Abstract
Imaging techniques are a cornerstone of contemporary biology. Over the last decades, advances in microscale imaging techniques have allowed fascinating new insights into cell and tissue morphology and internal anatomy of organisms across kingdoms. However, most studies so far provided snapshots of given reference taxa, describing organs and tissues under “idealized” conditions. Surprisingly, there is an almost complete lack of studies investigating how an organism′s internal morphology changes in response to environmental drivers. Consequently, ecology as a scientific discipline has so far almost neglected the possibilities arising from modern microscale imaging techniques. Here, we provide an overview of recent developments of X‐ray computed tomography as an affordable, simple method of high spatial resolution, allowing insights into three‐dimensional anatomy both in vivo and ex vivo. We review ecological studies using this technique to investigate the three‐dimensional internal structure of organisms. In addition, we provide practical comparisons between different preparation techniques for maximum contrast and tissue differentiation. In particular, we consider the novel modality of phase contrast by self‐interference of the X‐ray wave behind an object (i.e., phase contrast by free space propagation). Using the cricket Acheta domesticus (L.) as model organism, we found that the combination of FAE fixative and iodine staining provided the best results across different tissues. The drying technique also affected contrast and prevented artifacts in specific cases. Overall, we found that for the interests of ecological studies, X‐ray computed tomography is useful when the tissue or structure of interest has sufficient contrast that allows for an automatic or semiautomatic segmentation. In particular, we show that reconstruction schemes which exploit phase contrast can yield enhanced image quality. Combined with suitable specimen preparation and automated analysis, X‐ray CT can therefore become a promising quantitative 3D imaging technique to study organisms′ responses to environmental drivers, in both ecology and evolution.
Keywords: μ‐CT, anatomy, animal imaging, computed tomography, internal morphology
1. INTRODUCTION
Organisms respond to environmental drivers in a variety of ways, including changes in behavior, morphology, growth, or reproduction. Advances in imaging technology across scales have opened up new opportunities to estimate reproduction or to measure changes in morphology. Changes in internal morphology (such as complexity of neural tissues) are among the fastest (and often plastic) responses to environmental drivers, often preceding future changes in behavior or reproduction. Being able to fast‐track or even predict such responses will allow novel insights into physiological, behavioral, and evolutionary ecology.
A variety of automated techniques (such as confocal laser scanning microscopy, light sheet microscopy, nuclear magnetic resonance imaging, and microcomputed tomography) are available to study internal morphology or organisms, but these frequently require manual processing of low‐contrast regions in every section of the complete tissue, which precludes processing large numbers of samples. In contrast, classic histology (microtomy and episcopic microscopy) allows a variety of stains for tissue recognition, but requires destruction of the samples.
Yet, recent years have seen the development of novel imaging techniques such as microcomputed tomography (μ‐CT) that overcome these problems, allowing unprecedented insights into cell and tissue morphology and internal anatomy of organisms across kingdoms, from bacteria to vertebrates (Dhondt, Vanhaeren, Van Loo, Cnudde, & Inzé, 2010; Stender et al., 2014; Wipfler, Pohl, Yavorskaya, & Beutel, 2016). Fields such as taxonomy (Akkari, Enghoff, & Metscher, 2015; Faulwetter, Vasileiadou, Kouratoras, Dailianis, & Arvanitidis, 2013; Fernández, Kvist, Lenihan, Giribet, & Ziegler, 2014) and morphology (Mattei, Riccio, Avila, & Wolfner, 2015; Wipfler et al., 2016) have greatly benefited from μ‐CT technique. However, most studies to date have focused on in‐depth studies of single individuals, and organisms′ responses to environmental drivers were only rarely considered.
The effects of external drivers (such as global change components) on organisms can be studied by investigating the response of individuals (behavior, morphology, physiology) or populations (reproduction, survivorship) (Bale et al., 2002; Bidart‐Bouzat & Imeh‐Nathaniel, 2008). Morphological changes in response to environmental drivers are usually studied within the framework of trait‐based ecology (Deraison, Badenhausser, Loeuille, Scherber, & Gross, 2015). However, the traits considered are often related to external morphology or behavior, rather than to internal morphology of organisms.
Recently, studies (e.g., in pollinator ecology) have started to use modern imaging techniques, for example, to assess changes in brain morphology of bees and butterflies in response to environmental and social stimuli (Jones, Leonard, & Papaj, 2013; Maleszka, Barron, Helliwell, & Maleszka, 2009; Snell‐Rood, Papaj, & Gronenberg, 2009).
Here, we provide an overview of ecological studies using X‐ray CT, to study the three‐dimensional external and internal structures of organisms. In addition, we experimentally study a range of staining and fixation approaches useful for future studies and provide an outlook into questions that might be answered using micro‐CT in the future. Finally, we propose to especially exploit phase contrast, which has now become a reality also with advanced laboratory μ‐CT (Bartels, Hernandez, Krenkel, Moser, & Salditt, 2013; Töpperwien, Krenkel, Quade, & Salditt, 2016; Töpperwien et al., 2017)
2. TECHNIQUES FOR TOMOGRAPHIC RECONSTRUCTIONS
Tomography refers to imaging by sections or slices through a solid object, which can be achieved through several methodologies—usually requiring different sample preparations. This approach stands out because it allows imaging entire specimens (Jasanoff & Sun, 2002), does not require sample destruction (i.e., it is noninvasive), avoids tissue deformation (i.e., it retains natural stereogeometry), and is time‐saving (Jährling, Becker, Schönbauer, Schnorrer, & Dodt, 2010; Smith et al., 2016; Sombke, Lipke, Michalik, Uhl, & Harzsch, 2015). Tomographic imaging can be particularly advantageous in studies that require several assessments of the same individuals over time, as in developmental biology (Goodman & Chudekt, 1995; Hart, Bowtell, Köckenberger, Wenseleers, & Ratnieks, 2003), or when sectioning the same sample along different angles or axes is needed (Figure 1).
Figure 1.
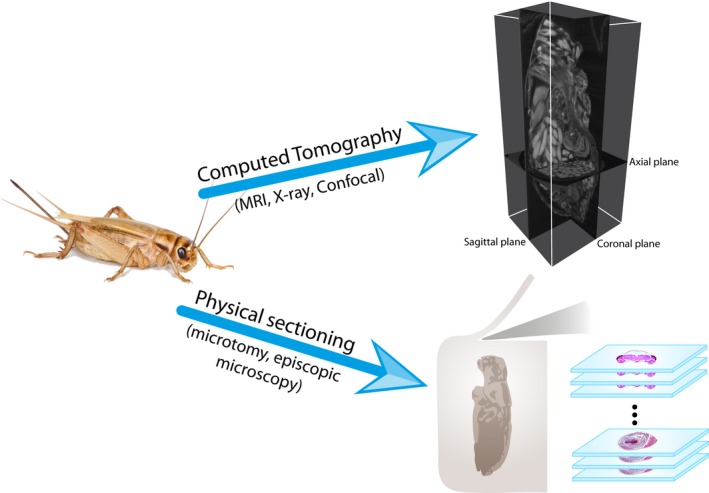
Conceptual figure showing procedural differences between computed tomography and physical sectioning of samples. CT (by means of several techniques) does not require sample destruction, and the resolution is identical in all orientations (isotropy) and enables visualization in different angles or axes. On the other hand, physical sectioning allows a wider variety of dying techniques for tissue recognition, but the plan is determined by the orientation of the sample; thus, also the z‐axis possesses a different (usually lower) resolution
One of the most interesting aspects of tomography is the possibility to generate three‐dimensional (3D) models. Such models, for example, have become popular within the area of plant phenotyping, where crop plant varieties are now routinely screened using a broad spectrum of imaging approaches (Fiorani & Schurr, 2013). Using classical histological procedures (e.g., microtomy and episcopic microscopy), such reconstructions can only be achieved going through every layer and manually selecting and aligning the desired tissue or organ, thereby commonly over‐ or underestimating the volume of soft tissues due to the lack of isotropic resolution (e.g., poor resolution in the z‐axis) (Sickert, Rodner, & Denzler, 2015; Smith et al., 2016). In contrast, with 3D imaging techniques, in particular μ‐CT, automatic surface (Friedrich & Beutel, 2008) and volume reconstructions can be quickly carried out, allowing to accurately determine surface areas and volumes for comparative studies (Hart et al., 2003), in addition to be visually attractive and self‐explanatory.
Apart from X‐ray CT, the most commonly used techniques for tomographic reconstructions are nuclear magnetic resonance imaging (MRI). Other techniques, such as ultramicroscopy, confocal laser scanning microscopy (CLSM), or light sheet microscopy, also allow high resolution down to the submicrometer range without physically sectioning the samples (Becker, Jährling, Kramer, Schnorrer, & Dodt, 2008). However, the latter two techniques are based on transmission of visible light (Jährling et al., 2010) and require the sample to go through a chemical clearance process. Moreover, these techniques are limited to tissues thinner than 500 μm, requiring elaborated sample preparation (Sombke et al., 2015).
MRI and X‐ray CT do not require complex sample manipulation, and can even be performed in vivo (Callaghan, 1991; Hart et al., 2003; Jasanoff & Sun, 2002), although this could compromise the quality of the images because of internal movements of organs and fluids (Hart et al., 2003). While some studies suggest that low doses (<500 Gray) of radiation have only negligible effect on insects survivorship, the long‐term effects of X‐rays on insects have remained poorly studied so far (Socha, Westneat, Harrison, Waters, & Lee, 2007; Westneat, Socha, & Lee, 2008).
Primarily due to the much higher spatial resolution of X‐ray CT, we consider it as a particularly well‐suited choice for future studies. Samples are certainly more easily prepared for MRI than X‐ray CT, in particular for those studies which require metal‐based staining for soft tissue imaging (Metscher, 2009a, 2013). Nevertheless, magnetic resonance scanning systems are limited in resolution (Metscher, 2013; Metzner et al., 2015) and often have prohibitive costs of operation (Ziegler et al., 2011). Furthermore, air spaces routinely found in biological samples can cause artifacts in MRI (Jasanoff & Sun, 2002; Wecker, Hörnschemeyer, & Hoehn, 2002; Ziegler et al., 2011).
3. X‐RAY‐BASED COMPUTED TOMOGRAPHY (X‐RAY CT AND μ‐CT)
The difference between X‐ray CT and μ‐CT is merely the level of detail: μ‐CT works at the micrometer range (Medical Subject Headings—MeSH) and has become an invaluable tool in the study of several organs and organ systems in arthropods (see a review in Westneat et al., 2008; Metscher, 2013; Sombke et al., 2015) and other invertebrates (Carbayo & Lenihan, 2016; Fernández et al., 2014). This technique allows spatial resolution in the 1–10 μm range (i.e., spanning the range from whole cells down to the level of single organelles) and a temporal resolution of less than 100 ms. Furthermore, due to recent improvements, it provides enough detail to successfully distinguish either cuticular structures or soft tissues as muscles and nervous system (Sena et al., 2015; Smith et al., 2016; Westneat et al., 2008).
Images obtained from X‐ray CT possess a homogeneous illumination with isotropic resolution at each slice, which allows consistent and precise volumetric estimates and some degree of automation of the process (Sickert et al., 2015; Smith et al., 2016). As reported by Seo, Lim, Seo, and Lee (2015), X‐ray CT is sensitive enough for internal modifications that in some cases cannot be traced through resin‐sectioned images.
More advanced variants of this technique have been demonstrated using synchrotron radiation (SR‐μCT), exploiting the high brilliance of the radiation. In practice, this can be used to generate better collimated (parallel) beams, higher flux density, and smaller bandpass by monochromatization (e.g., double silicon crystals). In combination with cone‐beam geometries, or focusing optics, submicron resolution has become possible (Hoshino, Uesugi, & Yagi, 2012; Sena et al., 2015; Westneat et al., 2008). Apart from resolution, the range of possible contrast mechanisms and levels has been significantly enhanced by use of SR. Note that by virtue of phase contrast, also nonabsorbing or weakly absorbing tissues can be visualized, based on the intrinsic phase shift which X‐rays undergo when traversing matter. In particular, phase contrast by free propagation has been exploited and has been demonstrated at submicron resolution (Cloetens et al., 1999; Lagomarsino et al., 1997; Paganin & Nugent, 1998).
Using highly focused radiation and a cone‐beam illumination geometry, a resolution range down to of 20–50 nm has even been demonstrated (Bartels, Krenkel, Haber, Wilke, & Salditt, 2015), allowing to examine details of cell organelles. Importantly, phase contrast based on free propagation is also compatible with the low partial coherence of laboratory sources, so that, subsequently, a translation from SR‐based phase‐contrast CT (SR‐PhC‐μCT) to advanced μ‐CT instrumentation was possible. The instrumental prerequisites and different geometries and phase retrieval approaches have been discussed and compared elsewhere (Bartels et al., 2013; Krenkel et al., 2015; Töpperwien, Krenkel, Müller, & Salditt, 2016; Töpperwien et al., 2017), in particular the adaptation of phase retrieval for the nonideal conditions of laboratory μ‐CT (Bartels et al., 2013; Krenkel et al., 2015; Töpperwien, Krenkel, et al., 2016; Töpperwien et al., 2017).
For this research, we have focused on studies using the more widely accessible μ‐CT versions based on laboratory radiation, both the common absorption‐based variant and the emerging phase‐contrast modality, which has also been exploited in the present experimental work.
In the context of the biological sciences, previously published reviews about X‐ray CT have focused on technical details and currently available techniques (Withers, 2007), current manufacturers and models (Schambach, Bag, Schilling, Groden, & Brockmann, 2010; for an updated list see Appendix S3), potentials and limitations of computed tomographic techniques on classical anatomy studies (Friedrich & Beutel, 2008), animal physiology (Westneat et al., 2008), and arthropod neuroanatomy (Sombke et al., 2015). To the extent of our knowledge, this is the first study addressing ecological studies involving CT for acquiring in vivo and ex vivo data.
4. APPLICATION OF X‐RAY COMPUTED TOMOGRAPHY IN ECOLOGICAL STUDIES
Although X‐ray CT scanners have been available since 1967 in clinics and laboratories (Hsieh, 2009), and μ‐CT has been available many years ago (Elliott & Dover, 1982; for an overview of the manufacturers see Schambach et al., 2010), few applications of these techniques in ecological studies can be found in the current literature. To date, these techniques have been more commonly used for taxonomy, phylogeny, and physiology (Beutel, Friedrich, Ge, & Yang, 2014; Fernández et al., 2014; Friedrich & Beutel, 2008; Metscher, 2013).
As a comprehensive historical analysis of the studies published on this topic was lacking, we conducted a literature search from 1974 (the date of the first paper) to 2017 using Thomson‐Reuters's ISI Web of Science (all databases). We used [“compute* tomogr*” OR “micro tomogr*” OR “micro CT”] as the primary search terms. To filter out the studies dealing with human and animal medicine and livestock production, we used secondary search terms including all animal phyla and plant and fungal divisions [Acanthocephala OR Acoelomorpha OR Annelida OR Arthropoda OR Brachiopoda OR Bryozoa OR Chaetognatha OR Chordata OR Cnidaria OR Ctenophora OR Cycliophora OR Echinodermata OR Entoprocta OR Gastrotricha OR Gnathostomulida OR Hemichordata OR Kinorhyncha OR Loricifera OR Micrognathozoa OR Mollusca OR Nematoda OR Nematomorpha OR Nemertea OR Onychophora OR Orthonectida OR Phoronida OR Placozoa OR Platyhelminthes OR Porifera OR Priapulida OR Rhombozoa OR Rotifera OR Sipuncula OR Tardigrada OR Xenacoelomorpha OR Anthocerotophyta OR Bryophyta OR Marchantiophyta OR Hepatophyta OR Lycopodiophyta OR Lycophyta OR Pteridophyta OR Pinophyta OR Coniferophyta OR Cycadophyta OR Ginkgophyta OR Gnetophyta OR Magnoliophyta OR Chytridiomycota OR Blastocladiomycota OR Zygomycota OR Glomeromycota OR Ascomycota OR Basidiomycota OR Microsporidia OR Neocallimastigomycota] and subsequently refined the search to the following research areas (in order of record count): Zoology, Environmental Sciences Ecology, Evolutionary Biology, Paleontology, Mycology, Plant Sciences, Marine Freshwater Biology, and Biodiversity Conservation.
Using only the primary search terms, we obtained 476,232 results on scientific publications, and secondary filtering resulted in 11,990 (2.52%) publications where X‐ray‐based tomography was used in the stricter sense of the natural science field biology (Figure 2). Because computed scanners were developed for medical applications, most of the obtained studies were published on related research areas—and single papers have reached up to 6,000 citations. Other outstanding fields represented in our results are engineering, mathematics, computer science, and physics.
Figure 2.
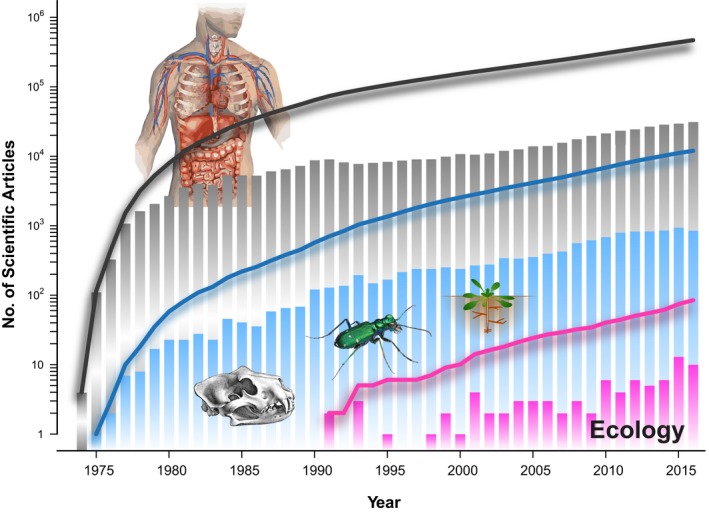
Historical analysis of the usage of x‐ray computed tomography in scientific studies by means of Thomson‐Reuters's ISI on the Web of Science (all databases). Dark gray: total number (primary search terms), blue: biological sciences (primary search terms plus secondary filtering to exclude studies dealing with human and animal medicine and livestock production), dark pink: manually refined selection of ecological studies. Bars represent the annual count of publications and lines the cumulative sum. Results showed on the y‐axis (presented in logarithmic scale) were obtained using a combination of search term and research areas explained in the text. Beetle image © Alex Wild, used by permission
Interestingly, ecology was included as “Environmental Sciences Ecology” in a middle ground among “The first 100 Research Areas” with 3,887 (0.82%) results, roughly one‐third from the biological field. Yet, the number of publications obtained through this filtering process still contained publications of several different areas. We therefore manually refined the selection of studies using X‐ray CT imaging in ecology, yielding a total of 81 (0.02% of total, but contributing 2.08% to the area of environmental sciences) studies retained in the final set (Appendix S1). A search for “Phase contrast tomography” and “Ecology” gave zero results.
Comparing the total number of publications (476,232) with those presented in Appendix S1 (81) already shows how underexplored X‐ray CT in animal and—even more in—plant ecology is. From the scarce examples of ecological studies, the most studied animals have been arthropods and annelids, and in a lesser extend other kingdoms such as plants and fungi (Figure 3). It is particularly evident that one of the most popular topics so far is soil ecology (Davey et al., 2011; Harrison, Gardner, Tollner, & Kinard, 1993; Tollner, 1991), and specifically the study of worm burrows (Amossé, Turberg, Kohler‐Milleret, Gobat, & Le Bayon, 2015; Auclerc, Capowiez, Guérold, & Nahmani, 2013; Capowiez, Monestiez, & Belzunces, 2001; Capowiez, Pierret, & Moran, 2003; Francis, Tabley, Butler, & Fraser, 2001; Jégou, Capowiez, & Cluzeau, 2001; Jégou, Cluzeau, Hallaire, Balesdent, & Tréhen, 2000; Jégou, Cluzeau, Wolf, Gandon, & Tréhen, 1998; Jégou, Hallaire, Cluzeau, & Tréhen, 1999; Jégou et al., 2002; Langmaack, Schrader, Rapp‐Bernhardt, & Kotzke, 1999; Pagenkemper et al., 2015; Pelosi, Grandeau, & Capowiez, 2017; Rogasik, Schrader, Onasch, Kiesel, & Gerke, 2014; Schrader, Rogasik, Onasch, & Jegou, 2007), mostly because of the ease of studying this type of sample. Soil can be considered a matrix where the components can be detected through X‐ray CT without any preparation (i.e., staining). In soil samples, air spaces such as pores or worm burrows can be easily distinguished and measured. Additionally, invertebrates can be tracked in soil cores without requiring particular sample preparation due to their lower density compared to soil components (Tollner, 1991).
Figure 3.
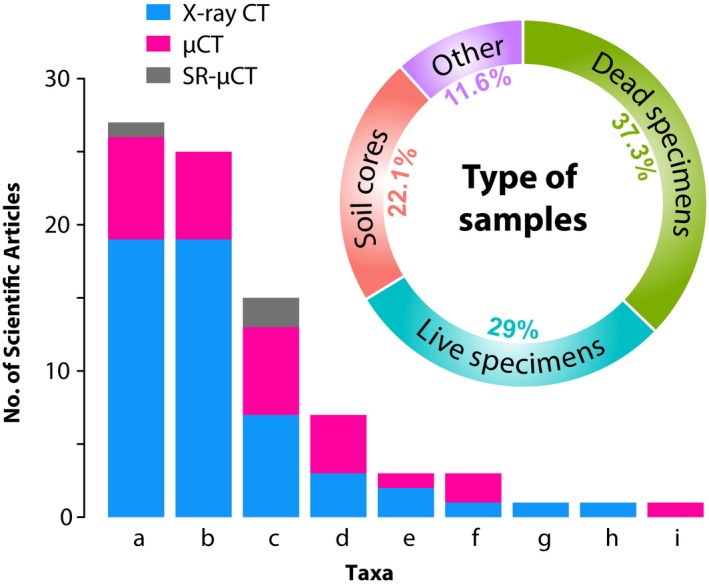
Quantitative analysis of ecological studies using x‐ray computed tomography. (a) Arthropoda, (b) Annelida, (c) Chordata, (d) Plantae (kingdom), (e) Cnidaria, (f) Mollusca, (g) Echinodermata, (h) Fungi (kingdom), (i) Porifera. μ‐CT: microcomputed tomography, SR‐μCT: synchrotron radiation microcomputed tomography
Another field with considerable usage of X‐ray CT is the study of social insects and their nest or gallery systems. Here, images are primarily acquired in vivo allowing to scan the same nest or colony several times during its developmental cycle (Eyer, Neumann, & Dietemann, 2016; Greco, Bell, Spooner‐Hart, & Holford, 2006; Greco, Spooner‐Hart, Beattie, Barchia, & Holford, 2011; Greco, Spooner‐Hart, & Holford, 2005; Rademacher, Fahlberg, Raddatz, Schneider, & Voigt, 2013). X‐ray CT has also proven to be a valuable tool for assessing processes in samples that do not allow direct visual evaluation without disturbing the organisms, such as parasitic relationships (Diez, Orensanz, Márquez, & Cremonte, 2013; Schwabe, Holtheuer, & Schories, 2014), seed‐feeding insects (Tarver et al., 2006), and growth strategies of animals (Cantin, Cohen, Karnauskas, Tarrant, & McCorkle, 2010; Fujiwara, Oji, Tanaka, & Kondo, 2005; Schönberg, 2001; Silbiger, Guadayol, Thomas, & Donahue, 2016), fungi (Van den Bulcke, Boone, Van Acker, & Van Hoorebeke, 2009), and plants (Dhondt et al., 2010; Ferreira et al., 2010; Gregory et al., 2003; Mairhofer et al., 2012; Mooney, Morris, & Berry, 2006; Perret, Al‐Belushi, & Deadman, 2007).
Establishment of trophic relationships in extant (Herrel et al., 2010; Kato et al., 2014; Pampush et al., 2016; Renaud et al., 2015; Self, 2015; Soons et al., 2015) and extinct species (Collareta et al., 2015; Gill et al., 2014) has also been possible by analysis of gut content, beak shape, or dental wear patterns.
In spite of the diversity of the topics explored in the ecological studies herein presented, there are scant examples of internal changes assessments, which in our opinion is one of the most promising avenues of the usage of X‐ray CT. Only some recent studies have shown changes in reproductive organs after mating and egg development in fruit flies (Mattei et al., 2015), and changes in volume of several organs of the trout after exposure to contaminated sediments (Brinkmann et al., 2016).
It is also interesting to note that, in terms of ecological applications, common X‐ray CT has been more widely used than μ‐CT (Figure 3). X‐ray CT scanners enable imaging of big samples (e.g., soil cores of 20 cm diameter and 70 height, pixel size around 200–400 μm) (Amossé et al., 2015; Pagenkemper et al., 2015), and although the rather low resolution allows accurate localization of internal structures in the nests of social insects (Fuchs, Schreyer, Feuerbach, & Korb, 2004; Greco et al., 2005, 2006), the identification of particular specimens is usually limited (Fuchs et al., 2004).
When scans of a single specimen are desired, resolutions of about 10 μm can be achieved during in vivo scanning (Postnov, De Clerck, Sasov, & Van Dyck, 2002; Yao et al., 2016). It is thus possible to reconstruct virtual cross sections through the specimens, even if respiratory, digestive, and/or cardiac systems are moving (De Clerck et al., 2004). However, volumetric assessments of particular organs in vivo have to be performed carefully as this technique can detect changes through time (e.g., several seconds) (Postnov et al., 2002; Westneat et al., 2008), which could cause measurement errors caused by variation in volume of structures due to normal physiological functions, such as ventilation and digestion. On the other hand, techniques for study real‐time dynamics (termed cine‐tomography) are being made available for 4D analysis (Rolo, Ershov, van de Kamp, & Baumbach, 2014).
When scanning live specimens, it is possible that organisms are negatively affected as a consequence of the absorbed radiation or overheating (Postnov et al., 2002). Depending on the resolution, area, and desired quality (signal‐to‐noise ratio), a scan might take from several minutes to hours (Dhondt et al., 2010). Several authors have claimed that repeated scans do not affect organisms (Postnov et al., 2002); in some studies, the same live specimens were scanned up to eight times (Halley, Burd, & Wells, 2005). When using live animals, the authors either did not assess possible side effects (Brinkmann et al., 2016) or just checked for a few hours or days after exposure (Dhondt et al., 2010).
In order to include the X‐ray CT analysis in vivo in ecological research, it is crucial to understand the effects of the radiation on the organisms before biological interpretation (Socha et al., 2007). In addition to the radiation, there are several steps of specimen preparation (such as staining and anesthesia) than can be potentially harmful. In some studies, tissue differentiation was achieved by staining live animals through dietary supplementation (e.g., with cadmium tungstate (CdWO4) and iodine) (Kim, Seo, Lim, & Lee, 2012; Socha et al., 2007), or injecting compounds into the circulatory system (Greco, Tong, Soleimani, Bell, & Schäfer, 2012). There are few studies about long‐term effects of X‐rays doses in invertebrates, and Kanao, Okamoto, Miyachi, and Nohara (2003) showed that low doses (0.5 Grays) caused transgenerational changes of emergence patterns in Drosophila melanogaster and this area would clearly need further study.
5. PREPARATION OF SAMPLES FOR X‐RAY CT
The preparation process preceding X‐ray CT scans is considerably shorter in comparison with classical histological techniques (Figure 1), in which obtaining a serial section of an average‐sized insect specimen (ca. 1 cm) can take several weeks (Friedrich & Beutel, 2008; Socha et al., 2007). In this review, we focus on the preparation of ex vivo specimens. This procedure usually comprises three simple steps: fixation, staining, and drying.
5.1. Fixation
Samples can be commonly fixed in ethanol or even embedded in resin (as in histology) (Metscher, 2013). For small vertebrates (e.g., laboratory mice, zebra fish, and embryos), it is also common to store specimen in formalin (Cnudde et al., 2008; Hoshino et al., 2012; Kim et al., 2012; Metscher, 2009a,b; Seo et al., 2015). Sombke et al. (2015) reported that fixation of several arthropod taxa in Bouin's solution provided better results in terms of tissue contrast when compared with ethanol and glutaraldehyde solution. To acquire images over short durations (e.g., to study mating), the sample can be flash‐frozen in liquid nitrogen (Mattei et al., 2015; Mouginot et al., 2015) and transferred to fixative solution afterward.
5.2. Staining
For soft tissues, X‐ray contrast can be enhanced using metal‐based stains, for instance, osmium tetroxide—widely used in transmission electron microscopy—and iodine. On the other hand, some structures can possess sufficient inherent contrast and do not require preparation or staining; this applies mainly to mineralized tissues such as bones or shells (Degenhardt, Wright, Horng, Padmanabhan, & Epstein, 2010; Metscher, 2013; Westneat et al., 2008). Proper differentiation of tissues within the sample is necessary for software‐based (semi‐) automatic recognition using thresholds of gray values (Friedrich & Beutel, 2008) or even for manually defining areas in each slice (Self, 2015). Metscher (2009a) suggested that stains are usually not tissue‐specific and the final quality depended mainly on the fixative. However, comparisons between different stains have shown that indeed some compounds can stain lipidic tissue more intensely (e.g., Lugol's solution) (Degenhardt et al., 2010); thus, recognition of different tissues can be greatly influenced by the chosen stain (Smith et al., 2016).
5.3. Drying
Samples can either be scanned in ethanol (inside plastic tubes that shall not interfere with the scanning process; plastic straws have proven to be good enough for this purpose) or dried and mounted/glued in custom‐made supports. Critical‐point drying (CPD, dehydration technique where water in biological tissue is replaced with CO2) gives good results preserving the fine structure of the sample (Beutel et al., 2014), keeping a high signal‐to‐noise ratio in the resulting images (Sombke et al., 2015). Chemical drying (e.g., using hexamethyldisilazane) is not recommendable because it causes tissue shrinkage and damage (e.g., in the brain of insects) (Sombke et al., 2015). At least air drying should be carried out to avoid shrinkage artifacts caused by water loss during the scanning process (Sena et al., 2015).
6. EXPERIMENTAL COMPARISON OF STAINING AND FIXATION APPROACHES
Although several combinations of methods for fixation, staining, and drying have previously been assessed and compared (Metscher, 2009a,b; Sombke et al., 2015), the authors generally used several species (vertebrate and invertebrate) and target tissues, and have carried out tomography without any phase retrieval. Here, we systematically compare different staining and fixation approaches, focusing on only one species, and consider in particular the more recent phase‐contrast modality. We combine techniques that previously proved successful, and provide results and comments about specific combinations in light of the desired results. As internal organs possess different densities (a very important characteristic when it comes to X‐ray‐based tomography) and chemical constitution (Nagy, 2001; Sterner & Elser, 2002), it is likely that one single perfect formulation is not possible in all cases. Here, we compare different methods to visualize structures in the head (mainly brain and muscles) and abdomen (ovaries and fat body) of Acheta domesticus (Linnaeus, 1758).
Commercially available A. domesticus adult females were euthanized in 70/30 solution ethanol/deionized water (although the specimens can also be fumigated with ethyl acetate or frozen as in Iwan, Kamimski, & Ras, 2015). Wings, legs, and antennae were removed with sharp dissecting scissors and discarded. The heads were removed from the bodies for separate scans, allowing better penetration of chemical compounds. For fixation, we used either ethanol (70/30 solution ethanol/deionized water), FAE (formaldehyde, acetic acid, and ethanol) or Bouin's solution (saturated aqueous picric acid, pure acetic acid, and formaldehyde), for 24 hr, following Beutel et al. (2014). For staining, either phosphotungstic acid (PTA) or iodine was used for 7 days and 24 hr, respectively, as described by Metscher (2009b). Every sample (e.g., head or abdomen) was scanned individually in random order either dry (air dried or critical‐point dried) or in ethanol. Every combination of fixative and staining was replicated three times (see Appendix S2 for detailed information about specimens’ preparation).
The samples (N = 48, Appendix S2) were scanned at the Institute for X‐ray Physics, University of Göttingen (Göttingen, Germany) using phase‐contrast μ‐CT techniques (for detailed specifications of the setup see Töpperwien, Krenkel, et al., 2016; Töpperwien, Krenkel, Quade, et al., 2016). Heads and abdomens were scanned using different detectors for a final pixel size effect of 2.6–2.7 (fiber‐coupled scintillator‐based sCMOS, Hamamatsu Photonics, Japan) and 10 μm (Dexela CMOS Flat Panel, PerkinElmer, Germany), respectively. In both cases, we acquired 1,000 projections in an angular range of 183–190°. After raw image correction (dark current subtraction and empty beam division), the projection images, which exhibited the typical edge enhancement as a manifestation of phase contrast, were treated using the Bronnikov‐aided correction (BAC) algorithm proposed by De Witte, Boone, Vlassenbroeck, Dierick, and Van Hoorebeke (2009). A phase retrieval assuming vanishing absorption was carried out based on Fourier filtering, followed by a correction step to represent the amplitude of the exit wave. As explained in Töpperwien, Krenkel, Quade, et al. (2016), Töpperwien et al. (2017), the BAC scheme inverts blur by diffraction, achieves higher sharpness in comparison with other schemes, while partially mixing amplitude and phase contrast to an effective contrast. The details of the data analysis used here closely follow the procedures published before (Töpperwien, Krenkel, Quade, et al. (2016); Töpperwien et al. (2017)).
Iodine proved to be the best staining agent in our tests, because of its faster tissue penetration and superior overall contrast across all tissues (Figure 4e‐h), also the usage of low concentrations assured minimum artifacts (Vickerton, Jarvis, & Jeffery, 2013). PTA failed to stain the complete sample (neither head nor abdomen), and the few stained regions had very strong contrast, precluding adequate tissue recognition (Figure 4a‐d). The unstained parts of the samples exposed to PTA had minor contrast, resembling tissues that had not been exposed to any stain (Degenhardt et al., 2010; Metscher, 2009a), thus indicating that the tissue penetration was insufficient and samples would have to be exposed for more days to attain the desired stain.
Figure 4.
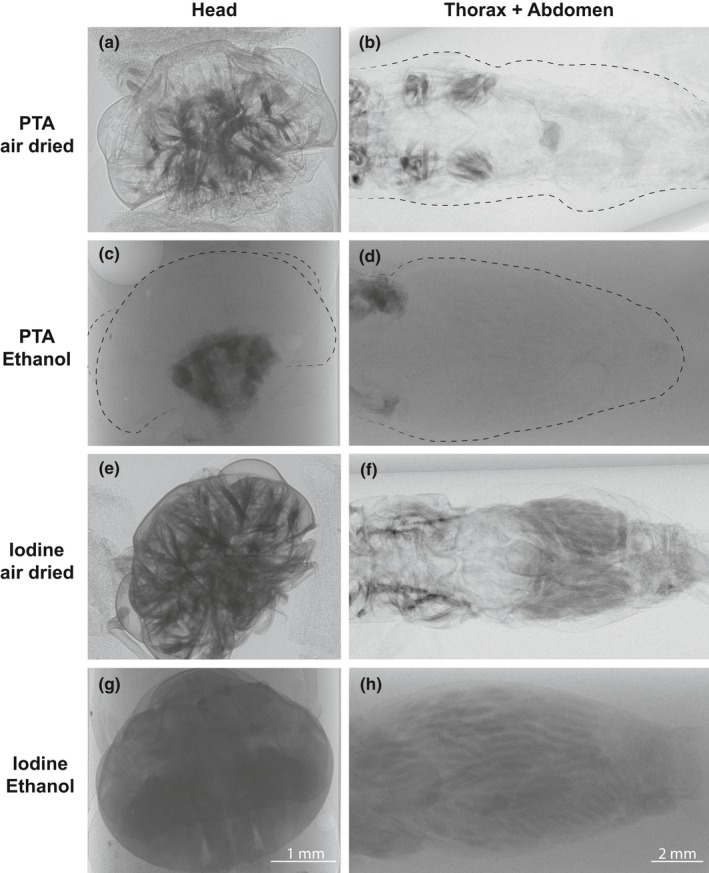
Experimental comparison of staining agents and scanning mediums using only ethanol as fixative in the cricket Acheta domesticus. After fixation for 24 hr in 70/30 solution ethanol/deionized water, samples were stained in phosphotungstic acid (PTA) or iodine (solution with ethanol) during 7 days and 24 hr, respectively. Posteriorly, the samples were scanned either in ethanol or air‐dried
PTA has shown to yield sufficient results to recognize of brain and muscle structures in arthropods (Smith et al., 2016; Swart, Wicklein, Sykes, Ahmed, & Krapp, 2016). However, because of its slow penetration rate—as evidenced by Smith et al. (2016) and also this study—it might be considered when the sample tissue is thin or removal of parts of the exoskeleton is possible (to facilitate stain perfusion). While osmium tetroxide (OsO4) has been a popular staining agent in previous X‐ray CT studies (Jahn et al., 2018; Kim et al., 2012; Metscher, 2009a,b; Ribi, Senden, Sakellariou, Limaye, & Zhang, 2008), we did not consider it because of its undesirable toxicity, high costs, limited tissue penetration, and failure to work properly in tissues preserved in alcohol (Hayat, 1970; Metscher, 2009a; Smith, Carson, & Ferguson, 1974; Smith et al., 2016).
With respect to the scanning medium, dried samples (air drying technique for this test) provided the best results. Many of the structures are already evident in projections prior to reconstruction (Figure 2e, f). Using ethanol as medium could possibly prevent artifacts (e.g., tissue shrinkage), but the contrast was greatly reduced and scanning time had to be tripled.
After having found an appropriate staining agent and scanning medium, we proceeded to test how the selection of the fixative and the drying technique affected the final result. Ethanol (as a fixative) provided undesirable results due to strong artifacts (e.g., air spaces and tissue shrinkage) especially when combined with air drying (Figure 5a, b). Although ethanol with CPD provided sufficient contrast, separation of tissue from the cuticle was still evident (Figure 5c, d). Samples fixed with Bouin's and air dried appeared overstained and also showed artifacts (Figure 5i, j). However, the usage of Bouin's in combination with CPD showed better results (Figure 5k, l). FAE fixative provided good contrast and quality in general when either air‐ or critical‐point dried; tissue conformation appeared natural and with few artifacts.
Figure 5.
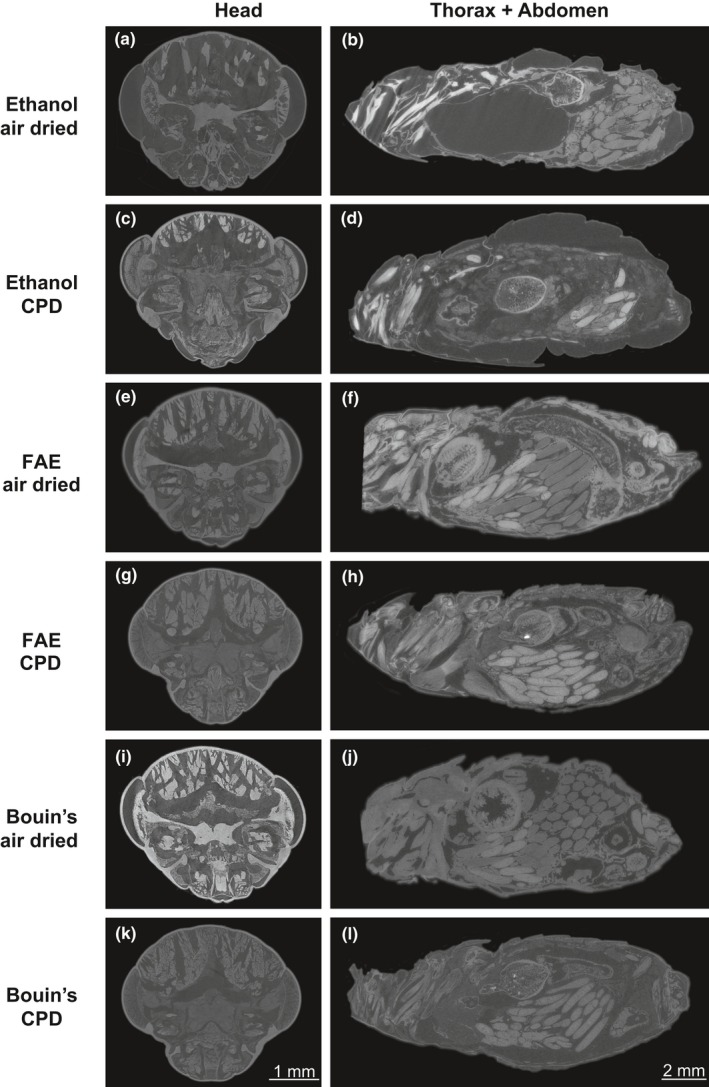
Experimental comparison of fixatives and drying techniques using iodine as chosen staining agent in the cricket Acheta domesticus. After fixation for 24 in either 70/30 solution ethanol/deionized water, FAE (formaldehyde, acetic acid, and ethanol), or Bouin's solution (saturated aqueous picric acid, pure acetic acid, and 10% formaldehyde solution), the samples were air‐dried or submitted to CPD (critical‐point drying) before being scanned. Heads are presented in coronal planes and abdomens in sagittal planes
As expected, regardless of the fixative used, CPD significantly increased stain complexity and quality (e.g., brain lobes in Figure 5g, h). This drying technique also avoided overstaining in combination Bouin's iodine. When brain and muscle morphology is the objective of the study, the sample should be critical‐point dried after fixation (with either FAE or Bouin's) and staining (iodine). If other tissues, such as ovaries, gut, or fat body, are targeted, FAE fixation should be prioritized and air drying provides good results. Air drying reduces the cost and time for sample preparation prior scanning.
6.1. Sample size, effort, and costs
In studies intending to include X‐ray CT imaging, the number of samples to be analyzed (i.e., prepared, scanned, reconstructed, and segmented) would depend to a large extent on the time necessary from obtaining the specimen/sample until data collection, and the costs associated with this process.
From several publications included in our historic review (Appendix S1, Figure 2), it is clear that sample size tended to be limited so far. For instance, Brinkmann et al. (2016) used just three specimens (rainbow trout) per treatment, although with a laborious segmentation process. Further, Greco et al. (2010) used five stingless‐bee hives to study the defensive mechanism against parasite beetles. On the other hand, when samples (e.g., soil cores) or specimens used allowed for an easier segmentation (because of higher contrast and therefore possible semiautomatic recognition), sample size tended to be bigger (N = 8–20, with more than 50 scans per study) (Himmi et al., 2016; Monaenkova et al., 2015; Pelosi et al., 2017; Silbiger et al., 2016).
Additionally, the sample size can be increased by batch‐preparing (fixation, staining, drying) and scanning the samples. Several specimens can be scanned at the same time as in Smith et al. (2016) and separated for individual analysis in the segmentation process.
On the other hand, the nature of the samples or specimens can also limit sample sizes in ecological studies. In some cases (e.g., fossils or rare species), few samples or specimens are available for analysis but the results are not less significant (Collareta et al., 2015; Gill et al., 2014).
With more manufacturers (Appendix S3) and institutions with facilities dedicated to X‐ray CT imaging, the cost of devices and scanning services will continue to decrease and this technology will become more accessible in the near future. Comparisons in terms of costs in the utilization or decision between techniques have been discussed previously (Cunningham, Rahman, Lautenschlager, Rayfield, & Donoghue, 2014). Recently, Silbiger et al. (2016) reported costs as low as $100 per scan.
Regarding the effort required to fully processes the samples, besides batch‐preparing, an optimization of the protocol for the model organism (or specific type of sample) is highly recommended. Pilot scans of the focal tissue pursuing an optimal fixation and staining time would enhance contrast, thus easing the segmentation process.
Individual researchers performing the segmentations have to undergo a training phase in order to reduce human‐introduced errors. After this phase, the segmentation and subsequent calculation of volumes can be performed in a short period of time (usually few hours when semiautomatic segmentation is possible).
The procedure used for specimen preparation in the present study is straightforward and can be carried out in any laboratory as we gave preference to nontoxic chemicals. Once the desired specimen is ready for preparation, it has to be fixated (2–12 hr depending on size), stained (24 hr when using iodine) and dried (approx. 1 hr for CPD). Time necessary to scan a sample was about 8.33 min (1,000 projections at an angular range of 190°, and 0.5 s of exposure time). The reconstruction process can be completed in ca. 20 min running the script in MATLAB (this last step is naturally dependent on computing power). After obtaining the final reconstructed file, this can be loaded in the software for segmentation (in this case, we used Seg3D) and the thresholding and cleaning of the region of interest (i.e., ovaries) took in average two hours per file. Overall, we estimate the current time budget needed per sample to be around 5 hr excluding fixation. As many processes can now be run on entire batches of samples, this is not more time‐consuming that other imaging techniques such as electron microscopy or fluorescence microscopy.
7. PROCESSING OF X‐RAY CT IMAGES
Once the samples are scanned, and the resulting projections (as in Figure 4) have undergone the phase retrieval step described above, tomographic reconstruction is carried out in order to obtain the slices (as presented in Figure 5). For instance, from 1,000 projections of a cricket head, 2,048 slices were acquired after reconstruction. Several algorithms are available for this task, for a thorough explanation of this process we encourage the reader to consult the study by Willemink et al. (2013). Here, raw data corrections and phase retrieval of projections were carried out in MATLAB (The MathWorks, Inc., Massachusetts, United States), and cone‐beam reconstruction was carried out using the ASTRA toolbox (van Aarle et al., 2016) interfaced with MATLAB.
The slices obtained after the reconstruction can be analyzed in many ways. For instance, inspection of tissue conformation in any anatomical plan, description of organs, and two‐dimensional measurements can be performed in any software for image visualization such as ImageJ (Schneider, Rasband, & Eliceiri, 2012). However, three‐dimensional measurements (i.e., volumes) are the most desired feature of X‐ray CT. In order to measure a single organ or structure contained in our scan, this has to be segmented; that is, the pixels associated with the structure have to be selected and labeled.
Segmentation is one of the most important steps in X‐ray CT data processing, and perhaps the most labor‐intensive part depending on the desired data. Several authors have compared the capabilities of the available software (Lautenschlager, 2016) and step‐by‐step protocols have been published recently (Abel, Laurini, & Richter, 2012; Fedorov et al., 2012; Smith et al., 2016; Supporting information), but still segmentation remains largely context‐dependent (Swart, Deaton, & Felgenhauer, 2006).
This process can be performed using commercially available (e.g., Avizo, AVGStudio), open‐source software (e.g., SPIERS, Seg3D, 3D Slicer) (Fedorov et al., 2012; Sutton, Garwood, Siveter, & Siveter, 2012; Tate, Burton, & Khan, 2016), and online applications (e.g., Biomedisa, Lösel & Heuveline, 2016). In addition to 3D reconstruction, some of these available software packages offer the possibility to measure linear features, areas, and volumes as well (Ravel & Orliac, 2015). Commonly, data (e.g., volumes) are obtained in voxels (from the contraction of vox “volume” and el for “element”), which is a unit of graphic information that defines a point in three‐dimensional space, each coordinate being defined in terms of its position, color, and density (Higgins, Williams, Nagel, & Higgins, 2006). The volume of the structure of tissue can be calculated through the voxel size (e.g., in this study, the voxel size for the cricket head was 2.7 μm3).
Several authors have pointed out how difficult the segmentation process can be (Greco et al., 2012; Gremse et al., 2015; Kim et al., 2012), mainly due to poorly contrasted regions (in many cases resulting from inadequate sample preparation). Although present‐day computers allow fast and accurate processing of tomographic data (Mattei et al., 2015), Metscher (2013) claimed that the development of novel machine‐learning algorithms operating in three‐dimensional space will be needed for 3D segmentation and semiautomatic comparisons of images (e.g., variation and growth of structures). Fast recognition of regions in the volumes would reduce the time and increase the accuracy of the quantitative analysis of a large number of datasets, thus allowing for more robust statistics (Sickert et al., 2015; Smith et al., 2016).
From our house crickets’ dataset, we chose the ovaries and spermathecae as focal tissues due to their high contrast in comparison with the surrounding tissues. By way of example, the segmentation was performed semiautomatically using Seg3D (Tate et al., 2016) from a single scan of an abdomen fixed with FAE, stained with iodine and dried using CDP (Figure 6). First, the slices were subjected to thresholding until the desired region was completely highlighted. Subsequently, a crop mask was applied to select the region of interest, and finally, undesired highlighted areas (belonging to other tissues) were erased manually using the brush tool. In the interactive Figure 6, the segmented ovaries (4.02e7 voxels, 0.398 mm3), spermatheca (1.44e6 voxels, 0.014 mm3), and stored sperm (1.04e6 voxels, 0.010 mm3) are presented. The ventral nervous cord (8.16e5 voxels, 8.08e−3 mm3), also included in Figure 6, is presented as an example of a manually segmented region (its low contrast precluded semiautomatic segmentation).
Figure 6.
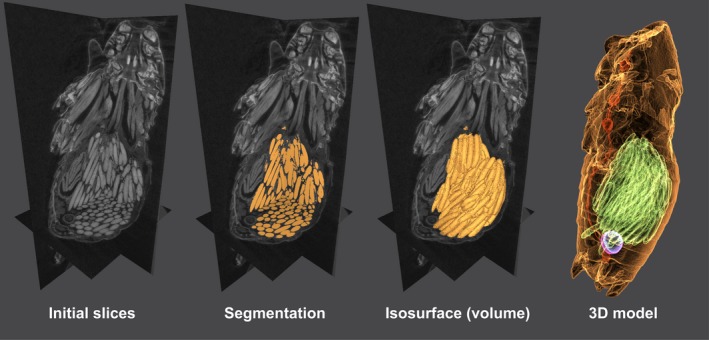
Segmentation process steps. Acheta domesticus female abdomen. Please click on the figure to activate the interactive 3D content and use the mouse to rotate the objects. Further functions (views, render modes, and model tree) are available in the menu
Care has to be taken when segmenting several specimens for quantitative purposes as variations as high as 20% have been reported (Parkinson, Badiei, & Fazzalari, 2008), however, using proper tissue preparation (staining and drying) and scanning techniques (e.g., phase contrast) this variation can be reduced. Besides, techniques for calibration are been developed, which would greatly increase the accuracy of the measurements (Léonard, Brown, Withers, Mummery, & McCarthy, 2014).
Due to the considerable size of the reconstructed scans (several gigabytes in most cases), it is usually not possible to include these as Supporting information to manuscripts. However, long‐lasting online repositories such as Digital Morphology Library (http://www.digimorph.org), Dryad Digital Repository (http://datadryad.org), MorphoBank (https://morphobank.org), MorphoSource (http://morphosource.org), and Science3D (https://www.science3d.org) provide storage facilities to ease the publication of datasets and accessibility to the scanned specimens. Outstanding examples of this possibility are the cybertypes of recently described species (Akkari et al., 2015) and online repositories of fossils (UMORF, https://umorf.ummp.lsa.umich.edu/).
8. PERSPECTIVES AND FINAL CONSIDERATIONS
As pointed out before in several reviews (Metscher, 2013; Socha et al., 2007; Westneat et al., 2008), X‐ray CT bears the potential to generate valuable morphological data from specimens and structures that would be impossible or expensive to acquire using other approaches (e.g., synchrotron or MRI facilities) (Brinkmann et al., 2016) or that would be time‐consuming (e.g., microtomy). Although reconstruction artifacts will inevitably be present in some images (Davis & Elliott, 2006), in this review we present several successful applications of this technique—and its variations—in ecological studies (Appendix S1). Our study outlines suitable combinations of techniques for preparation of specimen, providing sufficient contrast for image segmentation.
To date, X‐ray CT has proven to be a suitable technique to reveal the biology and ecology of elusive organisms (Jennings & Austin, 2011; Mouginot et al., 2015), diet analysis of extinct species (Collareta et al., 2015; Gill et al., 2014), and to examine nest/colony life, development, and structure (Fuchs et al., 2004; Greco et al., 2005, 2006). In terms of applied studies, monitoring of pest and invasive species (Fuchs et al., 2004; Halley et al., 2005; Harrison et al., 1993; Monaenkova et al., 2015; Soné, Mori, Ide, Setoguchi, & Yamanouchi, 1995; Tarver et al., 2006), interaction between hosts and parasites (Greco et al., 2010; Schwabe et al., 2014), forensic entomology (Johnson et al., 2012), and ecotoxicological research (Brinkmann et al., 2016; Holliday & Holliday, 2012; Lind et al., 2004; Pigneret et al., 2016; Yunusa, Braun, & Lawrie, 2009) have been also benefited greatly from this technique so far.
As the tissue detection in the technique explored in this review relies on metal‐based stains, X‐ray CT can also be considered as a method to detect, monitor, and even infer physiological pathways of metallic pollution in small animals (Bell et al., 2012). Furthermore, combination with other techniques—for instance fluorescence microscopy—may allow to identify processes within the tissues for future studies on ecophysiology and ecotoxicology (e.g., angiogenesis, apoptosis, inflammation) (Gremse et al., 2015; Handschuh, Baeumler, Schwaha, & Ruthensteiner, 2013; Metscher, 2009a). Possible detrimental effects of radiation and stain ingestion on living organisms (especially invertebrates) remain to be elucidated.
On the other hand, metal‐based staining is not a prerequisite, as evidence by the ethanol fixated and air‐dried sample (Figure 5a, b). Similar to synchrotron radiation, where the signal‐to‐noise is high enough to detect anatomical and histological details in unstained specimens, and similar to previous studies in other biological samples (Bartels et al., 2013; Krenkel et al., 2015; Töpperwien, Krenkel, Quade, et al., 2016), suitable drying procedures may thus be sufficient to yield sufficient image contrast also for insects and arthropods.
Overall, X‐ray computed tomography bears tremendous potential for future ecological research. We are just starting to unravel these possibilities.
CONFLICT OF INTEREST
None declared.
AUTHORS’ CONTRIBUTIONS
CS, YG, and DO conceived the idea of the manuscript. YG, TS, and MT designed the methodology and performed the scans. YG collected the literature data and led the writing of the manuscript. All authors gave final approval for publication.
DATA ACCESSIBILITY
Detailed information about the studies included in the historical review (Appendix S1), details of scans and specimens’ preparation (Appendix S2), updated list of manufacturers (Appendix S3), and a video of the scans used for Figure 6 (Video S4) can be found as online Supporting information. Representative scans (reconstructed files) of the techniques compared in this study are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.f9h43b0.
Supporting information
ACKNOWLEDGEMENTS
Financial support to YG was provided by Colciencias—Colombia (doctoral studies abroad program, 679‐2014). We thank to the technicians of the Electron microscopy division of IMPB Münster for performing the critical point drying of the samples. Additionally, we are grateful to two anonymous reviewers that contributed to the improvement of the manuscript.
Gutiérrez Y, Ott D, Töpperwien M, Salditt T, Scherber C. X‐ray computed tomography and its potential in ecological research: A review of studies and optimization of specimen preparation. Ecol Evol. 2018;8:7717–7732. 10.1002/ece3.4149
REFERENCES
- van Aarle, W. , Palenstijn, W. J. , Cant, J. , Janssens, E. , Bleichrodt, F. , Dabravolski, A. , … Sijbers, J. (2016). Fast and flexible X‐ray tomography using the ASTRA toolbox. Optics Express, 24, 25129–25147. 10.1364/OE.24.025129 [DOI] [PubMed] [Google Scholar]
- Abel, R. L. , Laurini, C. R. , & Richter, M. (2012). A palaeobiologist's guide to ‘virtual’ micro‐CT preparation. Palaeontologia Electronica, 15, 1–16. [Google Scholar]
- Akkari, N. , Enghoff, H. , & Metscher, B. D. (2015). A new dimension in documenting new species: high‐detail imaging for myriapod taxonomy and first 3D cybertype of a new millipede species (Diplopoda, Julida, Julidae). PLoS ONE, 10, e0135243 10.1371/journal.pone.0135243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amossé, J. , Turberg, P. , Kohler‐Milleret, R. , Gobat, J. M. , & Le Bayon, R. C. (2015). Effects of endogeic earthworms on the soil organic matter dynamics and the soil structure in urban and alluvial soil materials. Geoderma, 243–244, 50–57. 10.1016/j.geoderma.2014.12.007 [DOI] [Google Scholar]
- Auclerc, A. , Capowiez, Y. , Guérold, F. , & Nahmani, J. (2013). Application of X‐ray tomography to evaluate liming impact on earthworm burrowing activity in an acidic forest soil under laboratory conditions. Geoderma, 202–203, 45–50. 10.1016/j.geoderma.2013.03.011 [DOI] [Google Scholar]
- Bale, J. S. , Masters, G. J. , Hodkinson, I. D. , Awmack, C. , Bezemer, T. M. , Brown, V. K. , … Whittaker, J. B. (2002). Herbivory in global climate change research: direct effects of rising temperature on insect herbivores. Global Change Biology, 8, 1–16. 10.1046/j.1365-2486.2002.00451.x [DOI] [Google Scholar]
- Bartels, M. , Hernandez, V. H. , Krenkel, M. , Moser, T. , & Salditt, T. (2013). Phase contrast tomography of the mouse cochlea at microfocus X‐ray sources. Applied Physics Letters, 103, 83703 10.1063/1.4818737 [DOI] [Google Scholar]
- Bartels, M. , Krenkel, M. , Haber, J. , Wilke, R. N. , & Salditt, T. (2015). X‐ray holographic imaging of hydrated biological cells in solution. Physical Review Letters, 114, 48103 10.1103/PhysRevLett.114.048103 [DOI] [PubMed] [Google Scholar]
- Becker, K. , Jährling, N. , Kramer, E. R. , Schnorrer, F. , & Dodt, H. U. (2008). Ultramicroscopy: 3D reconstruction of large microscopical specimens. Journal of Biophotonics, 1, 36–42. 10.1002/(ISSN)1864-0648 [DOI] [PubMed] [Google Scholar]
- Bell, D. , Woolnough, L. , Mortimore, D. , Corps, N. , Hudson, D. M. , & Greco, M. K. (2012). A preliminary report on the use of bench‐top X‐ray micro‐computerised tomography to study the Malpighian tubules of the overwintering seven spotted ladybird Coccinella septempunctata L. (Coleoptera : Coccinellidae). Psyche, 2012, 1–6. [Google Scholar]
- Beutel, R.G. , Friedrich, F. , Ge, S. , & Yang, X. (2014). Traditional and modern techniques in insect morphology. Insect Morphology and Phylogeny, pp. 143–163.
- Bidart‐Bouzat, M. G. , & Imeh‐Nathaniel, A. (2008). Global change effects on plant chemical defenses against insect herbivores. Journal of Integrative Plant Biology, 50, 1339–1354. 10.1111/j.1744-7909.2008.00751.x [DOI] [PubMed] [Google Scholar]
- Brinkmann, M. , Rizzo, L. Y. , Lammers, T. , Gremse, F. , Schiwy, S. , Kiessling, F. , & Hollert, H. (2016). Micro‐computed tomography (μCT) as a novel method in ecotoxicology – determination of morphometric and somatic data in rainbow trout (Oncorhynchus mykiss). Science of the Total Environment, 543, 135–139. 10.1016/j.scitotenv.2015.11.020 [DOI] [PubMed] [Google Scholar]
- Callaghan, P. (1991). Principles of Nuclear Magnetic Resonance Microscopy. Oxford, UK: Oxford University Press. [Google Scholar]
- Cantin, N. E. , Cohen, A. L. , Karnauskas, K. B. , Tarrant, A. M. , & McCorkle, D. C. (2010). Ocean warming slows coral growth in the central red sea. Science, 329, 322–325. 10.1126/science.1190182 [DOI] [PubMed] [Google Scholar]
- Capowiez, Y. , Monestiez, P. , & Belzunces, L. (2001). Burrow systems made by Aporrectodea nocturna and Allolobophora chlorotica in artificial cores: Morphological differences and effects of interspecific interactions. Applied Soil Ecology, 16, 109–120. 10.1016/S0929-1393(00)00110-4 [DOI] [Google Scholar]
- Capowiez, Y. , Pierret, A. , & Moran, C. J. (2003). Characterisation of the three‐dimensional structure of earthworm burrow systems using image analysis and mathematical morphology. Biology and Fertility of Soils, 38, 301–310. 10.1007/s00374-003-0647-9 [DOI] [Google Scholar]
- Carbayo, F. , & Lenihan, J. W. (2016). Micro‐computed tomography scan and virtual histological slide data for the land planarian Obama otavioi (Platyhelminthes). GigaScience, 5, 13 10.1186/s13742-016-0119-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloetens, P. , Ludwig, W. , Baruchel, J. , Van Dyck, D. , Van Landuyt, J. , Guigay, J. P. , & Schlenker, M. (1999). Holotomography: Quantitative phase tomography with micrometer resolution using hard synchrotron radiation X‐rays. Applied Physics Letters, 75, 2912–2914. 10.1063/1.125225 [DOI] [Google Scholar]
- Cnudde, V. , Masschaele, B. , De Cock, H. E. , Olstad, K. , Vlaminck, L. , Vlassenbroeck, J. , … Jacobs, P. (2008). Virtual histology by means of high‐resolution X‐ray CT. Journal of Microscopy, 232, 476–485. 10.1111/j.1365-2818.2008.02142.x [DOI] [PubMed] [Google Scholar]
- Collareta, A. , Landini, W. , Lambert, O. , Post, K. , Tinelli, C. , Celma, C. Di , … Bianucci, G. (2015). Piscivory in a Miocene Cetotheriidae of Peru: first record of fossilized stomach content for an extinct baleen‐bearing whale. The Science of Nature, 102, 2–12. [DOI] [PubMed] [Google Scholar]
- Cunningham, J. A. , Rahman, I. A. , Lautenschlager, S. , Rayfield, E. J. , & Donoghue, P. C. J. (2014). A virtual world of paleontology. Trends in Ecology & Evolution, 29, 347–357. 10.1016/j.tree.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Davey, E. , Wigand, C. , Johnson, R. , Sundberg, K. , Morris, J. , & Roman, C. T. (2011). Use of computed tomography imaging for quantifying coarse roots, rhizomes, peat, and particle densities in marsh soils. Ecological Applications, 21, 2156–2171. 10.1890/10-2037.1 [DOI] [PubMed] [Google Scholar]
- Davis, G. R. , & Elliott, J. C. (2006). Artefacts in X‐ray microtomography of materials. Materials Science and Technology, 22, 1011–1018. 10.1179/174328406X114117 [DOI] [Google Scholar]
- De Clerck, N.M. , Meurrens, K. , Weiler, H. , Van Dyck, D. , Vanhoutte, G. , Terpstra, P. , & Postnov, A. A. (2004). High‐resolution X‐ray microtomography for the detection of lung tumors in living mice. Neoplasia (New York, N.Y.), 6, 374–379. 10.1593/neo.03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witte, Y. , Boone, M. , Vlassenbroeck, J. , Dierick, M. , & Van Hoorebeke, L. (2009). Bronnikov‐aided correction for X‐ray computed tomography. Journal of the Optical Society of America. A, 26, 890–894. 10.1364/JOSAA.26.000890 [DOI] [PubMed] [Google Scholar]
- Degenhardt, K. , Wright, A. C. , Horng, D. , Padmanabhan, A. , & Epstein, J. A. (2010). Rapid 3D phenotyping of cardiovascular development in mouse embryos by micro‐CT with iodine staining. Circulation: Cardiovascular Imaging, 3, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deraison, H. , Badenhausser, I. , Loeuille, N. , Scherber, C. , & Gross, N. (2015). Functional trait diversity across trophic levels determines herbivore impact on plant community biomass. Ecology Letters, 18, 1346–1355. 10.1111/ele.12529 [DOI] [PubMed] [Google Scholar]
- Dhondt, S. , Vanhaeren, H. , Van Loo, D. , Cnudde, V. , & Inzé, D. (2010). Plant structure visualization by high‐resolution X‐ray computed tomography. Trends in Plant Science, 15, 419–422. 10.1016/j.tplants.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Diez, M. E. , Orensanz, J. M. , Márquez, F. , & Cremonte, F. (2013). Shell damage in the Tehuelche scallop Aequipecten tehuelchus caused by Polydora rickettsi (Polychaeta: Spionidae) infestation. Journal of Invertebrate Pathology, 114, 107–113. 10.1016/j.jip.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Elliott, J. C. , & Dover, S. D. (1982). X‐ray microtomography. Journal of Microscopy, 126, 211–213. 10.1111/j.1365-2818.1982.tb00376.x [DOI] [PubMed] [Google Scholar]
- Eyer, M. , Neumann, P. , & Dietemann, V. (2016). A look into the cell: honey storage in honey bees, apis mellifera. PLoS ONE, 11, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulwetter, S. , Vasileiadou, A. , Kouratoras, M. , Dailianis, T. , & Arvanitidis, C. (2013). Micro‐computed tomography: Introducing new dimensions to taxonomy. ZooKeys, 1, 1–45. 10.3897/zookeys.263.4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, A. , Beichel, R. , Kalpathy‐cramer, J. , Finet, J. , Fillion‐robin, J. , Pujol, S. , … Kikinis, R. (2012). 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magnetic Resonance Imaging, 30, 1323–1341. 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, R. , Kvist, S. , Lenihan, J. , Giribet, G. , & Ziegler, A. (2014). Sine systemate chaos? A versatile tool for earthworm taxonomy: non‐destructive imaging of freshly fixed and museum specimens using micro‐computed tomography. PLoS ONE, 9, e96617 10.1371/journal.pone.0096617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, S. J. , Senning, M. , Sonnewald, S. , Kessling, P.‐M. , Goldstein, R. , & Sonnewald, U. (2010). Comparative transcriptome analysis coupled to X‐ray CT reveals sucrose supply and growth velocity as major determinants of potato tuber starch biosynthesis. BMC Genomics, 11, 93 10.1186/1471-2164-11-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani, F. , & Schurr, U. (2013). Future scenarios for plant phenotyping. Annual Review of Plant Biology, 64, 267–291. 10.1146/annurev-arplant-050312-120137 [DOI] [PubMed] [Google Scholar]
- Francis, G. S. , Tabley, F. J. , Butler, R. C. , & Fraser, P. M. (2001). The burrowing characteristics of three common earthworm species. Soil Research, 39, 1453–1465. 10.1071/SR00033 [DOI] [Google Scholar]
- Friedrich, F. , & Beutel, R.G. (2008). Micro‐computer tomography and a renaissance of insect morphology. Proceedings of SPIE, p. 70781U.
- Fuchs, A. , Schreyer, A. , Feuerbach, S. , & Korb, J. (2004). A new technique for termite monitoring using computer tomography and endoscopy. International Journal of Pest Management, 50, 63–66. 10.1080/0967087032000159300 [DOI] [Google Scholar]
- Fujiwara, S.‐I. , Oji, T. , Tanaka, Y. , & Kondo, Y. (2005). Relay strategy and adaptation to a muddy environment in Isselicrinus (Isselicrinidae: Crinoidea). Palaios, 20, 241–248. 10.2110/palo.2004.p04-25 [DOI] [Google Scholar]
- Gill, P. G. , Purnell, M. A. , Crumpton, N. , Brown, K. R. , Gostling, N. J. , Stampanoni, M. , & Rayfield, E. J. (2014). Dietary specializations and diversity in feeding ecology of the earliest stem mammals. Nature, 512, 303–305. 10.1038/nature13622 [DOI] [PubMed] [Google Scholar]
- Goodman, B. A. , & Chudekt, J. A. (1995). Nuclear magnetic resonance microscopy as a non‐invasive tool to study the development of Lepidopteran Pupae. Journal of Insect Physiology, 41, 419–424. 10.1016/0022-1910(94)00115-W [DOI] [Google Scholar]
- Greco, M. , Bell, M. , Spooner‐Hart, R. , & Holford, P. (2006). X‐ray computerized tomography as a new method for monitoring Amegilla holmesi nest structures, nesting behaviour, and adult female activity. Entomologia Experimentalis et Applicata, 120, 71–76. 10.1111/j.1570-7458.2006.00429.x [DOI] [Google Scholar]
- Greco, M. K. , Hoffmann, D. , Dollin, A. , Duncan, M. , Spooner‐Hart, R. , & Neumann, P. (2010). The alternative Pharaoh approach: Stingless bees mummify beetle parasites alive. Naturwissenschaften, 97, 319–323. 10.1007/s00114-009-0631-9 [DOI] [PubMed] [Google Scholar]
- Greco, M. K. , Spooner‐Hart, R. N. , Beattie, A. G. , Barchia, I. , & Holford, P. (2011). Australian stingless bees improve greenhouse Capsicum production. Journal of Apicultural Research, 50, 102–115. 10.3896/IBRA.1.50.2.02 [DOI] [Google Scholar]
- Greco, M. , Spooner‐Hart, R. , & Holford, P. (2005). A new technique for monitoring Trigona carbonaria nest contents, brood and activity using X‐ray computerized tomography. Journal of Apicultural Research, 44, 97–100. 10.1080/00218839.2005.11101156 [DOI] [Google Scholar]
- Greco, M. K. , Tong, J. , Soleimani, M. , Bell, D. , & Schäfer, M. O. (2012). Imaging live bee brains using minimally‐invasive diagnostic radioentomology. Journal of Insect Science (Online), 12, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, P. J. , Hutchison, D. J. , Read, D. B. , Jenneson, P. M. , Gilboy, W. B. , & Morton, E. J. (2003). Non‐invasive imaging of roots with high resolution X‐ray micro‐tomography. Plant and Soil, 255, 351–359. 10.1023/A:1026179919689 [DOI] [Google Scholar]
- Gremse, F. , Doleschel, D. , Zafarnia, S. , Babler, A. , Jahnen‐Dechent, W. , Lammers, T. , … Kiessling, F. (2015). Hybrid μCT‐FMT imaging and image analysis. Journal of Visualized Experiments, 52770, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley, J. D. , Burd, M. , & Wells, P. (2005). Excavation and architecture of Argentine ant nests. Insectes Sociaux, 52, 350–356. 10.1007/s00040-005-0818-9 [DOI] [Google Scholar]
- Handschuh, S. , Baeumler, N. , Schwaha, T. , & Ruthensteiner, B. (2013). A correlative approach for combining microCT, light and transmission electron microscopy in a single 3D scenario. Frontiers in Zoology, 10, 44 10.1186/1742-9994-10-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, R. D. , Gardner, W. A. , Tollner, W. E. , & Kinard, D. J. (1993). X‐ray computed tomography studies of the burrowing behavior of fourth‐instar pecan weevil (Coleoptera: Curculionidae). Journal of Economic Entomology, 86, 1714–1719. 10.1093/jee/86.6.1714 [DOI] [Google Scholar]
- Hart, A. G. , Bowtell, R. W. , Köckenberger, W. , Wenseleers, T. , & Ratnieks, F. L. W. (2003). Magnetic resonance imaging in entomology: a critical review. Journal of Insect Science, 3, 5. [PMC free article] [PubMed] [Google Scholar]
- Hayat, M. A. (1970). Principles and Techniques of Electron Microscopy. Biological Applications. New York, NY: Van Nostrand Reinhold Company. [Google Scholar]
- Herrel, A. , Soons, J. , Aerts, P. , Dirckx, J. , Boone, M. , Jacobs, P. , … Podos, J. (2010). Adaptation and function of the bills of Darwins finches: Divergence by feeding type and sex. Emu, 110, 39–47. 10.1071/MU09034 [DOI] [Google Scholar]
- Higgins, J. P. , Williams, G. , Nagel, J. S. , & Higgins, J. A. (2006). Left bundle‐branch block artifact on single photon emission computed tomography with technetium Tc 99 m (Tc‐99 m) agents: Mechanisms and a method to decrease false‐positive interpretations. American Heart Journal, 152, 619–626. 10.1016/j.ahj.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Himmi, S. K. , Yoshimura, T. , Yanase, Y. , Oya, M. , Torigoe, T. , Akada, M. , & Imadzu, S. (2016). Nest‐gallery development and caste composition of isolated foraging groups of the drywood termite incisitermes minor (Isoptera: Kalotermitidae). Insects, 7, 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, D. K. , & Holliday, C. M. (2012). The effects of the organopollutant PCB 126 on bone density in juvenile diamondback terrapins (Malaclemys terrapin). Aquatic Toxicology, 109, 228–233. 10.1016/j.aquatox.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Hoshino, M. , Uesugi, K. , & Yagi, N. (2012). Phase‐contrast X‐ray microtomography of mouse fetus. Biology Open, 1, 269–274. 10.1242/bio.2012430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, J. (2009). Introduction In Hsieh J., (Ed.), Computed tomography: principles, design, artifacts, and recent advances (p. 556). Bellingham, WA: SPIE Press. [Google Scholar]
- Iwan, D. , Kamimski, M. J. , & Ras, M. (2015). Arthropod structure & development the last breath : A m CT‐based method for investigating the tracheal system in Hexapoda. Arthropod Structure & Development, 44, 218–227. 10.1016/j.asd.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Jahn, H. , Oliveira, I. V. O. D. E. S. , Gross, V. , Martin, C. , Hipp, A. , Mayer, G. , & Hammel, J. U. (2018). Evaluation of contrasting techniques for X‐ray imaging of velvet worms (Onychophora). Journal of Microscopy, 270, 343–358. [DOI] [PubMed] [Google Scholar]
- Jährling, N. , Becker, K. , Schönbauer, C. , Schnorrer, F. , & Dodt, H.‐U. (2010). Three‐dimensional reconstruction and segmentation of intact Drosophila by ultramicroscopy. Frontiers in Systems Neuroscience, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasanoff, A. , & Sun, P. (2002). In vivo magnetic resonance microscopy of brain structure in unanesthetized flies. Journal of Magnetic Resonance, 158, 79–85. 10.1016/S1090-7807(02)00063-0 [DOI] [PubMed] [Google Scholar]
- Jégou, D. , Brunotte, J. , Rogasik, H. , Capowiez, Y. , Diestel, H. , Schrader, S. , & Cluzeau, D. (2002). Impact of soil compaction on earthworm burrow systems using X‐ray computed tomography: Preliminary study. European Journal of Soil Biology, 38, 329–336. 10.1016/S1164-5563(02)01148-2 [DOI] [Google Scholar]
- Jégou, D. , Capowiez, Y. , & Cluzeau, D. (2001). Interactions between earthworm species in artificial soil cores assessed through the 3D reconstruction of the burrow systems. Geoderma, 102, 123–137. 10.1016/S0016-7061(00)00107-5 [DOI] [Google Scholar]
- Jégou, D. , Cluzeau, D. , Hallaire, V. , Balesdent, J. , & Tréhen, P. (2000). Burrowing activity of the earthworms Lumbricus terrestris and Aporrectodea giardi and consequences on C transfers in soil. European Journal of Soil Biology, 36, 27–34. 10.1016/S1164-5563(00)01046-3 [DOI] [Google Scholar]
- Jégou, D. , Cluzeau, D. , Wolf, H. J. , Gandon, Y. , & Tréhen, P. (1998). Assessment of the burrow system of Lumbricus terrestris, Aporrectodea giardi, and Aporrectodea caliginosa using X‐ray computed tomography. Biology and Fertility of Soils, 26, 116–121. [Google Scholar]
- Jégou, D. , Hallaire, V. , Cluzeau, D. , & Tréhen, P. (1999). Characterization of the burrow system of the earthworms Lumbricus terrestris and Aporrectodea giardi using X‐ray computed tomography and image analysis. Biology and Fertility of Soils, 29, 314–318. [Google Scholar]
- Jennings, J. T. , & Austin, A. D. (2011). Novel use of a micro‐computed tomography scanner to trace larvae of wood boring insects. Australian Journal of Entomology, 50, 160–163. 10.1111/j.1440-6055.2010.00792.x [DOI] [Google Scholar]
- Johnson, A. , Archer, M. , Leigh‐Shaw, L. , Pais, M. , O'Donnell, C. , & Wallman, J. (2012). Examination of forensic entomology evidence using computed tomography scanning: Case studies and refinement of techniques for estimating maggot mass volumes in bodies. International Journal of Legal Medicine, 126, 693–702. 10.1007/s00414-012-0716-4 [DOI] [PubMed] [Google Scholar]
- Jones, B. M. , Leonard, S. , & Papaj, D. R. (2013). Plasticity of the worker bumblebee brain in relation to age and rearing environment. Brain, Behavior and Evolution, 82, 250–261. 10.1159/000355845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao, T. , Okamoto, T. , Miyachi, Y. , & Nohara, N. (2003). Parental exposure to low‐dose X‐rays in Drosophila melanogaster induces early emergence in offspring, which can be modulated by transplantation of polar cytoplasm. Mutation Research ‐ Fundamental and Molecular Mechanisms of Mutagenesis, 527, 1–6. 10.1016/S0027-5107(03)00051-4 [DOI] [PubMed] [Google Scholar]
- Kato, A. , Tang, N. , Borries, C. , Papakyrikos, A. M. , Hinde, K. , Miller, E. , … Smith, T. M. (2014). Intra‐ and interspecific variation in Macaque molar enamel thickness. American Journal of Physical Anthropology, 155, 447–459. 10.1002/ajpa.22593 [DOI] [PubMed] [Google Scholar]
- Kim, B. H. , Seo, E. S. , Lim, J. H. , & Lee, S. J. (2012). Synchrotron X‐ray microscopic computed tomography of the pump system of a female mosquito. Microscopy Research and Technique, 75, 1051–1058. 10.1002/jemt.22030 [DOI] [PubMed] [Google Scholar]
- Krenkel, M. , Markus, A. , Bartels, M. , Dullin, C. , Alves, F. , & Salditt, T. (2015). Phase‐contrast zoom tomography reveals precise locations of macrophages in mouse lungs. Scientific Reports, 5, 9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagomarsino, S. , Cedola, A. , Cloetens, P. , Di Fonzo, S. , Jark, W. , Soullié, G. , & Riekel, C. (1997). Phase contrast hard x‐ray microscopy with submicron resolution. Applied Physics Letters, 71, 2557–2559. 10.1063/1.119324 [DOI] [Google Scholar]
- Langmaack, M. , Schrader, S. , Rapp‐Bernhardt, U. , & Kotzke, K. (1999). Quantitative analysis of earthworm burrow systems with respect to biological soil‐structure regeneration after soil compaction. Biology and Fertility of Soils, 28, 219–229. 10.1007/s003740050486 [DOI] [Google Scholar]
- Lautenschlager, S. (2016). Reconstructing the past: methods and techniques for the digital restoration of fossils. Open Science, 3, 160342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léonard, F. , Brown, S. B. , Withers, P. J. , Mummery, P. M. , & McCarthy, M. B. (2014). A new method of performance verification for x‐ray computed tomography measurements. Measurement Science and Technology, 25, 65401 10.1088/0957-0233/25/6/065401 [DOI] [Google Scholar]
- Lind, P. M. , Milnes, M. R. , Lundberg, R. , Bermudez, D. , Örberg, J. , & Guillette, L. J. (2004). Abnormal bone composition female juvenile American alligators from a pesticide‐polluted lake (Lake Apopka, Florida). Environmental Health Perspectives, 112, 359–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lösel, P. , & Heuveline, V. (2016). Enhancing a diffusion algorithm for 4D image segmentation using local information. Medical Imaging: Image Processing, p. 97842L.
- Mairhofer, S. , Zappala, S. , Pridmore, T. , Mairhofer, S. , Zappala, S. , Tracy, S. R. , … Pridmore, T. (2012). RooTrak: automated recovery of three‐ dimensional plant root architecture in soil from X‐Ray microcomputed using visual tracking. Plant Physiology, 158, 561–569. 10.1104/pp.111.186221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszka, J. , Barron, A. B. , Helliwell, P. G. , & Maleszka, R. (2009). Effect of age, behaviour and social environment on honey bee brain plasticity. Journal of Comparative Physiology A, 195, 733–740. 10.1007/s00359-009-0449-0 [DOI] [PubMed] [Google Scholar]
- Mattei, A. L. , Riccio, M. L. , Avila, F. W. , & Wolfner, M. F. (2015). Integrated 3D view of postmating responses by the Drosophila melanogaster female reproductive tract, obtained by micro‐computed tomography scanning. Proceedings of the National Academy of Sciences of the United States of America, 112, 8475–8480. 10.1073/pnas.1505797112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher, B. D. (2009a). MicroCT for developmental biology: A versatile tool for high‐contrast 3D imaging at histological resolutions. Developmental Dynamics, 238, 632–640. 10.1002/dvdy.21857 [DOI] [PubMed] [Google Scholar]
- Metscher, B. D. (2009b). MicroCT for comparative morphology: simple staining methods allow high‐contrast 3D imaging of diverse non‐mineralized animal tissues. BMC Physiology, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher, B. D. (2013). Biological applications of X‐ray microtomography: imaging micro‐ anatomy, molecular expression and organismal diversity. Microscopy and Analysis, 27, 13–16. [Google Scholar]
- Metzner, R. , Eggert, A. , van Dusschoten, D. , Pflugfelder, D. , Gerth, S. , Schurr, U. , … Jahnke, S. (2015). Direct comparison of MRI and X‐ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods, 11, 17 10.1186/s13007-015-0060-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaenkova, D. , Gravish, N. , Rodriguez, G. , Kutner, R. , Goodisman, M. A. D. , & Goldman, D. I. (2015). Behavioral and mechanical determinants of collective subsurface nest excavation. Journal of Experimental Biology, 218, 1295–1305. 10.1242/jeb.113795 [DOI] [PubMed] [Google Scholar]
- Mooney, S. J. , Morris, C. , & Berry, P. M. (2006). Visualization and quantification of the effects of cereal root lodging on three‐dimensional soil macrostructure using X‐ray computed tomography. Soil Science, 171, 706–718. 10.1097/01.ss.0000228041.03142.d3 [DOI] [Google Scholar]
- Mouginot, P. , Prügel, J. , Thom, U. , Steinhoff, P. O. M. , Kupryjanowicz, J. , & Uhl, G. (2015). Securing paternity by mutilating female genitalia in spiders. Current Biology, 25, 2980–2984. 10.1016/j.cub.2015.09.074 [DOI] [PubMed] [Google Scholar]
- Nagy, T. R. (2001). The use of dual‐energy X‐ray absorptiometry for the measurement of body composition In Speakman J. R. (Ed.), Body Composition Analysis of Animals (pp. 211–229). New York, NY: Cambridge University Press; 10.1017/CBO9780511551741 [DOI] [Google Scholar]
- Paganin, D. , & Nugent, K. A. (1998). Noninterferometric phase imaging with partially coherent light. Physical Review Letters, 80, 2586 10.1103/PhysRevLett.80.2586 [DOI] [Google Scholar]
- Pagenkemper, S. K. , Athmann, M. , Uteau, D. , Kautz, T. , Peth, S. , & Horn, R. (2015). The effect of earthworm activity on soil bioporosity – Investigated with X‐ray computed tomography and endoscopy. Soil and Tillage Research, 146, 79–88. 10.1016/j.still.2014.05.007 [DOI] [Google Scholar]
- Pampush, J. D. , Spradley, J. P. , Morse, P. E. , Harrington, A. R. , Allen, K. L. , Boyer, D. M. , & Kay, R. F. (2016). Wear and its effects on dental topography measures in howling monkeys (Alouatta palliata). Physical Anthropology, 161, 705–721. 10.1002/ajpa.23077 [DOI] [PubMed] [Google Scholar]
- Parkinson, I. H. , Badiei, A. , & Fazzalari, N. L. (2008). Variation in segmentation of bone from micro‐CT imaging: implications for quantitative morphometric analysis. Australasian Physical & Engineering Science in Medicine, 31, 160–164. 10.1007/BF03178592 [DOI] [PubMed] [Google Scholar]
- Pelosi, C. , Grandeau, G. , & Capowiez, Y. (2017). Geoderma Temporal dynamics of earthworm‐related macroporosity in tilled and non‐tilled cropping systems. Geoderma, 289, 169–177. 10.1016/j.geoderma.2016.12.005 [DOI] [Google Scholar]
- Perret, J. S. , Al‐Belushi, M. E. , & Deadman, M. (2007). Non‐destructive visualization and quantification of roots using computed tomography. Soil Biology and Biochemistry, 39, 391–399. 10.1016/j.soilbio.2006.07.018 [DOI] [Google Scholar]
- Pigneret, M. , Mermillod‐blondin, F. , Volatier, L. , Romestaing, C. , Maire, E. , Adrien, J. , … Hervant, F. (2016). Urban pollution of sediments: Impact on the physiology and burrowing activity of tubificid worms and consequences on biogeochemical processes. Science of the Total Environment, 568, 196–207. 10.1016/j.scitotenv.2016.05.174 [DOI] [PubMed] [Google Scholar]
- Postnov, A. , De Clerck, N. , Sasov, A. , & Van Dyck, D. (2002). 3D in‐vivo X‐ray microtomography of living snails. Journal of Microscopy, 205, 201–204. 10.1046/j.0022-2720.2001.00986.x [DOI] [PubMed] [Google Scholar]
- Rademacher, E. , Fahlberg, A. , Raddatz, M. , Schneider, S. , & Voigt, K. (2013). Galenics: Studies of the toxicity and distribution of sugar substitutes on Apis mellifera. Apidologie, 44, 222–233. 10.1007/s13592-012-0174-5 [DOI] [Google Scholar]
- Ravel, A. , & Orliac, M. J. (2015). The inner ear morphology of the “condylarthran” Hyopsodus lepidus. Historical Biology, 27, 957 10.1080/08912963.2014.915823 [DOI] [Google Scholar]
- Renaud, S. , Gomes Rodrigues, H. , Ledevin, R. , Pisanu, B. , Chapuis, J.‐L. , & Hardouin, E. A. (2015). Fast morphological response of house mice to anthropogenic disturbances on a Sub‐Antarctic island. Biological Journal of the Linnean Society, 114, 513–526. 10.1111/bij.12454 [DOI] [Google Scholar]
- Ribi, W. , Senden, T. J. , Sakellariou, A. , Limaye, A. , & Zhang, S. (2008). Imaging honey bee brain anatomy with micro‐X‐ray‐computed tomography. Journal of Neuroscience Methods, 171, 93–97. 10.1016/j.jneumeth.2008.02.010 [DOI] [PubMed] [Google Scholar]
- Rogasik, H. , Schrader, S. , Onasch, I. , Kiesel, J. , & Gerke, H. H. (2014). Micro‐scale dry bulk density variation around earthworm (Lumbricus terrestris L.) burrows based on X‐ray computed tomography. Geoderma, 213, 471–477. 10.1016/j.geoderma.2013.08.034 [DOI] [Google Scholar]
- Rolo, S. , Ershov, A. , van de Kamp, T. , & Baumbach, T. (2014). In vivo X‐ray cine‐tomography for tracking morphological dynamics. Proceedings of the National Academy of Sciences, 111, 3921–3926. 10.1073/pnas.1308650111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach, S. J. , Bag, S. , Schilling, L. , Groden, C. , & Brockmann, M. A. (2010). Application of micro‐CT in small animal imaging. Methods, 50, 2–13. 10.1016/j.ymeth.2009.08.007 [DOI] [PubMed] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönberg, C. H. L. (2001). Estimating the extent of endolithic tissue of a Great Barrier Reef clionid sponge. Senckenbergiana Maritima, 31, 29–39. 10.1007/BF03042834 [DOI] [Google Scholar]
- Schrader, S. , Rogasik, H. , Onasch, I. , & Jegou, D. (2007). Assessment of soil structural differentiation around earthworm burrows by means of X‐ray computed tomography and scanning electron microscopy. Geoderma, 137, 378–387. 10.1016/j.geoderma.2006.08.030 [DOI] [Google Scholar]
- Schwabe, E. , Holtheuer, J. , & Schories, D. (2014). First record of a mesoparasite (Crustacea, Copepoda) infesting a polyplacophoran (Mollusca, Polyplacophora) in Chilean waters, with an overview of the family Chitonophilidae (Crustacea & Mollusca). Spixiana, 37, 165–182. [Google Scholar]
- Self, C. J. (2015). Dental root size in bats with diets of different hardness. Journal of Morphology, 276, 1065–1074. 10.1002/jmor.20400 [DOI] [PubMed] [Google Scholar]
- Sena, G. , Almeida, A. P. , Braz, D. , Nogueira, L. P. , Soares, J. , Azambuja, P. , … Barroso, R. C. (2015). On the possibilities of polychromatic synchrotron radiation microtomography for visualization of internal structures of Rhodnius prolixus. Radiation Physics and Chemistry, 115, 179–182. 10.1016/j.radphyschem.2015.07.006 [DOI] [Google Scholar]
- Seo, E. , Lim, J.‐H. , Seo, S. J. , & Lee, S. J. (2015). Whole‐body imaging of a hypercholesterolemic female Zebrafish by using synchrotron X‐ray micro‐CT. Zebrafish, 12, 11–20. 10.1089/zeb.2014.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickert, S. , Rodner, E. , & Denzler, J. (2015). Semantic Volume Segmentation with Iterative Context Integration. Proceedings of the 9th Open German‐Russian Workshop on Pattern Recognition and Image Understanding, pp. 1–6. Koblens, Germany.
- Silbiger, N. J. , Guadayol, O. , Thomas, F. O. M. , & Donahue, M. J. (2016). A novel μct analysis reveals different responses of bioerosion and secondary accretion to environmental variability. PLoS ONE, 11, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. B. , Bernhardt, G. , Raine, N. E. , Abel, R. L. , Sykes, D. , Ahmed, F. , … Gill, R. J. (2016). Exploring miniature insect brains using micro‐CT scanning techniques. Scientific Reports, 6, 21768 10.1038/srep21768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, I. C. , Carson, B. L. , & Ferguson, T. L. (1974). Osmium: An appraisal of environmental exposure. Environmental Health Perspectives, 8, 201–213. 10.1289/ehp.748201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell‐Rood, E. C. , Papaj, D. R. , & Gronenberg, W. (2009). Brain size: A global or induced cost of learning? Brain, Behavior and Evolution, 73, 111–128. 10.1159/000213647 [DOI] [PubMed] [Google Scholar]
- Socha, J. J. , Westneat, M. W. , Harrison, J. F. , Waters, J. S. , & Lee, W.‐K. (2007). Real‐time phase‐contrast x‐ray imaging: a new technique for the study of animal form and function. BMC Biology, 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sombke, A. , Lipke, E. , Michalik, P. , Uhl, G. , & Harzsch, S. (2015). Potential and limitations of X‐ray micro‐computed tomography in arthropod neuroanatomy: A methodological and comparative survey. Journal of Comparative Neurology, 523, 1281–1295. 10.1002/cne.23741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soné, K. , Mori, T. , Ide, M. , Setoguchi, M. , & Yamanouchi, K. (1995). Application of computer tomography to surveys of the galleries of the oak borer, platypus quercivorus (Maruyama) (Coleoptera: Platypodidae). Japanese Journal of Applied Entomology and Zoology, 39, 341–344. 10.1303/jjaez.39.341 [DOI] [Google Scholar]
- Soons, J. , Genbrugge, A. , Podos, J. , Adriaens, D. , Aerts, P. , Dirckx, J. , & Herrel, A. (2015). Is beak morphology in Darwin's finches tuned to loading demands? PLoS ONE, 10, e0129479 10.1371/journal.pone.0129479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender, A. S. , Marchuk, K. , Liu, C. , Sander, S. , Meyer, M. W. , Smith, E. A. , … Huang, B. (2014). Single cell optical imaging and spectroscopy. Chemical Reviews, 113, 2469–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, R.W. , & Elser, J.J. (2002). How to build an animal: the stoichiometry of Metazoans In Sterner R.W. & Elser J.J. (Eds.), Ecological stoichiometry. The biology of elements from molecules to the biosphere (pp. 135–178). Jersey, New Jersey: Princeton University Press. [Google Scholar]
- Sutton, M. D. , Garwood, R. J. , Siveter, D. J. , & Siveter, D. J. (2012). SPIERS and VAXML; A software toolkit for tomographic visualisation and a format for virtual specimen interchange. Palaeontologia Electronica, 15, 14. [Google Scholar]
- Swart, C. C. , Deaton, L. E. , & Felgenhauer, B. E. (2006). The salivary gland and salivary enzymes of the giant waterbugs (Heteroptera; Belostomatidae). Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 145, 114–122. 10.1016/j.cbpa.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Swart, P. , Wicklein, M. , Sykes, D. , Ahmed, F. , & Krapp, H. G. (2016). A quantitative comparison of micro‐CT preparations in Dipteran flies. Scientific Reports, 6, 39380 10.1038/srep39380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarver, M. R. , Shade, R. E. , Tarver, R. D. , Liang, Y. , Krishnamurthi, G. , Pittendrigh, B. R. , & Murdock, L. L. (2006). Use of micro‐CAT scans to understand cowpea seed resistance to Callosobruchus maculatus. Entomologia Experimentalis et Applicata, 118, 33–39. 10.1111/j.1570-7458.2006.00360.x [DOI] [Google Scholar]
- Tate, J. , Burton, B. , & Khan, A. (2016). Seg3D Basic Functionality. URL http://www.sci.utah.edu/devbuilds/seg3d_docs/Seg3DBasicFunctionality.pdf [accessed 18 April 2017]
- Tollner, E. W. (1991). X‐ray computed tomography applications in soil ecology studies. Agriculture, Ecosystems and Environment, 34, 251–260. 10.1016/0167-8809(91)90112-B [DOI] [Google Scholar]
- Töpperwien, M. , Krenkel, M. , Müller, K. , & Salditt, T. (2016). Phase‐contrast tomography of neuronal tissues: from laboratory‐to high resolution synchrotron CT. Society of Photo‐Optical Instrumentation Engineers (SPIE) Conference Series
- Töpperwien, M. , Krenkel, M. , Quade, F. , & Salditt, T. (2016). Laboratory‐based x‐ray phase‐contrast tomography enables 3D virtual histology. Proceedings SPIE Khounsary A. & van Dorssen G.E. (Eds.), pp. 1–10.
- Töpperwien, M. , Krenkel, M. , Vincenz, D. , Stöber, F. , Oelschlegel, A. M. , Goldschmidt, J. , & Salditt, T. (2017). Three‐dimensional mouse brain cytoarchitecture revealed by laboratory‐based x‐ray phase‐contrast tomography. Scientific Reports, 7, 42847 10.1038/srep42847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bulcke, J. , Boone, M. , Van Acker, J. , & Van Hoorebeke, L. (2009). Three‐dimensional X‐ray imaging and analysis of fungi on and in wood. Microscopy and Microanalysis: the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada, 15, 395–402. 10.1017/S1431927609990419 [DOI] [PubMed] [Google Scholar]
- Vickerton, P. , Jarvis, J. , & Jeffery, N. (2013). Concentration‐dependent specimen shrinkage in iodine‐enhanced microCT. Journal of Anatomy, 223, 185–193. 10.1111/joa.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecker, S. , Hörnschemeyer, T. , & Hoehn, M. (2002). Investigation of insect morphology by MRI: Assessment of spatial and temporal resolution. Magnetic Resonance Imaging, 20, 105–111. 10.1016/S0730-725X(01)00445-3 [DOI] [PubMed] [Google Scholar]
- Westneat, M. W. , Socha, J. J. , & Lee, W.‐K. (2008). Advances in biological structure, function, and physiology using synchrotron X‐ray imaging*. Annual Review of Physiology, 70, 119–142. 10.1146/annurev.physiol.70.113006.100434 [DOI] [PubMed] [Google Scholar]
- Willemink, M. J. , de Jong, P. A. , Leiner, T. , de Heer, L. M. , Nievelstein, R. A. J. , Budde, R. P. J. , & Schilham, A. M. R. (2013). Iterative reconstruction techniques for computed tomography Part 1: technical principles. European Radiology, 23, 1623–1631. 10.1007/s00330-012-2765-y [DOI] [PubMed] [Google Scholar]
- Wipfler, B. , Pohl, H. , Yavorskaya, M. I. , & Beutel, R. G. (2016). ScienceDirect A review of methods for analysing insect structures — the role of morphology in the age of phylogenomics. Current Opinion in Insect Science, 18, 60–68. 10.1016/j.cois.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Withers, P. J. (2007). X‐ray nanotomography. Materials Today, 10, 26–34. 10.1016/S1369-7021(07)70305-X [DOI] [Google Scholar]
- Yao, S. , Fan, J. , Zong, Y. , He, Y. , Zhou, G. , Sun, Z. , … Jiang, H. (2016). Equally sloped X‐ray microtomography of living insects with low radiation dose and improved resolution capability. Applied Physics Letters, 108, 123702 10.1063/1.4944727 [DOI] [Google Scholar]
- Yunusa, I. A. M. , Braun, M. , & Lawrie, R. (2009). Amendment of soil with coal fly ash modified the burrowing habits of two earthworm species. Applied Soil Ecology, 42, 63–68. 10.1016/j.apsoil.2009.02.002 [DOI] [Google Scholar]
- Ziegler, A. , Kunth, M. , Mueller, S. , Bock, C. , Pohmann, R. , Schröder, L. , … Giribet, G. (2011). Application of magnetic resonance imaging in zoology. Zoomorphology, 130, 227–254. 10.1007/s00435-011-0138-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed information about the studies included in the historical review (Appendix S1), details of scans and specimens’ preparation (Appendix S2), updated list of manufacturers (Appendix S3), and a video of the scans used for Figure 6 (Video S4) can be found as online Supporting information. Representative scans (reconstructed files) of the techniques compared in this study are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.f9h43b0.


