All organisms confront intermittent periods of fasting during which energy intake is insufficient to meet energy demands. Consequently, mechanisms have evolved for synthesizing and storing energy nutrients during times of caloric sufficiency that can be used during periods of caloric limitation. In mammals, the predominant storage form of energy is triglycerides. Triglycerides can be synthesized from fatty acids obtained through the diet. Alternatively, fatty acids can be produced through de novo synthesis from the metabolic intermediate acetyl-CoA. The latter pathway is termed lipogenesis. Although all cells can synthesize fatty acids, production of triglycerides for energy storage occurs principally in the liver and adipose in mammals. Lipogenesis is regulated through the acute control of key enzyme activities by means of allosteric and covalent modifications (1). Moreover, the production of many glycolytic and lipogenic enzymes is regulated in response to dietary status (1–4). This latter response is thought to be adaptive and occurs in large part through transcriptional regulation of genes encoding lipogenic enzymes.
Glucose, in particular, is a vital energy nutrient in mammals and a major source of acetyl-CoA for triglyceride production. The pancreatic hormones insulin and glucagon play a central role in controlling glucose and fatty acid homeostasis. Insulin promotes glucose utilization and storage as glycogen and triglycerides. Glucagon opposes these actions and promotes glucose production and triglyceride catabolism. In addition to these hormonal factors, glucose has also been implicated as an independent signal for controlling the synthesis of lipogenic enzymes. The intracellular mechanism by which glucose can generate a signal to affect transcription of lipogenic enzyme genes is poorly understood (2–4). However, in this issue of PNAS, Kawaguchi and colleagues present a significant step forward in unraveling this pathway (5). These authors have identified a hepatic transcription factor, designated carbohydrate response element-binding protein or ChREBP (6). They now demonstrate that this factor is regulated at two levels—nuclear entry and DNA binding—through phosphorylation-dependent mechanisms in response to signals from glucose and cAMP.
Feeding a diet high in simple carbohydrates to rats or mice results in increased transcription of at least 15 genes involved in glucose uptake, glycolysis, and lipogenesis
Feeding a diet high in simple carbohydrates to rats or mice results in increased transcription of at least 15 genes involved in glucose uptake, glycolysis, and lipogenesis (2). This dietary regimen leads to secretion of insulin from the pancreatic beta cell, as well as increased glucose metabolism in the liver. Studies using cultured primary hepatocytes have demonstrated that both insulin and increased glucose metabolism are necessary for the induction of most of these genes. A pathway by which insulin can impact this process has recently come to light through the investigation of the transcription factor sterol regulatory element binding protein (SREBP). SREBP was discovered as a transcription factor that controls genes involved in cholesterol uptake and biosynthesis (7). Most recently, SREBP, and particularly the SREBP-1c isoform, has been shown to control expression of genes involved in the lipogenic pathway too (8). The phenotype of transgenic mice that overexpress SREBP in the liver dramatically illustrated this point (9). These mice exhibit liver steatosis and increased mRNA levels of most lipogenic genes. SREBP binding sites have been found in the promoters of several lipogenic enzyme genes (e.g., ref. 10). SREBP-1c itself is rapidly induced by treatment of hepatocytes with insulin, providing a pathway for insulin mediation of enhanced lipogenic gene transcription (11, 12). Consistent with these observations, mice with a deletion of the SREBP-1 gene have an impaired ability to fully respond to a high-carbohydrate diet (13). However, these mice do retain some significant response to the diet, raising the question of what factor(s) might be responsible for induction in the absence of SREBP-1. The ChREBP gene product identified by Uyeda's (5) laboratory is a strong candidate to fulfill this role.
Earlier studies on the glucose pathway of lipogenic enzyme gene induction focused on an examination of the L-type pyruvate kinase and S14 genes. The expression of these two genes is induced transcriptionally in primary hepatocytes by changes in glucose levels in the media (in the presence of a fixed-insulin concentration). Through promoter-mapping studies, a glucose- or carbohydrate-response element was identified in both of these genes (14–16). This site consists of two E box-related motifs separated by a defined distance (16). Most recently, similar elements have been found in the promoters of the acetyl-CoA carboxylase and fatty acid synthase genes, the two central enzymes of fatty acid biosynthesis (17, 18). The presence of the E box (5′-CANNTG) motif in these response elements suggests that a member of the basic/helix–loop–helix (bHLH) family of transcription factors is responsible for regulation. However, the nature of this factor has remained elusive for many years. In a recently published article in PNAS, the Uyeda group (6) purified a transcription factor from liver based on its ability to bind to the carbohydrate-response element of the pyruvate kinase promoter. This factor turned out to be a novel bHLH/leucine zipper protein, which they designated ChREBP.
In this issue of PNAS, Kawaguchi et al. (5) have proceeded to examine how ChREBP activity is controlled in hepatocytes. By using a green fluorescent protein (GFP) fusion of ChREBP, the authors demonstrate that ChREBP is localized in the cytosolic compartment at low (fasting) glucose levels, but in the nucleus in the presence of high glucose. Intriguingly, this cellular localization can be controlled by the state of phosphorylation of a specific serine residue (S196) of ChREBP. This serine residue is located adjacent to the nuclear localization signal for ChREBP and hence in a logical position to control its partitioning between cytosol and nucleus. When S196 is mutated to an alanine residue, ChREBP is found predominantly in the nucleus, regardless of glucose concentration. Conversely, when mutated to an aspartate (a molecular mimic of phosphorylated serine), ChREBP is localized exclusively in the cytosol. These observations led the authors to propose that phosphorylation of S196 results in ChREBP retention in the cytosol and hence its inactivation. S196 is located in a consensus recognition site for protein kinase A (PKA), implicating the cAMP-dependent enzyme in its phosphorylation. Furthermore, treatment of cells with an inhibitor of PKA results in enhanced nuclear localization of ChREBP. These findings are consistent with the role of glucagon, which acting through its intracellular mediator cAMP, is known to inhibit synthesis of lipogenic enzymes induced by insulin and glucose.
The authors also note that two additional PKA consensus phosphorylation sites are present in the ChREBP sequence at S626 and T666. These residues are found immediately amino-terminal to the DNA-binding domain. When a recombinant fragment of ChREBP containing this domain is treated in vitro with PKA, binding to the L-PK carbohydrate regulatory element is inhibited. Subsequent treatment of this phosphorylated polypeptide with protein phosphatase 2A (PP2A) reverses the effect. Hence, the authors propose a second level of regulation for ChREBP involving an inhibition of its DNA binding activity on phosphorylation by PKA. Consistent with this proposal, a double mutation of S196 to alanine (to promote nuclear localization) and T666 to alanine (to promote DNA binding) is not inhibited by glucagon treatment of hepatocytes.
As a whole, these studies outline a pathway by which glucagon/cAMP can repress ChREBP action and the glucose-stimulated pathway. How does increased glucose metabolism activate ChREBP? The simplest model is that glucose acts by stimulating the dephosphorylation of the critical PKA-catalyzed phosphorylation events. To support this model, the authors show that treatment of hepatocytes with cantharidic acid, a phosphatase inhibitor with highest affinity for PP2A, inhibits the nuclear localization of ChREBP normally observed in high glucose. PP2A is well recognized for its role in mediating a diversity of signaling pathways (19). In fact, this same laboratory has previously described a form of PP2A in liver that is activated by xylulose-5-phosphate, a metabolic intermediate which is increased in hepatocytes incubated in high-glucose conditions (20). It will be interesting to learn whether this PP2A isoform acts on phosphorylated ChREBP in hepatocytes.
With the discovery of ChREBP and its potential role in glucose regulation, a number of important questions are now amenable to study. Of great interest is the intracellular signaling pathway that is responsible for activation by glucose. Studies in hepatocytes have demonstrated that increased glucose metabolism is necessary for the generation of this signal. However, it has been difficult to identify the key intermediate or step in the various pathways of glucose metabolism responsible for control. With the report of the end target of the pathway, researchers should now be able to work backwards in elucidating the molecular mechanism. Another key question is whether ChREBP binds to the glucose response element by itself or in conjunction with other components. Many members of the bHLH family act as heterodimers with specific partners. In fact, a recent article on WBSCR14 (the human homologue of ChREBP) suggests that it does not bind as a homodimer to an E box element (21). Instead, it heterodimerizes with Mlx, a member of the c-myc family, to form a stable heterodimer. Also, the potential role that ChREBP may play in the pathophysiologies associated with disorders of metabolic homeostasis—type II diabetes, obesity, and hypertension—will be of interest to explore.
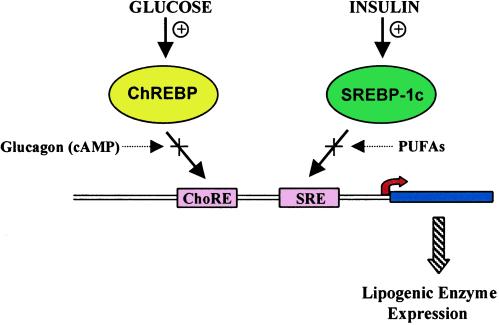
Together ChREBP and SREBP provide a pair of transcription factors that function coordinately to regulate lipogenesis (Fig. 1). Several of the lipogenic enzyme genes contain distinct binding sites for these two factors that function synergistically (18, 22). Why use two factors for control of this pathway? One consequence is that critical metabolic intermediates are not used for storage unless all conditions (insulin and glucose signals) are properly set. Another result is to provide a more integrated output for lipogenic enzyme production than would be possible by means of a single factor. A noteworthy observation in this regard is that the positive effects of insulin on SREBP can be overcome by polyunsaturated fatty acids, which are repressors of the lipogenic pathway (23, 24). Similarly, cAMP, which signals a catabolic state, functions to inhibit ChREBP. The ability of these factors to synergistically control transcription of lipogenic enzyme genes would hence result in an effective means of integrating multiple hormonal and nutritional inputs important in controlling energy status in mammals.
Figure 1.
Schematic of the roles of ChREBP and SREBP in regulation of lipogenic enzyme gene transcription. Most lipogenic enzyme genes (e.g., fatty acid synthase) have response elements for binding ChREBP (ChoRE) and SREBP (SRE). These two factors work synergistically to induce transcription of the lipogenic enzyme genes in the presence of glucose and insulin. Glucagon, through its intracellular mediator cAMP, and polyunsaturated fatty acids (PUFAs) act to inhibit the activity of ChREBP and SREBP, respectively. In this manner, the output of lipogenic enzyme gene production is integrated to multiple nutrient and hormonal signals.
Footnotes
See companion article on page 13710.
References
- 1.Hillgartner F B, Salati L M, Goodridge A G. Phys Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Towle H C, Kaytor E N, Shih H-M. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 3.Girard J, Ferre P, Foufelle F. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 4.Vaulont S, Vasseur-Cognet M, Kahn A. J Biol Chem. 2000;275:31555–31558. doi: 10.1074/jbc.R000016200. [DOI] [PubMed] [Google Scholar]
- 5.Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Proc Natl Acad Sci USA. 2001;98:13710–13715. doi: 10.1073/pnas.231370798. . (First Published November 6, 2001; 10.1073/pnas.231370798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamashita H, Takenoshita M, Sakurai M, Bruick R K, Henzel W J, Shillinglaw W, Arnot D, Uyeda K. Proc Natl Acad Sci USA. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. . (First Published July 24, 2001; 10.1073/pnas.10.1073/pnas.161284298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 8.Osborne T F. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 9.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magana M M, Osborne T F. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 11.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foretz M, Guichard C, Ferre P, Foufelle F. Proc Natl Acad Sci USA. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty A H, Osuga J-I, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, et al. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 14.Bergot M-O, Diaz-Guerra M-J M, Puzenat N, Raymondjean M, Kahn A. Nucleic Acids Res. 1992;20:1871–1878. doi: 10.1093/nar/20.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Thompson K S, Towle H C. J Biol Chem. 1993;268:12787–12795. [PubMed] [Google Scholar]
- 16.Shih H-M, Liu Z, Towle H C. J Biol Chem. 1995;270:21991–21997. doi: 10.1074/jbc.270.37.21991. [DOI] [PubMed] [Google Scholar]
- 17.O'Callaghan B L, Koo S-H, Wu Y, Freake H C, Towle H C. J Biol Chem. 2001;276:16033–16039. doi: 10.1074/jbc.M101557200. [DOI] [PubMed] [Google Scholar]
- 18.Rufo C, Teran-Garcia M, Makamura M, Koo S-H, Towle H C, Clarke S D. J Biol Chem. 2001;276:21969–21975. doi: 10.1074/jbc.M100461200. [DOI] [PubMed] [Google Scholar]
- 19.Janssens V, Goris J. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura M, Uyeda K. J Biol Chem. 1995;270:26341–26346. doi: 10.1074/jbc.270.44.26341. [DOI] [PubMed] [Google Scholar]
- 21.Cairo W, Merla G, Urbinati F, Ballabio A, Reymond A. Hum Mol Genet. 2001;10:617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- 22.Koo S-H, Dutcher A K, Towle H C. J Biol Chem. 2001;276:9437–9445. doi: 10.1074/jbc.M010029200. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Nakamura M T, Cho H P, Clarke S D. J Biol Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 24.Mater M K, Thelen A P, Pan D A, Jump D B. J Biol Chem. 1999;274:32725–32732. doi: 10.1074/jbc.274.46.32725. [DOI] [PubMed] [Google Scholar]