Abstract
CONTEXT:
Microorganisms produce a variety of pigments and many pigments from bacteria were reported to have therapeutic potential including anticancer effects.
AIM:
The aim of this study is to evaluate the anticancer potential a yellow pigment from newly isolated Pseudomonas stutzeri JGI 52.
MATERIALS AND METHODS:
Serial dilution method was adopted for the isolation of pigmented bacteria from soil sources. Pigment extraction was carried out from bacterial isolates using methanol as the solvent and the pigment was purified by thin layer chromatography. Through 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay, the effect of the pigment fraction on cancer cells was analyzed. Apoptosis induction was evaluated by caspase-3 activity assay, DNA fragmentation analysis, cell morphology observation by AO-EB staining under the fluorescence microscope, and cellular cytotoxicity was analysed by lactate dehydrogenase (LDH) release assay. Characterization of the purified pigment was by high-performance liquid chromatography and electrospray ionization-mass spectrometry analysis.
STATISTICAL ANALYSIS:
Significance of the results was confirmed by performing one-way analysis of variance.
RESULTS:
The pigment (PY3) from P. stutzeri inhibited the proliferation of HeLa, HepG2, and Jurkat cells and found to be less toxic to lymphocytes and CHO cells. PY3 exhibited apoptotic potential in the cancer cell lines, as evidenced by cleavage of DNA, LDH release, activation of caspase-3, and decrease in cell count. Results of mass spectra indicated the presence of “fucoxanthinol” which was earlier reported as an anticancer compound from seaweeds.
CONCLUSIONS:
This study revealed that the pigment PY3 from P. stutzeri has anticancer potential and induced cell death by apoptosis. It was found to have the carotenoid fucoxanthinol, responsible for its observed anticancer activity.
Keywords: Cytotoxicity, fucoxanthinol, HeLa, pigment, Pseudomonas stutzeri
Introduction
Cancer, one of the serious health problems afflicting mankind, has become a major cause for the death of millions of people around the globe, despite the accessibility to several treatment modalities such as chemotherapy, immunotherapy, radiation therapy, and surgery.[1] In the recent years, many commercially available anticancer drugs were originated either as chemical synthetic drugs or natural compounds derived from plants and microorganisms.[1,2,3] Besides being tremendously expensive, chemical drugs induce killing not only of cancer cells but also of normal cells, and majority are associated with life-threatening side effects and morbidity.[4] Natural compounds have contributed significantly toward drug discoveries aiming for cancer chemotherapy.[5] For example, anthracyclines (doxorubicin), bleomycin, dactinomycin (actinomycin), and mitomycin C were included in the class of antitumor antibiotics from microbes.[4] In summation, various examples of promising natural product-derived anticancer compounds from microbes were reported which are presently in advanced clinical development or were recently approved (e.g., epothilones and enediynes).[6] Hence, in our search for a novel therapeutic compound of microbial origin, in the present work, we isolated a compound (yellow in color) from Pseudomonas stutzeri JGI 52 and analyzed its anticancer effects against in vitro cancer cell lines and made an attempt to partially characterize the anticancer pigment.
Materials and Methods
Cell lines and growth conditions
Cervical cancer (HeLa), liver cancer (HepG2), leukemia (Jurkat), and normal ovarian (CHO) cell lines were procured from National Center for Cell Sciences (Pune, India) and maintained in Dulbecco's Modified Eagle's Medium (HiMedia, REF-AT065A), Modified Eagle's Medium (HiMedia. India, REF-AT017A), and RPMI-1640 (HiMedia, India, AT126A) media, supplemented with 10% fetal bovine serum (HiMedia, REF-RM112), 100 μg/ml of streptomycin, and 100 U/ml of penicillin. Cells were routinely subcultured and maintained at 37°C in a humified atmosphere of 5% CO2. Lymphocytes were collected from the blood of healthy individuals. Blood sampling was carried out by adopting the ethical guidelines for research (ICMR).
Isolation and identification of bacteria
Pigmented colonies of bacteria were isolated from soil samples of Bengaluru, India, by serial dilution method.[7] From the promising isolate, genomic DNA was extracted using bacterial genomic DNA isolation kit (Chromous Biotech, Bangalore). Identification of bacteria was performed by partial amplification of 16S rDNA using the forward primer, 5'-AGAGTTTGATCMTGGCTCAG-3', and reverse primer, 5'-TACGGYTACCTTGTTACGACTT-3' (ABI 3500xL Genetic Analyzer).
Extraction, purification, and determination of λ-max of the pigment
The yellow pigment from our bacterial isolate was extracted using methanol, following standard protocols.[8] The extract was fractionated by thin layer chromatography (TLC) on silica gel-coated TLC plates (60F 254, Merck) using acetone: ethyl acetate (1:1 v/v) as the solvent system. The fractions separated on TLC plates were visualized with the aid of an ultraviolet (UV) transilluminator (254 and 366 nm), and all the fractions were collected separately. The absorption maxima (λ-max) of the yellow-colored fraction (PY3) were determined by subjecting to spectral scanning (200–700 nm) in a UV-visible spectrophotometer (Shimadzu, UV 1800, Japan).
Assay of cytotoxicity
The cytotoxicity of the pigment fraction (PY3) against various cancer cell lines was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye conversion assay following the standard protocol.[9] In 96-well microtiter cell culture plates, cells were cultured in the absence (control) and presence of different concentration (0.25, 0.5, 1, 5, 10, and 20 μg/ml) of PY3 for 24, 48, and 72 h. After the indicated treatment period, cells were incubated for 3 h (dark chamber) in a media containing 20 μl of MTT dye (prepared in PBS). After incubation, the purple-colored crystals of MTT formazan were mixed with 100 μl of dimethyl sulfoxide for solubilization, and the optical density was recorded at 540 nm with the help of an ELISA reader (Lisa plus). The viability percentage was calculated using the formula, Percentage inhibition = (1 − Optical Density of sample/Optical Density of control) × 100.
Analysis of DNA fragmentation
Adhered cancer cell lines in exponential phase were treated with IC50 concentration of PY3 for 24 h, followed by trypsinization (Trypsin-HiMedia, REF-TCL007) and DNA extraction using the Mammalian genomic DNA isolation kit (HiPurA™, REF-MB506).[10] The extracted DNA was mixed with gel-loading dye and Tris-ethylenediaminetetraacetic acid (TE) buffer (20 μl each), loaded to the wells of agarose gel (0.8%), and electrophoresed. Using a UV transilluminator (Bangalore Genei), the gel was visualized.
Caspase-3 apoptotic assay
Caspase-3 activity was determined using the calorimetric assay kit for caspases (G-Biosciences Ltd., USA, Cat. No. 786-205A). Cleavage of synthetic substrate which has 7-amino-4-trifluoromethyl-coumarin (AFC) at the C-terminal was detected by this assay. AFC produces an optical change when liberated from the peptide which can be detected by colorimetric methods. HeLa cells were treated with PY3 fraction for 24 h. After 24 h, the cells were trypsinized and centrifuged, and the pelleted cells were resuspended in 50 μl of lysis buffer (supplied with the kit). The suspension was centrifuged and the supernatant was collected. About 50 μl of caspase assay buffer 1 M containing dithiothreitol was added to the lysate. About 5 μl of AFC conjugated substrate (50 μM final concentration) was added and incubated at 37°C for 2 h. Optical density (at 405 nm) was recorded at the beginning of the assay, followed by every 15 min, until a significant difference in absorbance was observed. The results were expressed as mean percentage caspase activity ± standard error of the mean for triplicates.
Lactate dehydrogenase analysis
Lactate dehydrogenase (LDH) is a colorimetric method of determination of cellular cytotoxicity. Due to the pigment PY3 treatment, cytotoxicity induced to cancer cells was quantitatively assayed by checking the activity of LDH, which is released upon cell lysis. Overnight-cultured cancer cells were treated with 20 μg/ml concentration of PY3 for 24 h, washed twice with PBS (pH 7.4), and collected by trypsinization and centrifugation for 10 min (1000 rpm). As per the instructions provided in the kit (from G-Biosciences Ltd., USA), the assay was performed. The optical density of treated, positive control, and negative control samples was recorded at 490 nm, which was used to calculate percentage cytotoxicity.
Chemical screening
The methanol extract and the yellow fraction (PY3) were subjected to tests pertaining to the detection of chemical constituents such as the glycosides, steroids, alkaloids, carotenoids, phenols, carbohydrates, protein, lipids, and flavonoids, as per the standard methodologies.[11,12]
High-performance liquid chromatography and electrospray ionization-mass spectrometry analysis of yellow fraction
High-performance liquid chromatography (HPLC) was employed to further purify the yellow fraction (PY3). A WATERS HPLC system having a binary pump (1525), a λ UV dual detector, and a column of C-18 octadecylsilane was used for the purification. Filtration of the sample and the mobile phase through 0.22 μm polyvinylidene fluoride filter was carried out before injecting them to the column. The sample was eluted using 1:2 ratio of methanol and water as the solvent, while keeping 1 mL/min as the standard flow rate. The fraction from HPLC was further subjected to electrospray ionization-mass spectrometry (ESI-MS). With the help of a Hewlett Packard HP 1100 MSD series mass spectrometer, the spectra were collected for a mass range of 50–1500 m/z.
Statistical analysis
All of the experiments were performed in triplicates. The statistical significance of the results obtained was verified by applying one-way analysis of variance calculations, making use of the GraphPad prism version 6.01 for windows (GraphPad Software, La Jolla, California, USA). P < 0.05 was taken as the standard for inferring statistical significance.
Results
When few pigmented bacterial colonies isolated from different soil sources were screened for their cytotoxic potential to HeLa, HepG2, and Jurkat cell lines, it was found that a yellow-colored bacterial isolate was having promising properties. This isolate was subjected to 16S rRNA sequence analysis. The sequence was subjected to BLAST, and the isolate was identified as Pseudomonas stutzeri JGI 52. In the GenBank, the sequence was deposited with the accession number KM386644.
Cytotoxic effect of PY3 on cervical, liver, and leukemia cell lines
TLC-purified fractions were tested for their cytotoxicity on cancer cells by MTT assay and the yellow-colored fraction named as PY3, exhibited highest cytotoxic activity. When the effect of PY3 on the viability of three cancer cell lines, i.e., HeLa, HepG2, and Jurkat was analyzed, it was found that the cell viability was inversely proportional to the increasing concentration and time of treatment of PY3. Viability of cells was reduced to < 50% in all the cell lines after 72 h and exposure to PY3 at 20 μg/ml concentration. The IC50 value of PY3 was 12.5, 13.9, and 14.5 μg/ml, respectively, on HeLa, HepG2, and Jurkat cell lines [Figure 1]. On the contrary, when PY3 was treated on to normal peripheral lymphocytes and CHO cells, the percentage viability was found to be >90% at all the tested concentrations, indicating that the pigment is nontoxic to normal cells.
Figure 1.
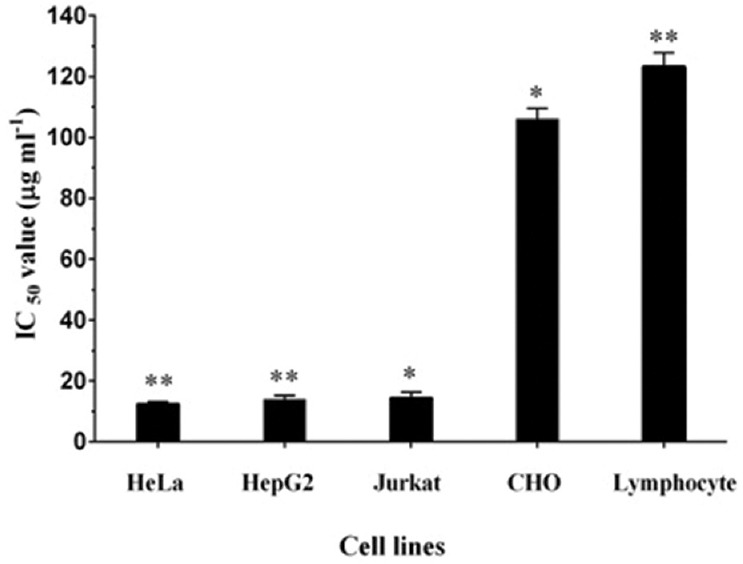
PY3 from Pseudomonas stutzeri inhibits the proliferation of HeLa, HepG2, and Jurkat cells with IC50 values of 12.48, 13.9, and 14.48 μg/ml. Each bar represents the mean of three replicates. *indicatesp< 0.05 and **indicatesP< 0.001 when compared to the untreated control
Further to determine the mechanism by which the percentage viability of the treated cells was brought down, DNA fragmentation analysis was performed. When the DNA extracted from PY3-treated HeLa and HepG2 cells was analyzed by 0.8% agarose gel electrophoresis, a DNA smear was observed on the gel, which indicated the fragmented or cleaved DNA which is one of the hallmarks of apoptotic cells [Figure 2]. However, a single band of DNA appeared in the gel when DNA from the untreated control cells was electrophoresed, confirming the presence of intact DNA in the untreated cells. DNA fragmentation indicates that DNA of the treated cells got cleaved into oligomers of 180–200 bp, and hence, we assume that PY3 had induced apoptosis in the cancer cells and thereby brought down the percentage viability.
Figure 2.
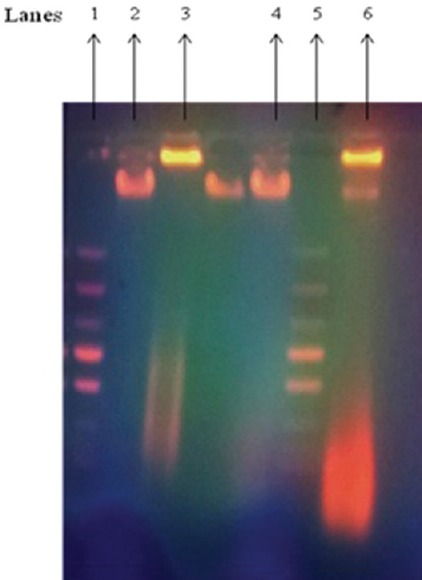
Effect of PY3 on DNA fragmentation in HeLa and HepG2 cells, Lanes 1. Ladder DNA, 2. DNA from untreated HeLa cells, 3. DNA from the PY3-treated HeLa cells, 4. DNA from untreated HepG2 cells, 5. Ladder DNA 6. DNA from PY3-treated HepG2 cells
Caspase-3 activation is one of the significant events happening during programmed cell death and it is the primary mediator of apoptosis. Lysates of HeLa and HepG2 cells after 48 h of exposure to PY3 had significantly increased activity of caspase-3, and there was about 6–7-fold increase in the activity of caspases when compared to that of the untreated control cells. The percentage increase in the caspase activity in HeLa and HepG2 cells was 78.92% and 65.4%, respectively, when caspase activity was analyzed from 0 to 45 min [Figure 3]. This further confirmed the induction of apoptosis by the pigment PY3, in the cervical and liver cancer cell lines.
Figure 3.
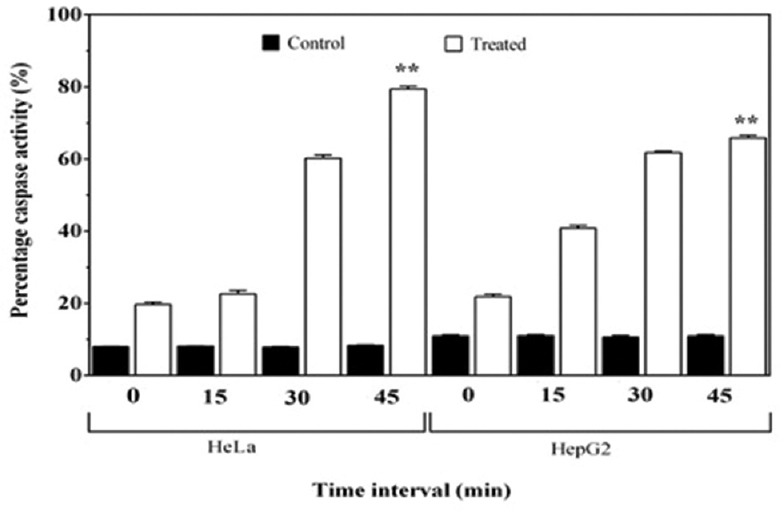
Comparison of percentage caspase activity between treated and control HepG2 and HeLa cells, with respect to time interval (0–45 min). Each bar represents the mean of three replicates. **indicatesP< 0.001 in comparison to the control
LDH release from the apoptotic cell is an indicator of cytotoxicity and LDH activity measured in the Triton-X-treated cells was used to estimate maximum LDH release. The cytotoxicity percentage in the HeLa and HepG2 cells was 62.63% and 55.02%, respectively, as compared with the positive control [Figure 4]. This indicates that PY3 was cytotoxic to the cells and induced the release of LDH when added to the cancer cell lines, i.e. HeLa and HepG2.
Figure 4.
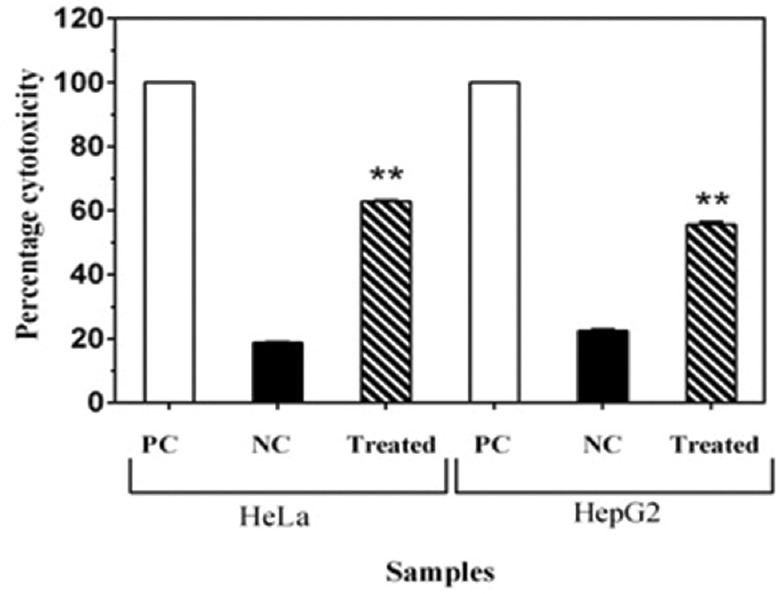
Percentage cytotoxic effects of PY3 on HeLa and HepG2 cells, as per lactate dehydrogenase leakage assay. PC = Positive control, NC = Negative control. Each bar indicates the mean of three replicates. **indicatesP< 0.001 level of significance
Characterization of the bioactive compound
As the yellow pigment PY3 was demonstrating promising anticancer effects, we wanted to characterize this compound and subjected the crude extract and PY3 to chemical screening to identify the active compound responsible for this bioactivity. Crude extract indicated the presence of carotenoid, phenol, and carbohydrates whereas the purified PY3 fraction indicated the presence of carotenoids [Table 1]. Further characterization of this fraction was carried out by performing LC-MS analysis. When PY3 was subjected to HPLC analysis, the larger peak at the retention time (RT) 15.3 min was found to have 100% relative abundance [Figure 5a]. The mass spectrometric analysis of this peak resulted in several fragment ions [Figure 5b]. One of the fragment ions was having an m/z value of 634.45. By library search and as per the database (https://metlin.scripps.edu/metabolites_list.php), this was found to corresponding to the compound fucoxanthinol with a molecular weight 616.42. As per the reports from earlier works, this carotenoid compound was having anticancer activity.[13,14] Hence, we assume that the presence of this compound might be responsible for the observed anticancer effects of this yellow pigment (PY3) from our isolate P. stutzeri JGI 52.
Table 1.
Preliminary chemical screening of crude extract and purified yellow fraction (PY3) from Pseudomonas stutzeri
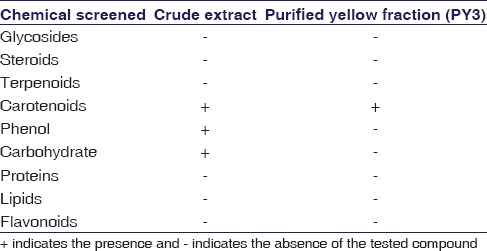
Figure 5.
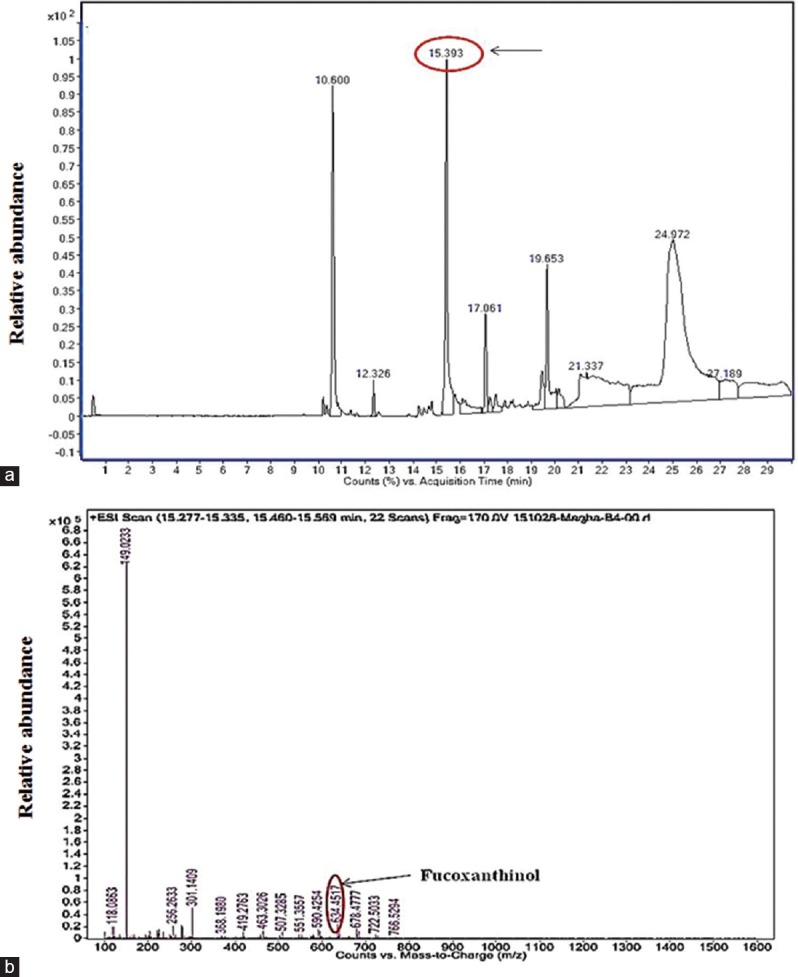
(a) High-performance liquid chromatography of PY3 showing larger peak at retention time 15.39 min (b) electrospray ionization-mass spectrum of the PY3 showing one of the fragment ions with an m/z value of 634.45 that indicates the presence of the compound fucoxanthinol
Discussion
Majority of the anticancer drugs involved in chemotherapy for cancer were reported as to mediate their effects by activating the apoptotic cascade in the cancer cells.[15] Search for newer molecule/approach capable of inducing apoptosis in cancer cells will lead to the development of effective strategies in cancer therapy. Taking this as our main goal, the current research work was aimed toward the isolation of novel microbes from soil sources and screening for their anticancer potentials. In this pursuit, we extracted a yellow pigment from a promising bacterial isolate identified as P. stutzeri JGI 52 and analyzed its anticancer effects on in vitro cancer cell lines. The results of our studies demonstrated that the yellow pigment (PY3) is strongly cytotoxic, induced apoptosis in HeLa, HepG2, and Jurkat cell lines and is nontoxic to nonmalignant cells (normal human lymphocytes and CHO cells). The dosage and time-dependent growth inhibitory effect of PY3 on all the cancer cells were observed. PY3 markedly decreased the viability (<50%) of all the cancer cells analyzed and had the IC50 value ranging from 12.48 to 14.48 μg/ml, whereas lymphocytes and CHO cells did not show any signs of toxicity at the same or higher concentrations (IC50 value >100 μg/ml).
Sensitivity of the cancer cell line to the anticancer compound is an essential component to be looked upon while choosing the chemotherapeutic agents. Here, in our study, all the cancer cell lines were sensitive to PY3, which induced apoptosis as shown by DNA fragmentation analysis. Furthermore, we demonstrated the onset of apoptosis that involves the activation of caspase-3 and release of LDH from cancer cells. In caspase-3 activity assay, we observed a 6–7-fold increment in the activity of caspases in the treated cells as compared to that of the control cells. Activation of caspase-3 is proved to be an important biomarker in cells undergoing apoptosis and is one of the major qualities (traits) to be considered while searching for anticancer drugs.
The yellow fraction (PY3) was subjected to HPLC and ESI-MS analysis for characterization. In HPLC chromatogram, we observed few major peaks and the one at RT 15.3 min was having 100% abundance. The mass spectrometric analysis of this larger peak resulted in several fragments and out of which the fragment ion with an m/z value of 634.45 was found to correspond to the compound “fucoxanthinol,” having a molecular weight 616.42. Fucoxanthinol is a metabolite of fucoxanthin, which belongs to a carotenoid class, present mostly in brown seaweeds.[13] As per our chemical screening test also it was confirmed as a carotenoid. Fucoxanthinol was reported to have many health effects such as antimutagenic, anti-diabetic, antiobesity, anti-inflammatory, and anticancer effects on different types of cancers such as the prostate, colon, liver, lung, and breast.[16,17] The inhibitory effects of fucoxanthinol against prostate cancer cell line PC-3 was reported in a previous study.[18] In another study, it was reported that fucoxanthinol had inhibitory effects against HepG2 cells, which was associated with cyclin D downregulation.[19] It was also reported that fucoxanthinol extracted from Undaria pinnatifida inhibited the cell viability of adult T-cell leukemia as well as human T-cell leukemia cells without any cytotoxic effects to normal blood cells.[20]
Conclusion
These studies revealed that fucoxanthinol is a potent anticancer agent that is effective against a variety of cancer cell lines by inducing apoptosis. Hence, we assume that the presence of this compound in the pigment of our isolated bacteria, P. stutzeri, might be responsible for the observed cytotoxic effects against cancer cell lines. This study shows its potential for developing into a cancer-chemotherapeutic drug, after extensive in vivo and clinical studies.
Financial support and sponsorship
This study was financially supported by DST INSPIRE Fellowship.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors gratefully acknowledge the financial support by the Department of Science and Technology (DST/INSPIRE Fellowship/2011/213). The support by management of Jain Group of Institutions is also gratefully acknowledged by the authors.
References
- 1.Ma X, Wang Z. Anticancer drug discovery in the future: An evolutionary perspective. Drug Discov Today. 2009;14:1136–42. doi: 10.1016/j.drudis.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Chabner BA, Roberts TG., Jr Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72. doi: 10.1038/nrc1529. [DOI] [PubMed] [Google Scholar]
- 3.DeVita VT, Jr, Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68:8643–53. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- 4.Cragg GM, Grothaus PG, Newman DJ. Impact of natural products on developing new anti-cancer agents. Chem Rev. 2009;109:3012–43. doi: 10.1021/cr900019j. [DOI] [PubMed] [Google Scholar]
- 5.Kinghorn AD. Drug discovery from natural products. In: Lemke TL, Williams DA, editors. Foye's Principles of Medicinal Chemistry. 6th ed. Philadelphia, USA: Lippincott Williams & Wilkins; 2008. pp. 12–25. [Google Scholar]
- 6.Newman DJ. The bryostatins. In: Cragg GM, Kingston DG, Newman DJ, editors. Anticancer Agents from Natural Products. Boca Raton, FL: CRC Press; 2005. pp. 137–50. [Google Scholar]
- 7.Aneja KR. Experiments in Microbiology, Plant Pathology and Biotechnology. 4th ed. New Delhi: New Age International (P) Ltd. Publishers; 2003. [Google Scholar]
- 8.Sasidharan P, Raja R, Karthik C, Ranandkumar S, Indra AP. Isolation and characterization of yellow pigment producing Exiguobacterium sps. J Biochem Technol. 2013;4:632–5. [Google Scholar]
- 9.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen MS, Chen D, Dou QP. Inhibition of proteasome activity by various fruits and vegetables is associated with cancer cell death. In Vivo. 2004;18:73–80. [PubMed] [Google Scholar]
- 11.Harborne JB. Phytochemical Methods. London, UK: Chapman and Hall Ltd.; 1973. pp. 49–188. [Google Scholar]
- 12.Wadood A, Ghufran M, Jamal SB, Naeem M, Khan A, Ghaffar R, et al. Phytochemical analysis of medicinal plants occurring in local area of Mardan. Biochem Anal Biochem. 2013;2:2–4. [Google Scholar]
- 13.Martin LJ. Fucoxanthin and its metabolite fucoxanthinol in cancer prevention and treatment. Mar Drugs. 2015;13:4784–98. doi: 10.3390/md13084784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zorofchian Moghadamtousi S, Karimian H, Khanabdali R, Razavi M, Firoozinia M, Zandi K, et al. Anticancer and antitumor potential of fucoidan and fucoxanthin, two main metabolites isolated from brown algae. ScientificWorldJournal. 2014;2014:768323. doi: 10.1155/2014/768323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufmann SH, Earnshaw WC. Induction of apoptosis by cancer chemotherapy. Exp Cell Res. 2000;256:42–9. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 16.Damonte EB, Matulewicz MC, Cerezo AS. Sulfated seaweed polysaccharides as antiviral agents. Curr Med Chem. 2004;11:2399–419. doi: 10.2174/0929867043364504. [DOI] [PubMed] [Google Scholar]
- 17.Cumashi A, Ushakova NA, Preobrazhenskaya ME, D'Incecco A, Piccoli A, Totani L, et al. A comparative study of the anti-inflammatory, anticoagulant, antiangiogenic, and antiadhesive activities of nine different fucoidans from brown seaweeds. Glycobiology. 2007;17:541–52. doi: 10.1093/glycob/cwm014. [DOI] [PubMed] [Google Scholar]
- 18.Asai A, Sugawara T, Ono H, Nagao A. Biotransformation of fucoxanthinol into amarouciaxanthin A in mice and HepG2 cells: Formation and cytotoxicity of fucoxanthin metabolites. Drug Metab Dispos. 2004;32:205–11. doi: 10.1124/dmd.32.2.205. [DOI] [PubMed] [Google Scholar]
- 19.Das SK, Hashimoto T, Kanazawa K. Growth inhibition of human hepatic carcinoma HepG2 cells by fucoxanthin is associated with down-regulation of cyclin D. Biochim Biophys Acta. 2008;1780:743–9. doi: 10.1016/j.bbagen.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita KI, et al. Adult T-cell leukemia: Antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci U S A. 1981;78:6476–80. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]


