Abstract
Aims
Calcific aortic valve disease (CAVD) is characterized by the osteogenic transition of valve interstitial cells (VICs). In CAVD, lysophosphatidic acid (LysoPA), a lipid mediator with potent osteogenic activity, is produced in the aortic valve (AV) and is degraded by membrane-associated phospholipid phosphatases (PLPPs). We thus hypothesized that a dysregulation of PLPPs could participate to the osteogenic reprograming of VICs during CAVD.
Methods and results
The expression of PLPPs was examined in human control and mineralized AVs and comprehensive analyses were performed to document the gene regulation and impact of PLPPs on the osteogenic transition of VICs. We found that PLPP3 gene and enzymatic activity were downregulated in mineralized AVs. Multidimensional gene profiling in 21 human AVs showed that expression of PLPP3 was inversely correlated with the level of 5-methylcytosine (5meC) located in an intronic mammalian interspersed repeat (MIR) element. Bisulphite pyrosequencing in a larger series of 67 AVs confirmed that 5meC in intron 1 was increased by 2.2-fold in CAVD compared with control AVs. In isolated cells, epigenome editing with clustered regularly interspersed short palindromic repeats-Cas9 system containing a deficient Cas9 fused with DNA methyltransferase (dCas9-DNMT) was used to increase 5meC in the intronic enhancer and showed that it reduced significantly the expression of PLPP3. Knockdown experiments showed that lower expression of PLPP3 in VICs promotes an osteogenic programme.
Conclusions
DNA methylation of a MIR-based enhancer downregulates the expression of PLPP3 and promotes the mineralization of the AV.
Keywords: Calcific aortic valve disease, Aortic stenosis, Epigenetics, PLPP3, PPAP2B, Valve interstitial cells, Lipid phosphate phosphatase, Phospholipid phosphatase, Enhancer, DNA methylation, H3K27me3, BMP2, RUNX2
1. Introduction
Calcific aortic valve disease (CAVD) is a prevalent heart valve disorder, which is characterized by the mineralization of the aortic valve (AV). During CAVD, valve interstitial cells (VICs), which have a high plasticity, undergo an osteogenic reprograming.1 So far, the process whereby VICs undergo a phenotypic switch towards osteogenic reprograming remains largely elusive. The epigenome is regarded as an important regulatory layer that allows cell reprograming.2 Previous work highlighted that altered DNA methylation during CAVS may participate to gene reprograming.3 Methylation of CpG sites as well as histone modifications exert a crucial control over the expression of genes by modifying the accessibility of DNA to the transcription factors and cofactors. In this regard, enhancers, which are regulatory elements that interact with promoters, are regulated through epigenetic processes.4 Enhancers may reside in large intergenic regions or may be located within introns. A large portion of the genome consists in transposable elements (TEs), which have been exapted throughout evolution as regulatory loci of the genome.5 For instance, mammalian interspersed repeat (MIR) elements are enriched in regions containing enhancers, which have low level of 5-methylcytosine (5meC) when active.6 In this regard, cytosine methylation of enhancers has been proposed to act as a memory during developmental processes to ensure tissue-specific gene expression pattern.7 During the development of complex trait disorders, such as CAVD, the gene expression pattern is considerably modified.8,9 Whether cytosine methylation-dependent memory of key enhancers is altered in CAVD is presently unknown.
Mendelian randomization studies have underlined that low-density lipoprotein cholesterol (LDL) and lipoprotein(a) [Lp(a)] are causally related to CAVD.10,11 Lp(a) is major carrier of oxidized phospholipids (OxPLs).12 In mice, the progression of CAVD is largely dependent on the production of lysophosphatidic acid (LysoPA), a small bioactive lipid derivative presents in human mineralized AVs and generated from OxPLs by autotaxin (ATX).13 ATX is a lysophospholipase D enzyme enriched in the Lp(a) fraction and secreted by VICs.13,14 Oxidized LDL (OxLDL) promotes the osteogenic reprograming of VICs through a LysoPA receptor 1-dependent pathway, which activates BMP2, a potent promoter of osteogenic transdifferentiation.15 Membrane-associated enzymes of the phospholipid phosphatases (PLPPs) family degrade bioactive lipid derivatives including LysoPA and may therefore modulate the osteogenic response of VICs.16 Whether PLPPs are expressed and dysregulated during CAVD is presently unknown. In this work, we identified a novel process whereby cytosine methylation of a MIR retrotransposon-based enhancer promotes a downregulation of phospholipid phosphatase 3 (PLPP3/PPAP2B) as well as an osteogenic programme during CAVD.
2. Methods
Expanded methods are in the Supplementary material online.
2.1 Procurement of tissues for analyses
We examined stenotic AVs (CAVS) that were explanted from patients at the time of AV replacement. Control non-calcified AVs with normal echocardiographic analyses were obtained during heart transplant procedures. The protocol was approved by the local ethical committee and informed consents were obtained from the subjects. The study conforms with the declaration of Helsinki.
2.2 Real-time polymerase chain reaction
RNA was extracted from valves explanted from patients or from cells during in vitro experiments. Total RNA was isolated with RNeasy micro kit from Qiagen (ON, Canada). The RNA extraction protocol was performed according to manufacturer’s instructions. The quality of total RNA was monitored by capillary electrophoresis (Experion, Biorad, ON, Canada). One μg of RNA was reverse transcribed using the Qscript cDNA supermix from Quanta (VWR, Canada). Quantitative real-time PCR (qPCR) was performed with perfecta sybr supermix from Quanta on the Rotor-Gene 6000 system (Corbett Robotics Inc, CA, USA). Primers for PLPP3 (cat no. QT00052836), RUNX2 (cat no. QT00020517), BGLAP (cat no. QT00232771), and BMP2 (cat no. QT00012544) were obtained from QIAGEN (ON, Canada). The expression of the hypoxanthine-guanine phosphoribosyltransferase (HPRT, cat no. QT00059066) gene (QIAGEN, ON, Canada) was used as a reference to normalize the results.
2.3 Western blotting
Tissue pieces were mixed with lysis buffer (150 mM NaCl, 20 mM Tris pH7.5, 10% glycerol, 5 mM EGTA, 0.5 mM EDTA, 2 mM sodium vanadate, 50 mM sodium fluoride, 1% triton X-100, 0.1% Sodium dodecyl sulphate (SDS), 80 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 1 mM PMSF and protease inhibitor cocktail). Mechanical lysis was performed by using a polytron, followed by centrifugation at 300 g for 10 min at 4°C, supernatants were harvested and protein loading buffer was added. Cells were harvested in protein loading buffer. Samples were boiled 5 min, proteins were loaded onto polyacrylamide gels followed by electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked with TBS-tween containing 5% non-fat dry milk and incubated with either PLPP3 (Novus Biologicals, ON, Canada) or β-ACTIN (Sigma-Aldrich, ON, Canada) primary antibodies overnight at 4°C. Membranes were then washed and incubated with HRP-labeled secondary antibodies (Cell Signaling Technology, MA, USA). Detection was done using clarity western ECL substrate (BioRad, ON, Canada). Images were acquired and quantification analyses were performed using a ChemiDocMP system (BioRad, ON, Canada).
2.4 Measurement of phosphate lipid phosphatase phosphohydrolase activity in tissue homogenates
Control non-mineralized and CAVD valve tissues were homogenized and harvested in lysis buffer 20 mM Tris HCl, pH7.5, 1 mM EGTA, 1 mM, 1 mM DTT, 1× PIC (protease inhibitor cocktail, SIGMA, ON, Canada). Homogenates were centrifuged at 500 g for 5 min and supernatant were used for protein assay. The reaction was allowed to proceed with 1ug of protein at 37°C for 30 min in 200 μl of reaction buffer consisting of 20 mM Tris HCl, pH7.5, 1 mM MgCl2, 1 mM DTT and 100 μM LPA. Malachite Green Phosphate Detection Kit was used to measure the phosphatase activity of PLPP3, according to the manufacturer's instructions.
2.5 Quantification of LysoPA by conventional thin-layer chromatography
CAVD valve tissues (10 mg for every specimen) were homogenized for 30 s in chloroform-methanol (2:1), 1% butylated hydroxytoluene (BHT), incubated 2 h at room temperature and centrifuged 5 min at 800 g at 4°C. Samples were then extracted in chloroform:methanol (1:1), 0.5% BHT, incubated at room temperature 2 h and centrifuged. The last extraction step was performed in chloroform:methanol (1:2), 0.5% BHT, incubated 2 hurs at room temperature and centrifuged. The chloroform phases were pooled and samples were evaporated to dryness. Extracts were dissolved in 100 µl chloroform:methanol (3:2), 0.5% BHT. A 20 μl was applied to a thin-layer chromatography plate (20 × 20 cm) and 20 µg of LPA standard was applied. Visualization was performed using 0.1% Amido Black 10 b in 1 M NaCl and tissues compared on relative scale by using band intensity by using iCY platform (http://icy.bioimageanalysis.org/).
2.6 Whole-genome gene expression and DNA methylation
Total RNA was extracted from 100 mg of tissue using the RNeasy Plus Universal Mini Kit (QIAGEN, ON, Canada) according to manufacturer’s protocol. Concentration was evaluated by UV measurement using the Nanovue spectrophotometer (GE Healthcare LifeScience, NJ, USA). RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologie, CA, USA). Expression was measured on the HumanHT-12 v4 Expression BeadChip from Illumina (CA, USA). Samples were removed based on outliers in pairwise correlation, principal components analysis and hierarchical clustering. Probes were excluded of the current analysis if the detection call rate was >0.2 (if the probes was detected in at least five patients). mRNA expression data were log2 transformed and quantile normalized prior to analysis, using R 3.0.0 statistical software and the Bioconductor package lumi. Methylation sites were interrogated on the Illumina Infinium HumanMethylation450 BeadChip testing 485, 513 CpG sites located in gene promoter, 5’UTR, first exon, gene body and 3’UTR regions, densely methylated regions and methylation islands.
2.7 Pyrosequencing
DNA was purified from AV samples using the QIAamp DNA mini kit (Qiagen).
Briefly, 200 ng of gDNA was treated with sodium bisulphite (EpiTech Bisulfite Kit, Qiagen) and amplified using Pyromark PCR kit (Qiagen). Amplicons were then pyrosequenced on the PyroMark Q24 (Qiagen). Primers were designed with PyroMark Assay Design v2.0.1.15 (Qiagen) (PLPP3-forward: 5’-AAGGGGTTTTGGAGAGTAT-3’, PLPP3-reverse: 5’-AAACTTCCAAAATTAAAATCAACCT -3’, PLPP3-sequencing: GTTTTGATGATAAGTATTGTG). The assay was designed to measure DNA methylation levels of cg02468627. cg02468627 co-localizes with rs111513408 (A/G) (MAF A = 1.1%); rare subjects with the minor allele A were excluded from the study.
2.8 Chromatin immunoprecipitation
Tissues were homogenized in 1 ml of phosphate-buffered saline (PBS)1× containing PIC (Sigma, ON, Canada). The homogenized samples were centrifuged 5 minutes at 800 g at 4°C. The supernatants were removed and pellets were resuspended in PBS1× containing PIC and centrifuged 5 min at 800 g at 4°C. Pellets were then resuspended in 500 μl lysis buffer (50 mM Hepes-KOH, pH7.5, 140 mM NaCl, 1 mM EDTA pH8.0, 1% triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, PIC) and sonicated to an average length of 400 base pairs. Antibodies against H3K27me3, H3K4me1 and H3K4me3 or isotype IgG (Cell Signaling, MA, USA) were incubated with proteins G dynabeads (Life technologies, Thermofisher, ON, Canada) for 6 h before DNA samples were added to the antibodies/dynabeads mixtures and incubated at 4°C overnight on a rotator. The next day, the samples were washed with low salt buffer (0.1% SDS, 1% triton X100, 2 mM EDTA, 20 mM Tris HCl pH8.0, and 150 mM NaCl), then with high salt buffer (0.1% SDS, 1% triton X100, 2 mM EDTA, 20 mM Tris HCl pH8.0, and 500 mM NaCl), followed with LiCl buffer (10 mM Tris-HCl, pH8.0, 250 mM LiCl, 1 mM EDTA, pH8.0, 1% NP-40) and finally with TE buffer (10 mM Tris-HCl, pH8.1, 1 mM EDTA, pH8.0). The complex was then eluted from dynabeads by adding 200 µl of elution buffer (1% SDS, 100 mM NaHCO3) at 65°C for 1 h. Eluted samples were reverse cross-linked at 65°C overnight. DNA fragments were purified using phenol-chloroform and samples were analysed by quantitative PCR assay using primers specific to the enhancer region: forward 5’-GTCTGTGCAGGAGCTGTATT-3’, reverse 5’-AAGAGCGGTGGGAGTTAAAG-3’.
2.9 Cell transfection
HEK293T cells were transfected by using the calcium phosphate technique. VICs were transfected using Nanojuice transfection reagent (EMD Millipore, VWR, QC, Canada). Following transfection of dCas9-DNMT3A constructs; pdCas9-DNMT3A-PuroR, no. 71667 and pdCas9-DNMT3A-PuroR (ANV), no. 71684 (Addegene, MA, USA) cells were selected with 2 µg/ml puromycin for 48 h (Wisent, QC, Canada).
2.10 Luciferase reporter assay
HEK-293T cells were transfected with the pGL4.10 luciferase vector (Promega, WI, USA) containing the PLPP3 minimal promoter sequence (−140:−700) with or without the enhancer region located in PLPP3 first intron (Gene synthesis and subcloning, Bio Basic, ON, Canada) along with a vector encoding for the renilla luciferase (Promega, WI, USA) as a reporter for transfection efficiency as well as with the dCas9-DMNT3 constructs where indicated. At 48-h post-transfection, cells were harvested and luciferase activity was measured using the Dual-Luciferase Reporter Assay System, according to manufacturer instructions (Promega, WI, USA).
2.11 In vitro analyses of calcification
Cells were incubated for 7 days with a mineralizing medium containing: DMEM + 5% FBS, 10−7 M insulin, 50 μg/ml ascorbic acid and NaH2PO4 at 2 mM. Cells were then incubated 24 h in 0.6 N HCl, supernatants was harvested and sent for quantification and cells were harvested in 1%SDS, 0.1 N NaOH for protein quantification.
2.12 Determination of calcium concentrations
Calcium content in cell cultures was determined by the Arsenazo III method (Synermed, Monterey Park, CA, USA), which relies on the specific reaction of Arsenazo III with calcium to produce a blue complex. Results are measured at 650 nm on the Modular P800 Elecsys of Roche Diagnostics apparatus (Roche Diagnostics, QC, Canada). This reaction is specific for calcium. Magnesium is prevented from forming a complex with the reactive. Results were normalized to protein contents for cell culture experiments.
2.13 Determination of alkaline phosphatase activity
VICs were washed with PBS, transferred in 200 μl of 0.2% NP-40, 1 mmol/l MgCl2 and then sonicated. Alkaline phosphatase (ALP) activity was assayed using p-nitrophenyl phosphate as substrate (Sigma, ON, Canada). Samples were incubated in presence of substrate for 30 minutes at 37°C. The ALP activity was then measured by absorbance reading at 410 nm. The assay was carried out in triplicate. Results were normalized to protein content.
2.14 Statistical analyses
Continuous data were expressed as mean ± SEM. Normality of distribution was tested with the Shapiro-Wilk test and data compared with Student t-test or Anova when two or more than two groups were compared respectively. For data with a non-normal distribution or a n < 10 data were compared between groups with non-parametric Wilcoxon-Mann-Whitney or Kruskal-Wallis test when two or more than two groups were compared respectively. Post-hoc Steel-Dwass multiple comparisons test were performed when the P value of the Kruskal-Wallis test was <0.05. Correlation analyses were performed with Pearson’s coefficient. Categorical data were expressed as percentage and compared with Fischer’s exact test. For in vitro experiments with VICs, n represents the number of experiment performed with different donors. A P-value < 0.05 was considered significant. Statistical analysis was performed with a commercially available software package JMP 13.0 or Prism 6.0.
3. Results
3.1 PLPP3 is downregulated in mineralized AVs
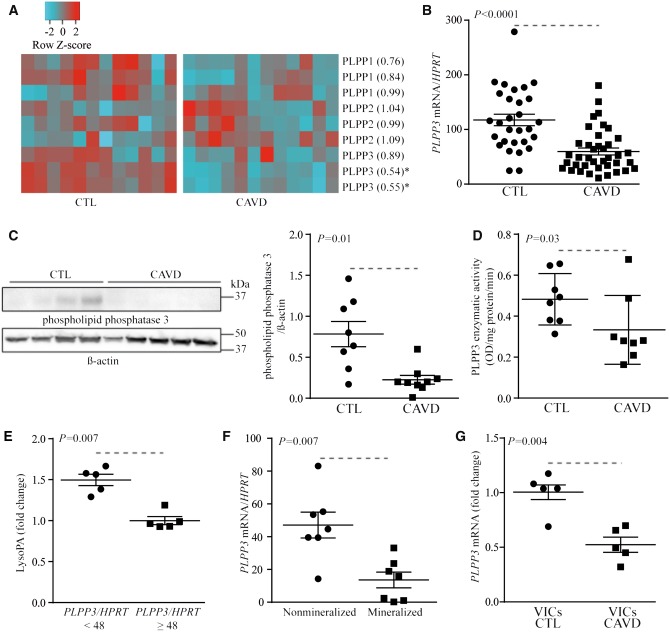
We first examined the expression of PLPPs by using whole-genome transcription dataset using array technology and performed in 12 mineralized and 12 non-mineralized control AVs (population previously described in17). Demographic and clinical data for patients included in the transcriptomic experiment are presented in Supplementary material online, Table S1. Among the different PLPPs, PLPP3/PPAP2B was reduced significantly in mineralized AVs for two probes testing this gene, whereas the expression levels of other PLPPs were not modified (Figure 1A). To support these results, PLPP3 mRNA levels were measured in an independent series of 39 mineralized and 29 control non-mineralized AVs (Table 1). Mineralized and non-mineralized AVs were obtained from AV replacement and heart transplant surgeries respectively. The level of PLPP3 mRNA was decreased by 49% in mineralized AVs (p< 0.0001; age- and sex-adjusted P-value= 0.002) (Figure 1B). There is one outlier in the control group, re-analysis without this outlier gave a similar P-value (age- and sex-adjusted P-value = 0.003). To further corroborate these findings, we next measured the PLPP3 protein level in eight control and nine mineralized AVs. By using western blotting, we documented that the level of PLPP3 was decreased by 71% in mineralized AVs (Figure 1C). In the same line, PLPP3 enzymatic activity, which was measured by an assay measuring the release of phosphate from LysoPA, was reduced by 31% in CAVD compared with control non-mineralized AVs (Figure 1D). In 10 mineralized AVs, we measured semi-quantitatively the level of LysoPA, a substrate for PLPP3, by using conventional thin-layer chromatography.13 In the AVs that were used for LysoPA measurement, the level of transcript encoding for PLPP3 was also measured. Noteworthy, the level of LysoPA was increased by 1.5-fold in AVs with less PLPP3 mRNA expression (divided at the median) (Figure 1E). Next, to address whether the level of mineralization may be associated with PLPP3 expression we measured the amount of transcripts in macroscopically dissected pathologic AVs into non-mineralized and mineralized areas. In 7 AVs from patients with CAVD, we found that the level of PLPP3 mRNA was significantly decreased in the mineralized portion compared to non-mineralized area (Figure 1F). In isolated VICs, we found that the mRNA level encoding for PLPP3 was significantly reduced in cells isolated from mineralized AVs when compared with VICs isolated from control non-mineralized valves, even after cell passages (≥3 cell passages) (Figure 1G). These data indicate that mineralization of the AV is associated with a downregulation of PLPP3, which could be linked to an epigenetic memory in CAVD.
Figure 1.
Expression of PLPP3 is reduced in CAVD. (A) Transcriptome of PLPPs in CTL (n = 12) and CAVD specimens (n = 12), the number in parentheses following the gene name represents the relative change in expression (*indicates P < 0.05). (B) mRNA levels for PLPP3 in CTL (n = 29) and CAVD (n = 39) (P < 0.0001). (C) Western blotting for PLPP3 showing a representative experiment and the quantification in CTL (n = 8) and CAVD (n = 9) (P = 0.01). (D) PLPP enzyme activity in CTL (n = 8) and CAVD (n = 8) (P = 0.03). (E) LysoPA relative levels according to PLPP3 mRNA levels (divided at median) (n = 10) (P = 0.007). (F) PLPP3 mRNA levels in pathologic aortic valves (CAVD) according to the state of mineralization (n = 7) (P = 0.007). G) PLPP3 mRNA levels in VICs isolated from CTL (n = 5) and CAVD (n = 5) (P = 0.004). Values are mean ± SEM, (B) Student t-test, (C–G) Wilcoxon-Mann-Whitney statistical analyses; CTL, control; CAVD, calcific aortic valve disease.
Table 1.
Clinical characteristics of patients for qPCR analyses
| Control valves (n = 29) | CAVD (n = 39) | P-value | |
|---|---|---|---|
| Age | 49 ± 3 | 69 ± 2 | <0.0001 |
| Male (%) | 76 | 51 | 0.14 |
| Smoking (%) | 3 | 8 | 0.4 |
| Hypertention (%) | 34 | 76 | 0.001 |
| Diabetes (%) | 21 | 31 | 0.41 |
| Coronary heart disease (%) | 34 | 38 | 0.8 |
| Bicuspid AVs (%) | 0 | 51 | <0.0001 |
| BMI (kg/m2) | 28.8 ± 1.1 | 28.6 ± 0.9 | 0.73 |
| Waist circumference (cm2) | 97.2 ± 5.7 | 101.0 ± 2.5 | 0.59 |
| Statins (%) | 62 | 100 | <0.001 |
| AV area (cm2) | – | 0.77 ± 0, 04 | – |
| Aortic peak gradient (mmHg) | – | 74.2 ± 5.4 | – |
| Aortic mean gradient (mmHg) | – | 45.6 ± 3.5 | – |
| Triglycerides (mmol/l) | 1.80 ± 0.26 | 1.75 ± 0, 18 | 0.9 |
| LDL (mmol/l) | 2.13 ± 0.22 | 2.29 ± 0, 14 | 0.49 |
| HDL (mmol/l) | 1.04 ± 0.08 | 1.31 ± 0.072 | 0.04 |
| Creatinine (μmol/l) | 111.5 ± 7.3 | 87.6 ± 3.6 | 0.006 |
| Creatinine clearance (ml/min) | 77.1 ± 7.2 | 67.1 ± 3.8 | 0.27 |
Values are mean ± S.E.M. or %; BMI, body mass index; LDL, low-density lipoprotein; HDL, high-density lipoprotein.
3.2 Dysregulation of DNA methylation is associated with the level of PLPP3
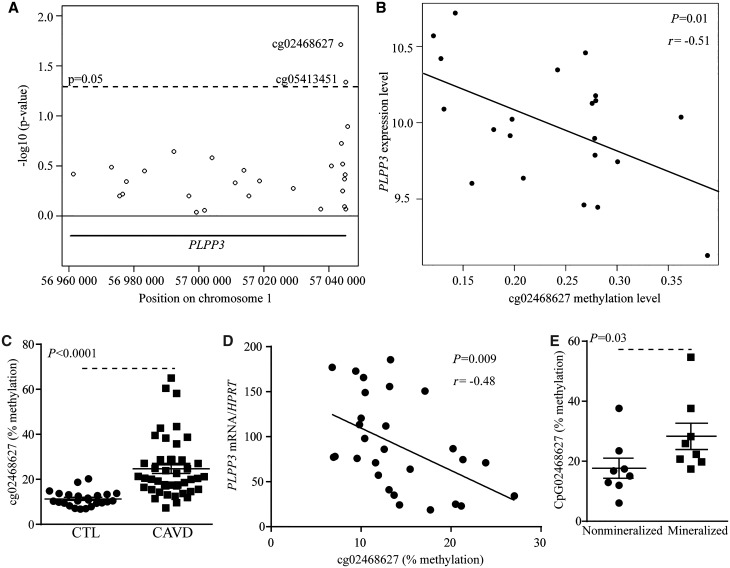
To determine if an epigenetic process could explain the dysregulation of PLPP3 in CAVD, we profiled 21 mineralized AVs for whole-genome 5meC level using the 450 K Illumina array.18 The data were analysed considering a region spanning the PLPP3 gene containing 28 CpG sites. Cytosine methylation levels of two of these CpG sites were significantly associated with PLPP3 mRNA levels, already measured using gene expression arrays (Figure 2A). Cytosine methylation in cg02468627, located in intron 1, was the most significant site and was inversely correlated with the expression of PLPP3 (r = −0.51, P = 0.01) (Figure 2B). To confirm these findings, we measured 5meC level in cg02468627 by using bisulphite treatment and pyrosequencing in an independent series of 24 control non-mineralized and 43 mineralized AVs (Table 2). We found that the level of 5meC of the CpG targeted by the cg02468627 probeset was increased by 2.2-fold in CAVD (Figure 2C). In 28 AVs, enough tissue was available to measure both 5meC (bisulphite pyrosequencing) at the same locus (cg02468627) and PLPP3 mRNA levels. In these AVs, the level of 5meC was inversely related with the amount of PLPP3 mRNA (r = −0.48, P = 0.009) (Figure 2D). Considering that we found a lower level of PLPP3 in the mineralized portion of CAVD, we also measured the level of 5meC of cg02468627 in macroscopically dissected pathologic AVs from non-mineralized and mineralized portions. In eight AVs, by using bisulphite pyrosequencing, we found that level of 5meC at cg02468627 was increased in the mineralized area compared with non-mineralized portion (Figure 2E). Hence, these data support that the level of 5meC in PLPP3 intron 1 increases during the mineralization process of the AV and is inversely associated with its mRNA levels.
Figure 2.
5meC level at cg02468627 inversely correlates with PLPP3 expression. (A) Genomic coordinates (hg19) of CpG sites (open circles) and their relationships with PLPP3 expression represented as –log10 (P-value). (B) Correlation analysis between the expression of PLPP3 and CpG methylation level (cg02468627) (n = 21) in CAVD (r = −0.51, P = 0.01). Bisulphite pyrosequencing of cg02468627 in CTL (n = 24) and CAVD (n = 43) (P < 0.0001) (C), and correlation analysis between methylation of cg02468627 with PLPP3 expression (n = 28) (r = −0.48, P = 0.009) (D). (E) Bisulphite pyrosequencing levels in pathologic aortic valves according to the state of mineralization (n = 8) (P = 0.03). Values are mean ± S.E.M.; (A) linear regression model, (B and D) Pearson’s coefficient, (C) student t-tests, (E) Wilcoxon-Mann-Whitney analysis; CTL, control; CAVD, calcific aortic valve disease.
Table 2.
Clinical characteristics of patients for bisulphite pyrosequencing
| Control valves (n = 24) | CAVD (n = 43) | P-value | |
|---|---|---|---|
| Age | 50 ± 3 | 68 ± 1 | <0.0001 |
| Male (%) | 79 | 67 | 0.4 |
| Smoking (% | 4 | 14 | 0.25 |
| Hypertention (%) | 38 | 72 | 0.009 |
| Diabetes (%) | 13 | 23 | 0.35 |
| Coronary heart disease (%) | 25 | 44 | 0.18 |
| Bicuspid AVs (%) | 0 | 33 | <0.0001 |
| BMI (kg/m2) | 27.0 ± 1.2 | 28.9 ± 0.8 | 0.08 |
| Waist circumference (cm2) | 92.5 ± 4.8 | 104.2 ± 2.2 | 0.07 |
| Statins (%) | 67 | 97 | 0.009 |
| AV area (cm2) | – | 0.77 ± 0.04 | – |
| Aortic peak gradient (mmHg) | – | 66.5 ± 4.3 | – |
| Aortic mean gradient (mmHg) | – | 41.4 ± 2.8 | – |
| Triglycerides (mmol/L) | 1.42 ± 0.16 | 1.49 ± 0.12 | 0.06 |
| LDL (mmol/l) | 2.04 ± 0.21 | 2.10 ± 0.12 | 0.057 |
| HDL (mmol/l) | 1.04 ± 0.07 | 1.37 ± 0.05 | <0.0001 |
| Creatinine (μmol/l) | 107.5 ± 7.76 | 78.0 ± 2.6 | 0.0007 |
| Creatinine clearance (ml/min) | 82.6 ± 8.2 | 81.1 ± 4.0 | 0.68 |
Values are mean ± S.E.M. or %; BMI, body mass index; LDL, low-density lipoprotein; HDL, high-density lipoprotein.
3.3 Intron 1 contains a conserved MIR transposon
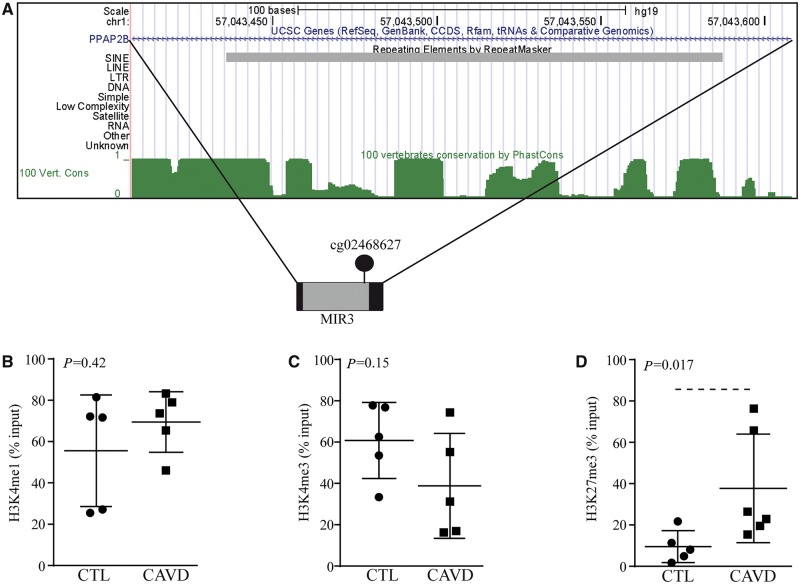
Considering that cytosine methylation of intron 1 was associated with the expression of PLPP3, we examined the gene body sequence for TEs, which often contains regulatory motifs. The intronic cg02468627 site is located within a MIR sequence and the locus is highly conserved as shown by PhastCons analysis (Figure 3A). MIRs are ancient TEs, which are enriched in enhancers and within introns of genes involved in response to biotic stimulus and inflammation.19 Histone marks were thus analysed to determine whether this intronic sequence was an enhancer. We measured in control and mineralized AVs the level of methylation (mono and tri) on lysine 4 of histone 3 (H3K4me1 and H3K4me3) by using quantitative chromatin immunoprecipitation assay (qChIP). In control and mineralized AVs, we found a high level of H3K4me1, a mark associated with enhancers, in the intron 1 region containing MIR (Figure 3B). Also, the level of H3K4me3, which is a mark associated with promoters and enhancers, was elevated in this region with a trend for lower levels in CAVD (P = 0.15) (Figure 3C). These data are consistent with a strong candidate enhancer, which is rich in both H3K4me1 and H3K4me3.20 Crosstalk between 5meC and histone methylation has been underlined and might be involved in the control of this region.21 Polycomb repressive complex 2 (PRC2), which brings H3 trimethylation on lysine 27 (H3K27me3), is positively related to 5meC and is associated with a repressive state of chromatin.21 By using qChIP, we documented that H3K27me3 level was weak in control AVs, whereas it was increased by 28% in the intron 1 region of CAVD (Figure 3D). These data are consistent with a strong enhancer in PLPP3 intron 1 in control AVs, which is, however, repressed in mineralized AVs.
Figure 3.
Intron 1 of PLPP3 contains an enhancer. (A) UCSC browser image (hg19) showing cg02468627 in highly conserved region containing a MIR3 sequence. (B–D) H3K4me1 (CTL n = 5, CAVD n = 5) (P = 0.42) (B), H3K4me3 (CTL n = 5, CAVD n = 5) (P = 0.15) (C), and H3K27me3 (CTL n = 5, CAVD n = 6) (P = 0.017) (D) levels at MIR3 locus (determined by quantitative ChIP). Values are mean ± S.E.M., Wilcoxon-Mann-Whitney analyses; CTL, control; CAVD, calcific aortic valve disease.
3.4 DNA methylation of intronic enhancer controls the expression of PLPP3
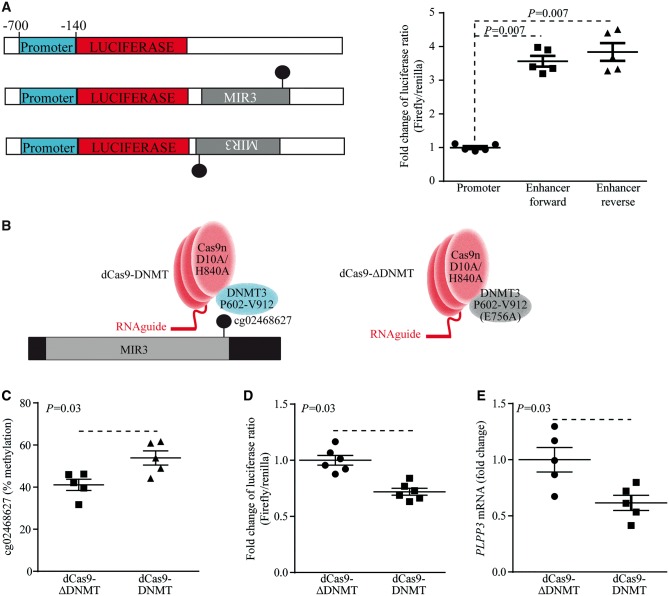
To confirm whether the intronic region is an enhancer for PLPP3, the intron 1 region containing MIR was cloned along with a minimal promoter from the PLPP3 gene (−700/−140) and was coupled with the luciferase reporter gene (Figure 4A). The different constructions were transfected in HEK293T cells. In both forward and reverse orientations, the cloned intronic sequence increased the reporter activity by 3.6- and 3.8-fold respectively (Figure 4A). These data thus indicate that intron 1 contains an enhancer, which has higher 5meC level in CAVD and may regulate the expression of PLPP3. Next, to buttress this finding epigenome editing was performed with the clustered regularly interspersed short palindromic repeats-Cas9 system to target DNA methyltransferase (DNMT) to sequence-specific site.22 Guide RNA (sgRNA) targeting the intron 1 enhancer was cloned in a vector containing a puromycin resistance gene and a nuclease-deficient Cas9 mutant (dCas9) fused with the active portion of DNMT3A (dCas9-DNMT) (Figure 4B). The control consisted in a vector containing identical target-specific sgRNA and a dCas9 fused with a catalytically inactive mutant DNMT3A (dCas9-ΔDNMT). Cells were transfected with the construction and selected with a puromycin resistance gene. When compared with the dCas9-ΔDNMT control vector, the transfection of dCas9-DNMT in HEK293T cells increased the 5meC level of cg02468627 by 13%, which was documented after 4 days by using bisulphite pyrosequencing (Figure 4C). The vectors encoding for dCas9-DNMT and dCas9-ΔDNMT were transfected in cells along with the enhancer reporter. When compared with the control inactive dCas9-ΔDNMT vector, the methyltransferase active vector dCas9-DNMT reduced the reporter activity by 28% (Figure 4D). In isolated human VICs, the transfection with dCas9-DNMT reduced PLPP3 mRNA level by 38% (Figure 4E). Taken together, these data indicate that increased 5meC in intronic enhancer downregulates the expression of PLPP3.
Figure 4.
Intronic enhancer regulates PLPP3 expression. (A) Reporter assay constructions and activities (HEK293T) (n = 5) (P = 0.007). (B) Scheme showing dCas9-DNMT and dCas9-ΔDNMT for epigenome editing. C) Bisulphite pyrosequencing of intronic enhancer performed in HEK293T after epigenome editing (n = 5) (P = 0.03). (D) Reporter assay in response to epigenome editing (HEK293T) (n = 6) (P = 0.03). (E) Expression of PLPP3 in VICs in response to epigenome editing (n = 5) (P = 0.03). Values are mean ± S.E.M., Wilcoxon-Mann-Whitney analyses.
3.5 PLPP3 promotes an osteogenic programme in VICs
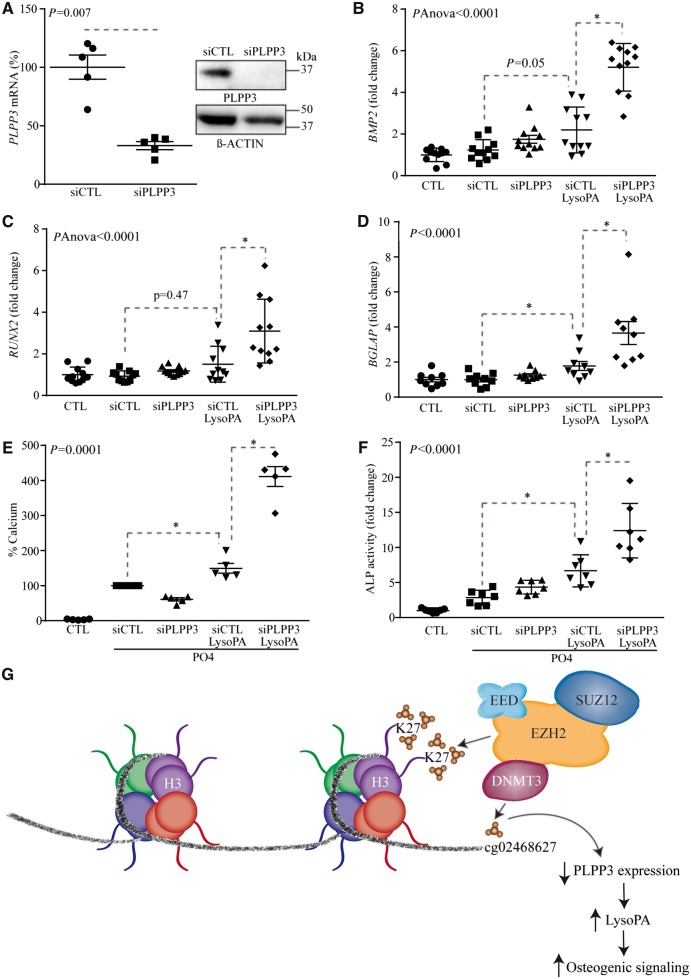
LysoPA is a strong promoter of osteogenic activity in VICs. We thus hypothesized that a lower PLPP3 expression level could exacerbate LysoPA-induced osteogenic activity in VICs. Human primary VICs isolated from non-mineralized AVs obtained from heart transplants were treated with LysoPA (10 µM) during 24 h and osteogenic genes were measured by RT-qPCR with and without a small interfering RNA (siRNA)-mediated knockdown of PLPP3 (Figure 5A). Short interfering RNA for PLPP3 (Figure 5A) increased the expression of BMP2 by 2.2-fold in response to LysoPA (Figure 5B). BMP2 is a strong promoter of osteogenesis and promotes the expression of RUNX2, a bone-related transcription factor overexpressed in human CAVD. A treatment of VICs with a siRNA targeting PLPP3 increased LysoPA-induced expression of RUNX2 and BGLAP, a downstream target of RUNX2, by 2.2- and 2- fold respectively (Figure 5C and D). We next evaluated the impact of PLPP3 on the mineralization process. VIC cultures were treated with a mineralizing medium containing LysoPA for 7 days and the amount of calcium deposited extracellularly was measured by the Arsenazo III method, which is specific and does not cross-react with other divalent ions, and results were normalized according to the protein levels.18 In this assay, the knockdown of PLPP3 increased LysoPA-induced mineralization of cell cultures by 2.8-fold (Figure 5E). Also, the activity of ALP, a marker of osteogenic activity, was increased by 1.9-fold in response to LysoPA after the silencing of PLPP3 (Figure 5F).
Figure 5.
Lower expression of PLPP3 enhances the osteogenic transition of VICs. (A) siRNA-mediated knockdown on mRNA (n = 5) (P = 0.007) and protein levels. (B–D) PLPP3 siRNA on lysoPA-mediated gene expression of BMP2 (n = 11) (pAnova < 0.0001) (B), RUNX2 (n = 11) (pAnova < 0.0001) (C), and BGLAP (n = 9) (P < 0.0001) (D). (E and F) PLPP3 knockdown on lysoPA-induced mineralization of VIC cultures (n = 5) (day 7) (P = 0.0001) (E), and ALP activity (n = 7) (day 7) (P < 0.0001) (F). (G) Proposed working model showing that EZH2 (enzyme component of the PRC2 complex that increases H3K27me3) may associate with DNMT3 (de novo methylation of CpG) and promote silencing of PLPP3 intronic enhancer during the mineralization of the AV. Values are mean ± S.E.M.; (A) Wilcoxon-Mann-Whitney analysis, (B and C) Anova, post-hoc Tukey, (D–F) Kruskal-Wallis, post-hoc Steel; *P < 0.05; LysoPA: 10 µM; PO4 is mineralizing medium (PO4 2 mM); percentage of calcium in (E) is indicated and represents a surrogate of mineralization (hydroxyapatite of calcium) that is deposited by VICs.
4. Discussion
In this work, we highlighted a novel process whereby the osteogenic transition of VICs is promoted by a downregulation of PLPP3. We found that a CpG site located in an intronic MIR-based enhancer is methylated and has a repressive chromatin state consisting in H3K27me3 in CAVD. Cytosine methylation of intronic enhancer actively participates to the downregulation of PLPP3 during the mineralization of the AV. Epigenome editing of MIR-based enhancer by using dCas9-DNMT reduced the expression of PLPP3 in VICs. In turn, reduced expression of PLPP3 in human VICs promoted the expression of osteogenic genes and the mineralization of cell cultures. Taken together, these data support that epigenetic silencing of an intronic enhancer in PLPP3 promotes an osteogenic programme in the AV (Figure 5G).
4.1 Acquired dysregulation of PLPP3 expression during mineralization of the AV
Comprehensive analyses in human AVs showed that in addition to mRNA, PLPP3 protein level, and enzymatic activity were also downregulated in pathologic specimen. In order to decipher the process whereby PLPP3 was dysregulated in CAVD we generated a multidimensional dataset including gene expression and genome-wide DNA methylation array. This analysis showed that 5meC level in cg02468627, located in intron 1 of PLPP3, was associated with gene expression level in CAVD. By using bisulphite pyrosequencing in a different set of 24 control and 43 mineralized AVs we confirmed that 5meC level at cg02468627 was increased by 2.2-fold in CAVD and was inversely associated with the expression of PLPP3. Of interest, analyses performed among pathologic leaflets showed that the mineralized portion had reduced expression of PLPP3. Incidentally, 5meC was also increased in mineralized portion compared with non-mineralized area of pathologic AVs. These data thus underscored that during the mineralization process of the AV, cytosine methylation in intron 1 is increased and this concomitantly with a reduction in PLPP3 expression.
4.2 Intron 1 contains a MIR-based enhancer for PLPP3
TEs, which comprise >45% of the genome, have been exapted for different control functions.5 To this effect, TEs of the MIR family are enriched in enhancers.6 Enhancers, which are located in large intergenic regions or within the introns, exert crucial regulatory role on the expression of genes. Previous work has shown that rs72664324, a gene variant located in intron 5 of PLPP3 and in high linkage disequilibrium with rs17114036, a single nucleotide polymorphism with a genome-wide significant association with coronary artery disease, is located in an enhancer that regulates the expression of PLPP3.23 In this work, we discovered a novel enhancer located in intron 1 of PLPP3, which is epigenetically silenced during CAVD. In reporter assay, MIR located in intron 1 increased the activity of a minimal promoter by 3.6-fold in an orientation-independent manner, which is consistent with an enhancer. In AVs, this locus has high levels of H3K4me1 and H3K4me3, which are epigenetic marks of strong enhancer.20
4.3 5meC and H3K27me3 at intronic enhancer in CAVD
Analyses of intronic enhancer at PLPP3 showed that a high level of 5meC was accompanied by an increased H3K27me3 level in CAVD. H3K27me3 is a repressive mark deposited by PRC2 often in the context of increased 5meC.21 We have used epigenome editing with dCas9-DNMT to probe the role of 5meC at intronic MIR-based enhancer.22 Of interest, dCas9-DNMT increased 5meC at cg02468627 by 13% and decreased enhancer reporter activity by 41%. In VICs, dCas9-DNMT reduced significantly the expression of PLPP3. Taken together, these data strongly suggest that increased methylation of a MIR-based enhancer in intron 1 exerts a control on the expression of PLPP3. Whether or not trimethylation on lysine 27 of histone 3 (H3K27me3) at the same locus is necessary or exacerbate the knockdown of PLPP3 is, however, unknown and warrants further investigation.
4.4 PLPP3 is a regulator of osteogenic transition
Studies have shown that Lp(a), which transports a large fraction of OxPLs in the bloodstream, is causally associated with CAVD.10 Investigations have underscored that ATX binds to Lp(a) and promotes the production of LysoPA in the AVs.13 LysoPA produced by ATX is a strong promoter of osteogenic transition in VICs. We recently identified that LysoPA derived from the OxLDL promotes in VICs the expression of BMP2, a potent morphogen that drives an osteogenic programme.15 In this work, we hypothesized that LysoPA signalling in the AV could be modulated by the expression of PLPPs. We found that PLPP3 was downregulated in VICs during CAVD and exacerbated the osteogenic transition of cells in response to LysoPA. In this regard, a knockdown of PLPP3 with siRNA increased LysoPA-induced expression of key drivers of mineralization such as BMP2 and RUNX2. Consistently, LysoPA-induced mineralization of VIC culture was exacerbated by the knockdown of PLPP3. Hence, these data indicates that PLPP3 is a key modulator of osteogenic fate in VICs.
4.5 Limitations
In this work, we delineated the role of a key regulator of LysoPA signalling. These data were obtained by using comprehensive analyses of human AVs and in vitro experiments conducted with human primary VICs. Whether the expression of PLPP3 could be modulated and whether it could impact on the development of CAVD in vivo is actually unknown. However, recent works in a preclinical mouse model of CAVD showed that circulating level of LysoPA is increased and the blockade of this pathway prevented the progression of AV mineralization.15 Also, it is worth highlighting that some variables, difficult to control for, may have affected the results. For instance, DNA methylation in tissues could differ according to cell types24 and following the plating of VICs, a heterogeneous cell population, the epigenetic memory could change.7 Hence, further work is needed to elucidate the processes whereby PLPP3 is epigenetically silenced in CAVD. In the end, it may help design novel therapies that would modulate LysoPA signalling.
5. Conclusion
The expression of PLPP3 is decreased in CAVD and promotes the osteogenic transition of VICs. We discovered a novel intronic enhancer, which has increased 5meC level in CAVD and exerts an important control over the expression of PLPP3. Gene reprogramming of VICs at PLPP3 enhancer may hold promise for the development of novel epigenetic-based therapies.
Authors’ contributions
G.M., M.C.B., and P.M. conceived and designed experiments. G.M. performed cell culture experiments and qChIP. G.M. and M.C.B. performed genome editing experiments. G.M. performed luciferase reporter assays. V.G.O. and L.B. performed pysosequencing experiments. M.J.N. performed mineralization experiments. G.M., F.H., D.A., and G.R. performed qPCRs. N.G. and Y.B. generated multidimensional datasets including transcriptomic and 5meC array. G.M. and M.R. generated tables. M.C.B. generated the figures. P.M. drafted the article. All the authors critically reviewed the article and provided scientific input.
Supplementary Material
Acknowledgements
This work was supported by CIHR grants to P.M. (MOP114893, MOP245048, MOP341860, and MOP365029) the Heart and Stroke Foundation of Canada and the Quebec Heart and Lung Institute Fund. LB is a junior research scholar from the Fonds de la recherche du Québec en santé (FRQS) and a member of the FRQS-funded Centre de recherche du CHUS (affiliated to the Centre hospitalier universitaire de Sherbrooke). VGO received a doctoral research award from FRQS. Y.B. holds a Canada Research Chair in Genomics of Heart and Lung Diseases. P.M. holds a FRQS Research Chair on the Pathobiology of Calcific Aortic Valve Disease.
Conflict of interest: none declared.
Footnotes
Time for primary review: 36 days
References
- 1. Mathieu P, Boulanger MC.. Basic mechanisms of calcific aortic valve disease. Can J Cardiol 2014;30:982–993. [DOI] [PubMed] [Google Scholar]
- 2. Migicovsky Z, Kovalchuk I.. Epigenetic memory in mammals. Front Genet 2011;2:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagy E, Back M.. Epigenetic regulation of 5-lipoxygenase in the phenotypic plasticity of valvular interstitial cells associated with aortic valve stenosis. FEBS Lett 2012;586:1325–1329. [DOI] [PubMed] [Google Scholar]
- 4. Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B.. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 2013;45:1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 2008;9:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jjingo D, Conley AB, Wang J, Marino-Ramirez L, Lunyak VV, Jordan IK.. Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mob DNA 2014;5:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee HJ, Hore TA, Reik W.. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell 2014;14:710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosse Y, Miqdad A, Fournier D, Pepin A, Pibarot P, Mathieu P.. Refining molecular pathways leading to calcific aortic valve stenosis by studying gene expression profile of normal and calcified stenotic human aortic valves. Circ Cardiovasc Genet 2009;2:489–498. [DOI] [PubMed] [Google Scholar]
- 9. Guauque-Olarte S, Droit A, Tremblay-Marchand J, Gaudreault N, Kalavrouziotis D, Dagenais F, Seidman JG, Body SC, Pibarot P, Mathieu P, Bossé Y.. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol Genomics 2016;48:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thanassoulis G, Campbell CY, Owens DS, Smith JG, Smith AV, Peloso GM, Kerr KF, Pechlivanis S, Budoff MJ, Harris TB, Malhotra R, O'Brien KD, Kamstrup PR, Nordestgaard BG, Tybjaerg-Hansen A, Allison MA, Aspelund T, Criqui MH, Heckbert SR, Hwang SJ, Liu Y, Sjogren M, van der Pals J, Kalsch H, Muhleisen TW, Nothen MM, Cupples LA, Caslake M, Di AE, Danesh J, Rotter JI, Sigurdsson S, Wong Q, Erbel R, Kathiresan S, Melander O, Gudnason V, O'Donnell CJ, Post WS.. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med 2013;368:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith JG, Luk K, Schulz C-A, Engert JC, Do R, Hindy G, Rukh G, Dufresne L, Almgren P, Owens DS, Harris TB, Peloso GM, Kerr KF, Wong Q, Smith AV, Budoff MJ, Rotter JI, Cupples LA, Rich S, Kathiresan S, Orho-Melander M, Gudnason V, O’donnell CJ, Post WS, Thanassoulis G.. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. Jama 2014;312:1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathieu P, Arsenault BJ, Boulanger MC, Bosse Y, Koschinsky ML.. Pathobiology of Lp(a) in calcific aortic valve disease. Expert Rev Cardiovasc Ther 2017;15:797–807. [DOI] [PubMed] [Google Scholar]
- 13. Bouchareb R, Mahmut A, Nsaibia MJ, Boulanger MC, Dahou A, Lepine JL, Laflamme MH, Hadji F, Couture C, Trahan S, Page S, Bosse Y, Pibarot P, Scipione CA, Romagnuolo R, Koschinsky ML, Arsenault BJ, Marette A, Mathieu P.. Autotaxin derived from lipoprotein(a) and valve interstitial cells promotes inflammation and mineralization of the aortic valve. Circulation 2015;132:677–690. [DOI] [PubMed] [Google Scholar]
- 14. Nsaibia MJ, Mahmut A, Boulanger MC, Arsenault BJ, Bouchareb R, Simard S, Witztum JL, Clavel MA, Pibarot P, Bosse Y, Tsimikas S, Mathieu P.. Autotaxin interacts with lipoprotein(a) and oxidized phospholipids in predicting the risk of calcific aortic valve stenosis in patients with coronary artery disease. J Intern Med 2016;280:509–517. [DOI] [PubMed] [Google Scholar]
- 15. Nsaibia MJ, Boulanger MC, Bouchareb R, Mkannez G, Le QK, Hadji F, Argaud D, Dahou A, Bosse Y, Koschinsky ML, Pibarot P, Arsenault BJ, Marette A, Mathieu P.. OxLDL-derived lysophosphatidic acid promotes the progression of aortic valve stenosis through a LPAR1-RhoA-NF-kappaB pathway. Cardiovasc Res 2017;113:1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brindley DN, Pilquil C.. Lipid phosphate phosphatases and signaling. J Lipid Res 2009;50 Suppl:S225–S230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guauque-Olarte S, Messika-Zeitoun D, Droit A, Lamontagne M, Tremblay-Marchand J, Lavoie-Charland E, Gaudreault N, Arsenault BJ, Dubé M-P, Tardif J-C, Body SC, Seidman JG, Boileau C, Mathieu P, Pibarot P, Bossé Y.. Calcium signaling pathway genes RUNX2 and CACNA1C are associated with calcific aortic valve disease. Circ Cardiovasc Genet 2015;8:812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hadji F, Boulanger M-C, Guay S-P, Gaudreault N, Amellah S, Mkannez G, Bouchareb R, Marchand JT, Nsaibia MJ, Guauque-Olarte S, Pibarot P, Bouchard L, Bossé Y, Mathieu P.. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation 2016;134:1848–1862. [DOI] [PubMed] [Google Scholar]
- 19. Sironi M, Menozzi G, Comi GP, Cereda M, Cagliani R, Bresolin N, Pozzoli U.. Gene function and expression level influence the insertion/fixation dynamics of distinct transposon families in mammalian introns. Genome Biol 2006;7:R120.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE.. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van EA, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di CL, de LY, Fuks F.. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006;439:871–874. [DOI] [PubMed] [Google Scholar]
- 22. Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, Zoldos V.. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016;44:5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reschen ME, Gaulton KJ, Lin D, Soilleux EJ, Morris AJ, Smyth SS, O'Callaghan CA.. Lipid-induced epigenomic changes in human macrophages identify a coronary artery disease-associated variant that regulates PPAP2B Expression through Altered C/EBP-beta binding. PLoS Genet 2015;11:e1005061.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan S, Lai H, Shen Y, Breeze C, Beck S, Hong T, Wang C, Teschendorff AE.. DNA methylome analysis reveals distinct epigenetic patterns of ascending aortic dissection and bicuspid aortic valve. Cardiovasc Res 2017;113:692–704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.