative units (dNTPs/fork) to depict how RNR activity is expected to increase dNTP pools to a maximum level (2.0) that is diluted by the number of concurrent, active forks. (A and B) Under slow growth, RNR activity rises and then falls to the baseline required to maintain synthesis. (C and D) Faster growth requires a second round of replication. Note that further rounds of overlapping replication do not significantly alter predicted dNTPs/fork, the hypothesized driver of mutation rate variability.