Abstract
Hepatocellular carcinoma (HCC) is a male-dominant cancer and androgen receptor (AR) has been linked to the pathogenesis of HCC. However, AR expression and its precise role in HCC remain controversial. Moreover, previous anti-androgen and -AR clinical trials in HCC failed to demonstrate clinical benefits. In this study, we found that AR is overexpressed in the nucleus of approximately 37% HCC tumors, which is significantly associated with advanced disease stage and poor survival. AR overexpression in HCC cells markedly alters AR-dependent transcriptome, stimulates oncogenic growth and determines therapeutic response to enzalutamide, a second generation of AR antagonist. However, AR inhibition evokes feedback activation of AKT-mTOR signaling, a central regulator for cell growth and survival. On the other hand, mTOR promotes nuclear AR protein expression by restraining ubiquitin-dependent AR degradation and enhancing AR nuclear localization, providing a mechanistic explanation for nuclear AR overexpression in HCC. Finally, co-targeting AR and mTOR shows significant synergistic anti-HCC activity and decreases tumor burden by inducing apoptosis in vivo. Conclusion: Nuclear AR overexpression is associated with the progression and prognosis of HCC. However, enzalutamide alone has limited therapeutic utility due to feedback activation of AKT-mTOR pathway. Moreover, mTOR drives nuclear AR overexpression. Co-targeting AR and mTOR is a promising therapeutic strategy for HCC.
Keywords: Hepatocellular carcinoma (HCC), mTOR, Signal transduction, Androgen receptor (AR), Liver, Enzalutamide, Rapamycin, Targeted therapy
Introduction
Hepatocellular carcinoma (HCC) is the predominant form of liver cancer (1). It is one of the most common malignancies worldwide and a leading cause of cancer-related death, with 782,500 new cases and 745,500 deaths worldwide in 2012 (2, 3). HCC is most prevalent in Eastern Asian and Sub-Saharan African countries due to chronic hepatitis B virus (HBV) infections (2, 3). Despite the downward trend in the overall cancer incident and cancer-related mortality in developed countries, HCC has seen a significant increase in both HCC incidence and death (4). The mortality rate for liver cancer nearly doubled in the USA from 1980 to 2014, which is in sharp contrast to the 20% decrease in overall cancer-related death during the same period (5). Conventional treatments for early HCCs include surgery, chemotherapy and liver transplantation. Surgery and liver transplantation can be curative for ~ 30% early stage patients (6). Unfortunately, most HCC cases are diagnosed at unresectable advanced stages. Sorafenib has been the only systemic drug for advanced HCC until the recent approval of regorafinib, another multi-kinase inhibitor. Both sorafenib and regorafinib have a low durable response rate and survival benefit, with a median increase in overall survival (OS) by only 2–3 months (7, 8). New systemic drugs and therapeutic strategies are urgently needed to improve the clinical outcome of HCC.
Androgen receptor (AR) is a member of the steroid hormone receptor superfamily. In response to stimulation by androgens such as testosterone, AR translocates into the nucleus where it binds to androgen response elements (AREs) and regulates cell growth and survival genes (9). AR plays a critical role in the development and progression of prostate cancer, a malignancy in the male reproductive system (9). HCC exhibits a significant gender preference, with a male to female ratio of 2:1 to 7:1 as previously surveyed (2). Accumulating evidences have linked AR the male-dominant trait of liver cancer (10, 11). AR promotes HBV viral RNA expression (12, 13), suggesting that it plays a role in the HBV-driven hepatocarcinogenesis. AR knockout in mice reduces the number and burden of carcinogen-induced liver tumors (14), indicating that AR is necessary for full cancer development in this animal model. Because prostate cancer is dependent on AR in all stages of the disease, AR antagonists have been developed for AR-targeted therapy. Bicalutamide is a first generation AR inhibitor. Enzalutamide is a second-generation AR inhibitor that was approved by FDA for prostate cancer in 2012 (15). It is structurally related to bicalutamide with improved affinity and potency, and inhibits certain bicalutamide-resistant AR mutants. Unfortunately, early clinical trials on anti-androgen and bicalutamide therapies in liver cancer met with disappointing results, producing no apparent clinical benefits (16, 17).
Mechanistic target of rapamycin (mTOR) protein is a key member of the PI3K-AKT-mTOR signaling pathway that is frequently hyper-activated in many human malignancies, driving oncogenic growth and proliferation (18). In response to mitogenic stimuli by growth factors and nutrients, mTOR promotes protein synthesis and ribosome biogenesis (19). mTOR is an established cancer therapeutic target: the macrolide rapamycin and rapamycin analogs everolimus and temsirolimus are FDA-approved drug for advanced breast, renal and neuroendocrine tumors. Hyperactive AKT-mTOR signaling is a common phenomenon in HCC (20, 21). Activation of AKT and mTOR is critical for hepatocarcinogenesis in various common HCC models in mice (22–25). However, recent clinical trial did not achieve desirable endpoints on the rapamycin analog everolimus in advanced HCC patients (26, 27). Therefore identifying new treatment strategies such as combinational therapy is necessary for improving the efficacy of rapamycin and rapamycin analogs in order to bring mTOR-targeted therapies into the clinic.
Newer AR inhibitors such as enzalutamide have improved specificity and potency, leading to renewed enthusiasm to explore the utility of anti-AR therapeutics in HCC. A human trial has recently initiated on enzalutamide without or with sorafenib for treatment of advanced HCC patients (NCT02642913). However, it remains resolved why bicalutamide failed earlier HCC clinical trials. In order to evaluate the clinical potential of AR-targeting and provide guidance to current and future trials, it will be necessary to fully understand the mechanism of AR signaling and anti-AR therapeutic response in HCC. To this end, we investigated the significance and mechanism of AR overexpression and AR-mTOR crosstalk in HCC, and their roles in AR-targeted therapy. Our results indicate that AR expression and feedback activation of AKT-mTOR signaling are major factors for enzalutamide treatment response, and that combination of AR and mTOR inhibitors is a promising therapeutic strategy for HCC.
Materials and Methods
Human HCC patients and tissue samples
142 clinical tissue samples of HCC were collected from HCC patients who underwent hepatic resection from January, 2004 to August, 2012 at the Sun Yat-Sen University Cancer Center. All the cases were diagnosed by pathological examination. The median follow-up is 48 months (4 to 111 months). None of the patients received chemotherapy or radiotherapy treatment before surgery. The demographic and clinicopathological parameters of HCC patients are listed in Table 1. This study was reviewed and approved by the Ethics Committees of Sun Yat-Sen University Cancer Center. Informed consents signed by the patients were collected. An HCC transcriptome dataset available from the Oncomine cancer microarray database (https://www.oncomine.org/) was used to analyze AR mRNA and AR target gene expression. This dataset contains mRNA expression information of 10,802 genes from 104 human primary HCC tumors and 76 normal liver tissues, of which 93 tumors are HBV+ (28). We analyzed the mRNA expression of AR and AR target genes using Student’s t-test. HCC proteomic data derived from 184 primary human HCC tumors (113 Caucasian Americans, 52 Asian Americans and 12 African Americans) were downloaded from The Cancer Proteome Atlas (http://tcpaportal.org/tcpa/) (29). Normalized phosphoprotein levels of AKT (S473) and phospho-p70S6K (T389) were used to represent AKT-mTOR signaling. The correlation between the level of AR protein and p-AKT(473) or p-p70S6K(389) was analyzed using Pearson correlation test as previously described (30).
Animal studies
Mouse procedures and protocols were approved by Institutional Animal Care and Use Committee of Sun Yat-Sen University. For xenograft tumor formation assay, male BALB/c athymic nude mice 4 weeks of age were injected with 2 × 106 MHCC-97L of tumor cells in 0.1 ml PBS subcutaneously into the flank of each mouse. When average tumor volume reached 300 mm3, tumor-bearing animals were randomized and treatment was initiated. 10 mg/kg enzalutamide was solubilized in 100 μl of CMC Na+1% / Tween-80 1% andadministered intragastrically daily. 10 mg/kg rapamycin was dissolved in 100 μl of 30% PEG300 and 5% Tween-80, and given by intraperitoneally daily for consecutive days, followed by 2 days without drug; Enzalutamide and rapamycin were freshly prepared daily just before administration. The control group was given the vehicle used for administration. Tumors were measured every 3 days and tumor sizes were calculated from the formula: Tumor volume =Length ×Width2× (π /6). After mice were euthanized, xenograft tumors were collected and fixed in paraffin block. The synergism of enzalutamide and rapamycin was calculated using the Chou-Talalay method with the Calcusyn software (Biosoft, Cambridge, UK). A combination index (CI) less than 1 indicates the drug combination has synergism. The results show that the CI value for the combination of the two drugs is 0.28, indicating that they have synergistic effect.
Statistical analysis
Statistical data analysis was performed by SPSS 11.0 software (SPSS Inc., USA) and Graph Pad Prism 5.0 for windows (Graph Pad Prism, Inc., San Diego, CA, USA). The experimental data were expressed as mean ± SD or SEM. Receiver operating characteristic (ROC) curve analysis was used to estimate the cutoff value for high and low AR expression in HCC patients. Kaplan-Meier plots and log-rank test were performed to compare cancer specific survival rates between patients with high and low AR expression. Univariate and multivariate survival analyses were performed using a Cox proportional hazards regression model. Repeated measures ANOVA was performed to analyze xenograft tumor size and proliferation of cultured cells. One-way ANOVA test and student’s t-test were conducted to evaluate the variables of different groups. Paired t-test was used to analyze xenograft tumor size and immunostaining scores of AR in paired clinical samples. A nonparametric Spearman correlation test was used to compare the correlation between AR expression and enzalutamide IC50 values. p < 0.05 was considered as statistical significance (indicated in corresponding figures with one asterisk for p < 0.05, and two asterisks for p < 0.01).
For additional materials and methods see Supporting Information
Results
AR is overexpressed in HCC, which is associated with disease progression and is an independent predictor of overall survival (OS)
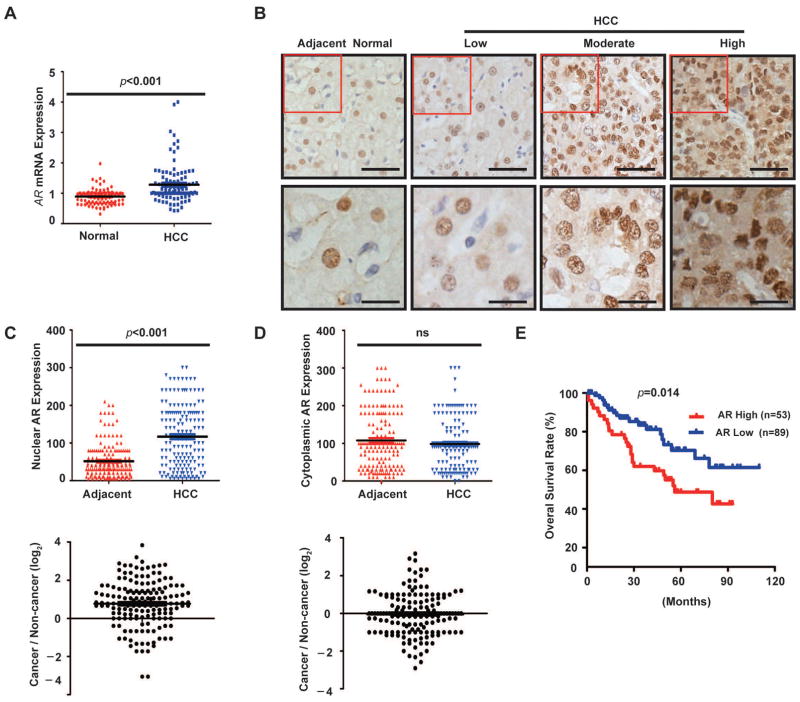
Although there were several studies on AR mRNA or protein expression in human HCCs, the results and clinical significance were contradictory (31, 32). Limiting factors include small sample sizes and non-paired tumor/normal tissues, and the use of radiolabeled ligands to detect AR in tissues. We analyzed an HCC gene expression dataset available in Oncomine from a previous transcriptome study of 104 primary HCC and 76 normal liver tissues (28), which shows that AR mRNA expression is highly variable in tumors, but not in normal liver tissues (Fig. 1A). Overall, AR mRNA level is higher in tumors. To verify this finding, we examined AR protein expression by immunohistochemistry (IHC) in a cohort of 142 paired human HCCs and adjacent non-cancerous liver tissues. AR is expressed in both normal hepatocytes and tumor cells with predominant nuclear localization (Fig. 1B), indicating that AR is largely in the activated form. Nuclear AR protein is significantly overexpressed in tumors compared with the adjacent non-cancerous liver tissues (p < 0.001) (Fig. 1C), with 37% (53/142) cases with 2-fold or higher nuclear AR in tumors (Fig. 1C, lower panel). In contrast, there is no significant difference in cytoplasmic AR expression between tumor and adjacent liver tissues (Fig. 1D).
Fig. 1. Nuclear AR is overexpressed in HCC, which is associated with tumor progression and poor prognosis.
(A) AR mRNA is overexpressed in a subset of HCC. AR mRNA expression data is downloaded from an OncoMine microarray dataset that includes 104 HCC and 76 normal liver tissues. The results were analyzed by student’s t-test (bar represents mean value).
(B) Representative AR immunohistochemistry (IHC) staining in primary HCC and adjacent noncancerous liver tissues (scale bar 50 μm).
(C) Scatter plots of nuclear AR IHC scores in HCC and adjacent noncancerous liver tissues. Bar represents mean value (N = 142, paired t-test). The lower panel shows the ratio of nuclear AR in paired tumor/adjacent noncancerous tissues.
(D) Scatter plot of cytoplasmic AR IHC scores in HCC and adjacent noncancerous liver tissues. Bar represents mean value (N = 142, sample-paired t-test). The lower panel shows the ratio of cytoplasmic AR in paired tumor/adjacent noncancerous tissues.
(E) Kaplan-Meier analysis of overall survival in patients with high and low nuclear AR expression.
We next examined the relationship between nuclear AR protein expression and clinicopathologic parameters. Advanced TMN stage, multiple tumor nodules and high HBV viral copies are positively correlated with high nuclear AR expression (Table 1), indicating that AR is associated with HCC disease progression. There is no statistically significant correlation between AR protein expression and age, gender, tumor size and grade, HBsAg and AFP (Table 1). Kaplan-Meier analysis demonstrates that high nuclear AR staining is significantly correlated with poorer overall survival (OS) of HCC patients (p < 0.001) (Fig. 1E). Furthermore, multivariate analysis shows that nuclear AR overexpression in tumors is an independent predictor for poor OS (HR= 2.423, 95% CI = 1.053–5.577, P = 0.037) (Table S1). These results indicate that nuclear AR protein is highly expressed in a subset of HCC tumors characteristic of advanced disease stage and poor prognosis.
AR overexpression alters AR-dependent transcriptome in HCC
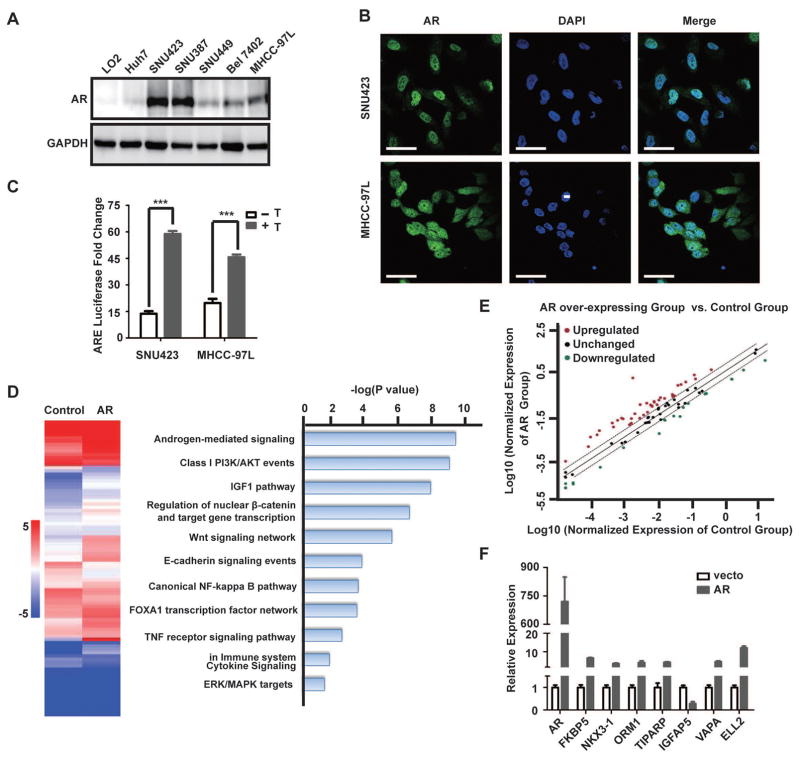
We also examined AR protein expression in a panel of immortalized hepatocyte and HCC cell lines. Compared with the immortalized LO2 hepatocyte cell line, AR is significantly higher in SNU423 and SNU387 cells and moderately higher in BEL7402 and MHCC-97L cells (Fig. 2A). Immunofluorescence (IF) staining indicates that AR is primarily localized in the nucleus of these HCC cells (Fig. 2B). These results indicate that HCC cell lines share the same heterogeneity in AR expression as primary liver tumors, and that they are useful in vitro models for studying AR functions. AR is a transcription factor that regulates expression of genes with diverse functions, especially those related to cell growth in prostate cancer (33, 34). Therefore, we investigated the transcriptional function of AR in HCC using a luciferase reporter driven by an androgen-responsive element (ARE). Treatment of SNU423 and MHCC-97L cells with testosterone stimulates AR reporter activity in these AR-positive HCC cells (Fig. 2C), demonstrating that AR regulates target gene expression in HCC cells.
Fig. 2. AR overexpression promotes AR-dependent transcriptome in HCC cells.
(A) AR protein expression in a panel of immortalized liver and HCC cell lines as determined by immunoblot. GAPDH serves as a loading control (top panel).
(B) AR is predominantly localized in the nucleus of SNU423 and MHCC-97L cells. Shown is immunofluorescent (IF) staining of AR in SNU423 and MHCC-97L cells. The nuclei were counterstained by DAPI. Scar bar, 50 μm.
(C) Luciferase reporter driven by androgen response element (ARE) was measured in the presence or absence of testosterone for 24 h in SNU423 and MHCC-97L cell lines. Data (mean ± SD, n = 3) were analyzed by unpaired two-tail t test; *** p < 0.001.
(D) AR overexpression significantly alters AR-dependent transcriptome in HCC cells. SNU449 cells were infected with a lentiviral AR-Flag or a control lentivirus, and analyzed for the expression of key AR target genes using the Human Androgen Receptor Signaling Targets PCR Array. Left panel shows a heat map of key AR target genes in control and AR overexpressing cells. Right panel shows biological pathways enrichment in differentially expressed key AR target genes. Data were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID).
(E) Scatter plot identifies differentially expressed AR target genes due to AR overexpression. Red and green dots indicate with significantly increased or decreased AR target genes, respectively, in AR overexpressing cells versus control cells.
(F) qRT-PCR analysis of eight randomly selected AR target genes to verify results from the Human Androgen Receptor Signaling Targets Array. GAPDH was used as an internal control. Data (mean ± SD, n = 3) were analyzed by Student’s T test; * p < 0.05.
To assess the transcriptional function of AR in HCC, we overexpressed AR in SNU449 cells (Fig. S1), a cell line with relatively low AR level. The global transcriptional effect of AR overexpression was examined using the Human Androgen Receptor Signaling Targets PCR Array that consists of 84 key AR target genes (Qiagen). AR overexpression leads to up-regulation of 38 AR target genes and down-regulation of 18 AR target genes by > 2-fold (Fig. 2D and 2E), representing 67% key AR target genes. Analysis of 8 randomly selected AR target genes by qRT-PCR verified the AR target gene PCR array results (Fig. 2F). Significantly altered AR target genes are enriched in Androgen, PI3K, IGF1 and Wnt and β-catenin signaling (Fig. 2D), pathways that are well known for their crucial role in liver cancer cell proliferation and survival, which is consistent with a tumor-promoting role of AR overexpression in HCC. In the same HCC transcriptome dataset (28), the expression of AR target genes is significantly altered. For example, APPBP2, Rab4A, SMS, ZNF189, SLC26A2 and TPD52, which were up-regulated by AR overexpression (Table S2), are significantly overexpressed in primary HCC samples compared with the normal liver tissues (Fig. S2). Similarly, VIPR1, MME and TMPRSS2, which were down-regulated by AR (Table S2), are under-expressed in primary HCC tumor samples compared with the normal liver tissues (Fig. S2). Together, these results support the notion that AR pathway is activated in human primary HCCs.
AR overexpression renders AR-dependent and enzalutamide-sensitive growth of HCC cells
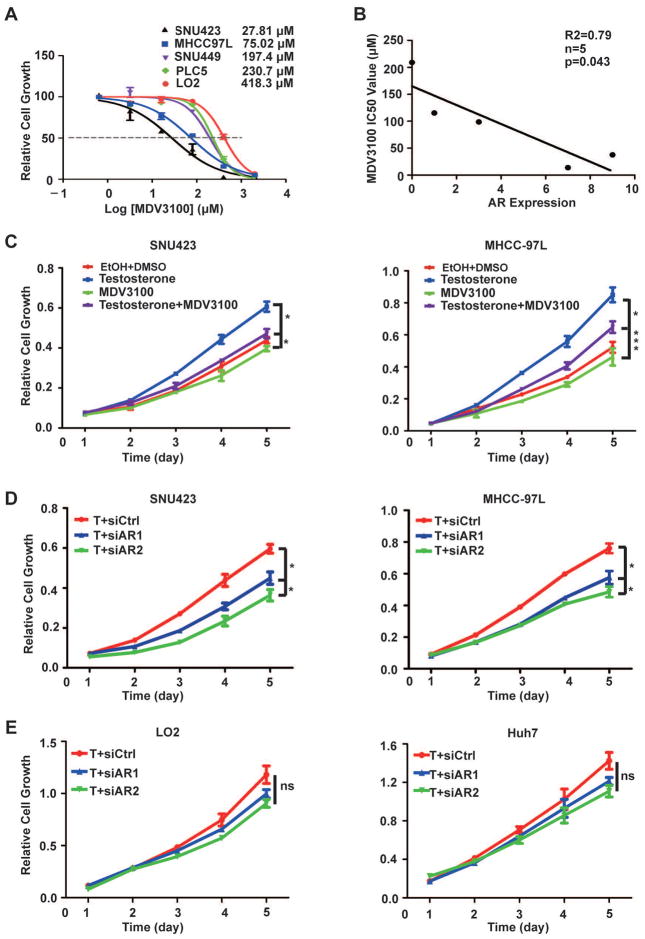
To investigate the role of AR in HCC growth, we treated a panel of immortalized liver and HCC cell lines with enzalutamide (MDV3100). The IC50 for enzalutamide is highly variable, ranging from 27.8 to 418.3 μM (Fig. 3A). There is a striking inverse relationship between AR protein expression and enzalutamide IC50 (R = 0.79, p < 0.043) (Fig. 3B), indicating that the growth of HCC cells with high AR expression is more AR-dependent and hence more sensitive to AR blockage. Consistently, enzalutamide blocks testosterone stimulated growth of SNU423 and MHCC97L cells (Fig. 3C). We further tested the effect of AR knockdown by siRNA (siAR). Four siARs were tested and found to produce significant AR knockdown (Fig. S1B). Two siARs were selected for further studies, resulting in more growth inhibition in the high AR-expressing SNU423 and MHCC97L cells (Fig. 3D) than the low AR-expressing LO2 and Huh7 cells (Fig. 3E). These results suggest that AR overexpression renders AR-dependent and enzalutamide-sensitive growth in HCC cells.
Fig. 3. AR overexpression renders AR-dependent growth and enzalutamide sensitivity in HCC cells.
(A) SNU423, MHCC97L, SNU449, PLC5 and LO2 cells were treated with different concentrations of enzalutamide (MDV3100) treatment for 4 days. Shown is the dose response curve. Data represent mean ± SD (n = 4).
(B) Correlation analysis of AR expression and enzalutamide (MDV3100) IC50 for SNU423, MHCC97L, SNU449, PLC5 and LO2 cells. Correlation was evaluated by a nonparametric Spearman test. The number of cell lines (n), coefficient of correlation (r), and p value (p) are as indicated.
(C) Enzalutamide attenuates testosterone-stimulated growth of AR-positive HCC cells. SNU423 and MHCC97L cells were treated with testosterone in the presence or absence of enzalutamide (MDV3100) for 5 days. Cell growth was measured daily by the CCK8 assay. Data were analyzed by Repeated measures ANOVA (mean ± SD, n = 4, * p < 0.05).
(D) AR knockdown inhibits testosterone-stimulated growth of SNU423 and MHCC97L cells. SNU423 and MHCC-97L cells were transfected with AR-specific siRNA (siAR) or a control siRNA (siCtrl) in the presence of testosterone. Cell growth was measured by CCK8 assay. Data were analyzed by Repeated measures ANOVA (mean ± SD, n = 4, * p < 0.05).
(E) AR knockdown expression has little effect on the growth of LO2 and Huh7 cells. LO2 and Huh7 cells were transfected with siAR or siCtrl in the presence of testosterone. Cell growth was measured by CCK8 assay. Data were analyzed by Repeated measures ANOVA (mean ± SD, n = 4, * ns, not significant).
Inhibition of AR leads to feedback activation of AKT-mTOR signaling through FKBP5
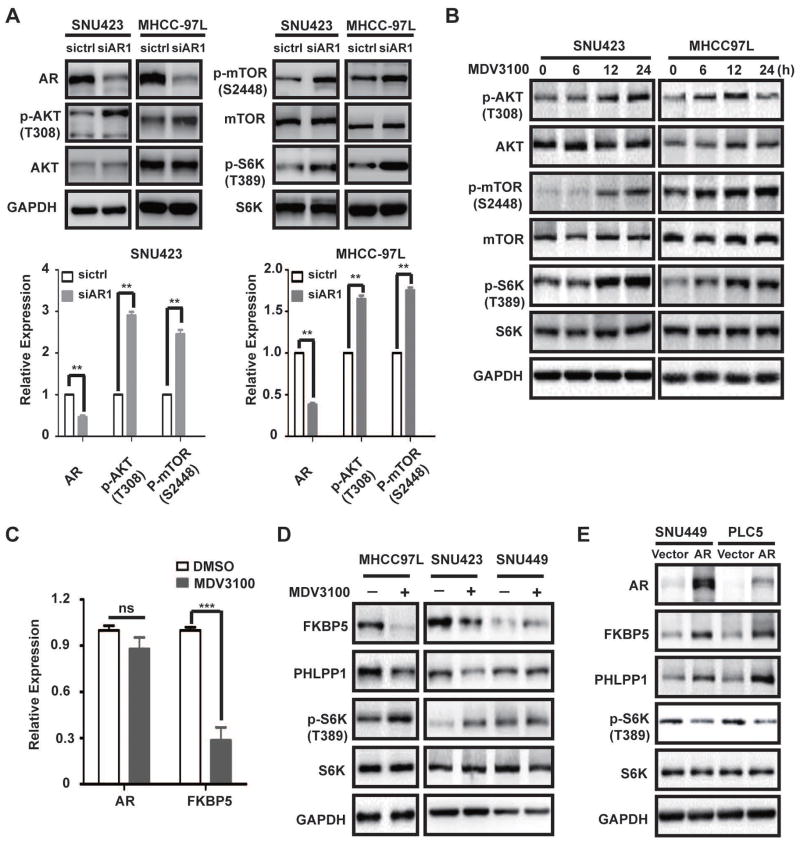
The fact that AR blockage only produces a modest growth inhibition raises an interesting possibility of a feedback mechanism that limits the impact of AR loss. Indeed, AR knockdown in SNU423 and MHCC-97L cells leads to elevated phosphorylation of AKT, mTOR and S6K1, indicating that AKT-mTOR pathway is activated (Fig. 4A and S3A). Similarly, pharmacological inhibition of AR by enzalutamide treatment also results in activation of AKT-mTOR signaling in SNU423 and MHCC97L cells (Fig. 4B and S3B). Moreover, AR inhibition by siRNA-mediated knockdown or enzalutamide results in increased phosphorylation of GSK3, a known AKT substrate (35). Thus blockage of AR activity triggers feedback activation of AKT-mTOR signaling. In contrast, AR inhibition has little effect on c-MYC expression or ERK phosphorylation (Fig. S3), suggesting that blockage of AR pathway selectively activates AKT-mTOR signaling in HCC.
Fig. 4. Inhibition of AR leads to feedback activation of AKT-mTOR signaling through FKBP5 in HCC cells.
(A) AR knockdown leads to activation of AKT-mTOR signaling in high AR-expressing cells. AR is knocked down in SNU423 and MHCC-97L cells. The effect on AKT-mTOR signaling was examined by immunoblot analysis of phosphorylation of AKT, mTOR and S6K. GAPDH was used as a loading control. Lower panels, quantification of the results (Mean ± SD, ** p < 0.01).
(B) Enzalutamide treatment leads to activation of AKT-mTOR signaling in high AR-expressing cells. SNU423 and MHCC-97L cells were treated with enzalutamide (MDV3100) for different times and examined for PI3K-AKT-mTOR signaling by immunoblot.
(C) Enzalutamide inhibits the expression of FKBP5 mRNA. SNU423 cell was treated with enzalutamide (MDV3100) for 48 h. The expression of FKBP5 was analyzed by qRT-PCR. Data represent mean ± SD in triplicates and were analyzed using Student T-test, *** p < 0.001.
(D) AR inhibition leads to down-regulation of FKBP5 and PHLPP1 proteins in high AR-expressing cells, not low AR-expressing cells. SNU423, MHCC-97L and SNU449 cells were treated with enzalutamide (MDV3100) for 48 h. The effect on mTOR signaling, and the expression of FKBP5 and PHLPP1 was analyzed by immunoblot. GAPDH was used as a loading control.
(E) AR overexpression promotes FKBP5 expression and attenuates AKT-mTOR signaling. AR was transiently overexpressed in SNU449 and PLC5 cells. The effect on mTOR signaling and the expression of FKBP5 and PHLPP1 was analyzed by immunoblot.
However, it takes 6–12 hours for AKT-mTOR signaling to become activated by enzalutamide (Fig. 4B), suggesting that the feedback mechanism involves a transcriptional mechanism. FKBP5 is an AR target gene in prostate cancer that is known to inhibit AKT activity by promoting AKT dephosphorylation through scaffolding AKT with PHLPP, an AKT phosphatase (36–38). Interestingly, AR also promotes FKBP5 expression in HCC cells (Fig. 2F). Enzalutamide causes down-regulation of FKBP5 mRNA and protein expression in MHCC-97L and SNU423 cells (Fig. 4C and 4D). Curiously, the level of PHLPP1 is also decreased by enzalutamide (Fig. 4D), though the mechanism is currently not understood. In contrast, enzalutamide does not affect the expression of FKBP5 and PHLPP1 in the low AR-expressing SNU449 cell line (Fig. 4D). Moreover, ectopic AR expression in SNU449 and PLC5 cell lines stimulates FKBP5 and PHLPP1 expression, with concurrent suppression of AKT/mTOR signaling (Fig. 4E). These observations demonstrate that blockage of AR leads to activation of AKT-mTOR signaling, which is at least in part due to down-regulation of the AR target gene FKBP5 and AKT phosphatase PHLPP1.
mTOR promotes AR transcriptional activity by enhancing AR stability and nuclear localization
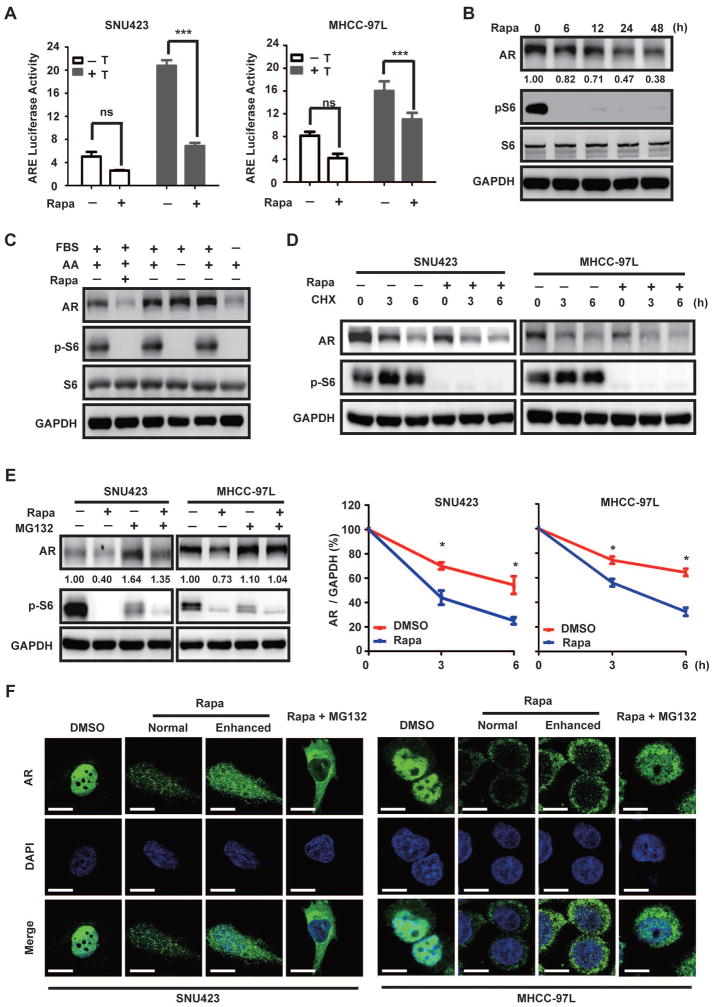
Inhibition of mTOR by rapamycin is known to stimulate AR transcriptional activity in prostate cancer (39). Surprisingly, however, we found that rapamycin treatment in MHCC-97L and SNU423 cells leads to marked inhibition of AR transcriptional activity under both basal and testosterone-stimulated conditions (Fig. 5A). Rapamycin treatment significantly decreases AR protein level (Fig. 5B). mTOR is regulated by both growth factors and amino acids. However, starvation from fetal bovine serum (FBS), not amino acids, causes AR down-regulation (Fig. 5C), indicating that regulation of AR by mTOR responds specifically to growth factor, not nutrient signaling. Under the condition when new protein synthesis is blocked by cycloheximide (CHX), AR is more rapidly degraded in rapamycin-treated cells than control cells (Fig. 5D), demonstrating that rapamycin accelerates AR degradation. Moreover, treatment with the proteasome inhibitor MG132 abrogates rapamycin-induced AR degradation (Fig. 5E). Thus, mTOR negatively regulates the proteasome-dependent AR degradation in HCC. We have also examined AR localization by immunofluorescence (IF) staining. As shown earlier, AR is predominantly localized in the nucleus in SNU423 and MHCC-97L cells. Consistent with proteasome-dependent degradation of AR, rapamycin treatment leads to a marked decrease in AR staining (Fig. 5F). Interestingly, AR becomes predominantly cytoplasmic in rapamycin-treated HCC cells (Fig. 5F). Rapamycin also inhibits AR nuclear localization in the presence of MG132. These results demonstrate that mTOR positively regulates both AR stability and nuclear localization.
Fig. 5. mTOR promotes AR transcriptional activity by enhancing AR stability and nuclear localization.
(A) Rapamycin suppresses AR transcriptional activity in HCC. The activity of ARE luciferase reporter was assayed in the absence or presence of rapamycin without or with testosterone for 24 h in SNU423 and MHCC9-7L cells. Luciferase activity is expressed relative to the Renilla control. Data (mean ± SD, n = 3) were analyzed by Student’s T test; *** p < 0.001.
(B) Rapamycin decreases AR protein level. SNU423 cells were treated with rapamycin for different times and measured for AR protein by immunoblot. Numbers represent relative quantification of AR protein (representative of three independent experiments, arbitrary unit).
(C) AR protein level is regulated by growth factors, not amino acids. SNU423 cells were starved from serum or amino acids for 24 h and analyzed for AR protein level by immunoblot.
(D) Rapamycin accelerates AR protein turnover. SNU423 and MHCC-97L cells were treated with or without rapamycin in the presence of cycloheximide (CHX). The ratio of AR/GAPDH is used to calculate AR stability. Lower panel shows quantification of the results. Data (mean ± SD, n = 4) were analyzed by Student’s T test; *p < 0.05.
(E) Rapamycin induces proteasome-dependent AR degradation. SNU423 and MHCC-97L cells were treated without or with rapamycin in the presence of the proteasome inhibitor MG132. The numbers show the relative AR protein amount representative of three independent experiments.
(F) Rapamycin inhibits AR nuclear localization in HCC cells. SNU423 and MHCC-97L cells were treated without or with rapamycin for 24 h. AR localization was analyzed by IF and the nuclei were counterstained by DAPI. Scar bar 10 μm. Normal, normal exposure; Enhanced, images were enhanced to show details.
Co-targeting AR and mTOR shows synergistic action against HCC and reduces tumor burden by inducing apoptosis in vivo
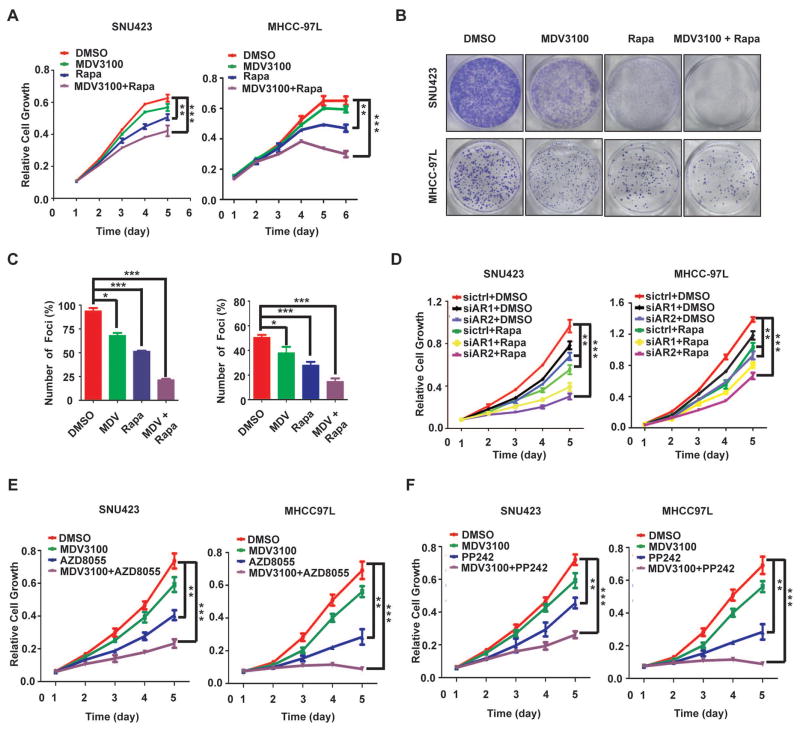
AR inhibition by enzalutamide or siRNA displays modest anticancer activity in HCC cells (Fig. 6A–D), which is at least in part due to the feedback activation of AKT-mTOR signaling (Fig. 4). We therefore explored the utility of combinating AR and mTOR inhibitors. Enzalutamide and rapamycin together indeed produce more potent inhibition of HCC cell growth and proliferation than each drug individually (Fig. 6A–C and S4). Moreover, the drug combination leads to a marked increase in apoptotic cell death (Fig. S5). Similar effect was observed with AR knockdown in the presence of rapamycin (Fig. 6D). mTOR kinase inhibitors target mTOR kinase domain and inhibit both mTORC1 and mTORC2 (40). They exhibit excellent anticancer activity in preclinical models and are currently under clinical trials for various malignancies (40). We hence tested two mTOR kinase inhibitors, AZD8055 and PP242. Each agent significantly improves the anticancer activity of enzalutamide (Fig. 6E–F). These results indicate that combination of enzalutamide with rapamycin or mTOR kinase inhibitor yields more potent anti-HCC activity in vitro than each drug alone.
Fig. 6. Co-targeting AR and mTOR improves anti-HCC activity in vitro.
(A) Enzalutamide and rapamycin inhibit HCC cell proliferation. SNU423 and MHCC-97L cells were treated with drug carrier (DMSO), enzalutamide (MDV3100), rapamycin, or the combination of two drugs for different times, and measured for cell proliferation by the CCK8. Data (mean ± SD, n = 4) were analyzed by Repeated measures ANOVA; *** p < 0.001.
(B) Enzalutamide and rapamycin inhibit oncogenic growth of HCC cells. Foci formation assay was performed with SNU423 and MHCC-97L cells treated with DMSO, enzalutamide (MDV3100), rapamycin, or their combination.
(C) Quantification of the above results. Data (mean ± SD, n = 4) were analyzed by Repeated measures ANOVA; ***p < 0.001.
(D) Same as Fig. 6A except AR siRNA was used. Data (mean ± SD, n = 4) were analyzed by Repeated measures ANOVA; *** p < 0.001.
(E) Enzalutamide and AZD8055 display strong anti-HCC activity. SNU423 and MHCC-97L cells were treated with DMSO, enzalutamide (MDV3100), AZD8055, or the combination of two drugs for different times, and measured for cell proliferation by the CCK8. Data (mean ± SD, n = 4) were analyzed by Repeated measures ANOVA; *** p < 0.001.
(F) Same as Fig. 6E except PP242 was used. Data (mean ± SD, n = 4) were analyzed by Repeated measures ANOVA; *** p < 0.001.
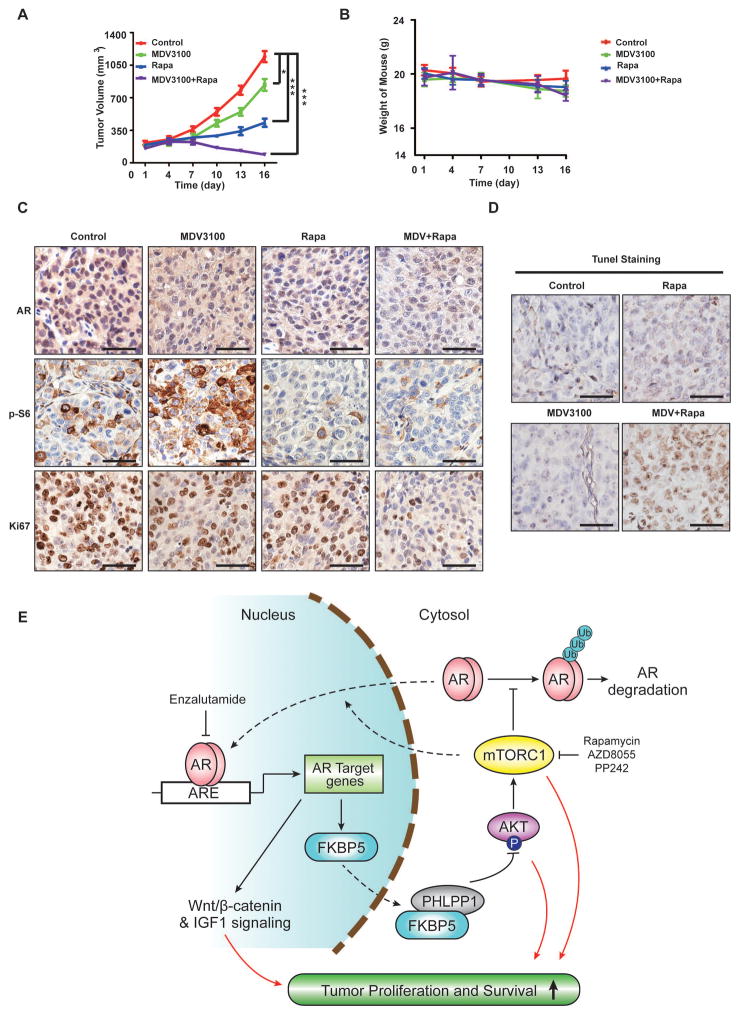
We next assessed the in vivo antitumor activity of enzalutamide alone or in combination with rapamycin in xenograft tumors derived from MHCC-97L cells. Enzalutamide alone has statistically significant, albeit modest antitumor activity, while rapamycin alone is more potent, as measured by slower increase in tumor burden (Fig. 7A). Remarkably, combination of enzalutamide and rapamycin not only blunts tumor growth, but also significantly reduces tumor burden (Fig. 7A). No significant weight loss occurs in different treatment groups (Fig. 7B), indicating that each drug and their combination are well tolerated. As observed in vitro, rapamycin reduces AR protein expression, while enzalutamide causes mTOR activation as indicated by increased p-S6 staining (Fig. 7C). The drug combination much more potently inhibits tumor cell proliferation as judged by Ki-67 staining (Fig. 7C and S6A). Consistent with the tumor burden reduction, the drug combination, not each drug individually, triggers elevated apoptosis as judged by positive TUNEL staining (Fig. 7D and S6B). These results indicate that the combination of enzalutamide and rapamycin generates cytotoxic antitumor activity against HCC, which is much more robust in vivo than in vitro. A combination index (CI) of enzalutamide and rapamycin as calculated by the Chou-Talalay method using the Calcusyn software (Biosoft, Cambridge, UK) yielded a value of 0.28. A CI value of less than, equal to and greater than 1 indicates that two drugs are synergistic, additive and antagonistic, respectively. Our result demonstrates that enzalutamide and rapamycin have strong synergism against HCC.
Fig. 7. Co-targeting AR and mTOR produces robust anti-HCC activity in vivo.
(A) Combination of enzalutamide and rapamycin significantly reduces HCC tumor burden. Mice bearing MHCC-97L xenograft tumors were treated with a drug carrier, enzalutamide (MDV3100), rapamycin or their combination. Tumor volume was measured at different times. Data (mean ± SD, n = 10) were analyzed by Repeated measures ANOVA; *** p < 0.001.
(B) Targeting AR and mTOR is well tolerated in mice. Mouse weight was measured every 3 day in the above experiment. Data (mean ± SD, n = 10) were analyzed by Student’s T test; *** p < 0.001.
(C) Xenograft tumors from the above experiment were analyzed for AR, p-S6 and Ki67 by IHC staining. Shown are representative IHC stained tumor sections. Scar bar, 50 μm.
(D) Co-targeting AR and mTOR triggers strong apoptotic cell death in HCC tumors. HCC xenograft tumors in different treatment groups were examined for apoptotic cell death by TUNEL staining. Scar bar, 50 μm. Right panel shows quantification of the TUNEL staining. Data (mean ± SD, n = 5) were analyzed by Student’s T test; *** p < 0.001.
(E) A working model shows the crosstalk between mTOR and AR pathways in the pathogenesis and therapy of HCC.
Discussion
HCC is a male-dominant malignancy and AR has been implicated to play a central role in this gender preference. AR regulates the transcription of HBV RNAs (12, 13) and is necessary for full tumor development in carcinogen-induced HCC in mice (14). Previous studies on AR expression in HCC were controversial, which is likely to be due to the small sample sizes (32, 41, 42). Analysis of available microarray data revealed that AR mRNA is overexpressed in a subset of tumors. Consistently, in a cohort of 142 paired HCC tumors and adjacent non-cancerous liver tissues, nuclear AR protein is significantly overexpressed in approximately one third of the HCC tumors. Importantly, AR overexpression is strongly correlated with advanced tumor stages and poor survival. AR regulates a large panel of genes important for cellular growth, proliferation and survival in prostate cancer (14). Here we show that AR overexpression significantly alters 67% of the key AR target genes in HCC cells, and promotes oncogenic growth and proliferation. Similarly, the expression of AR target genes is also considerably altered in primary human HCC tissues (Fig. S2). Together with the previous studies in AR knockout mouse models, these observations indicate that AR also has a broad oncogenic role in HCC.
We found that mTOR positively regulates AR transcriptional activity in HCC. This is in contrast to prostate cancer in which mTOR inhibits AR (39). It will be an interesting future direction to investigate how mTOR-AR crosstalk is differentially wired in the two different cancer types. mTOR promotes AR function through at least two distinct mechanisms: mTOR enhances the stability of AR protein by antagonizing proteasome-dependent AR degradation and AR nuclear localization. mTOR is commonly activated in human HCC (20, 21). This is predicted to promote elevated nuclear AR protein level as a result of stabilized AR protein and AR nuclear localization. Moreover, AR positively regulates its own expression (43), mTOR-dependent increase in AR protein stability and nuclear localization will further enhance AR level. Indeed, analysis of a TCGA Proteomic Dataset indicates a positive correlation between AKT-mTOR signaling and AR protein expression in a cohort of 184 human primary HCC tumors (Fig. S7). The frequent activation mTOR signaling provides a plausible molecular mechanism for nuclear AR overexpression in HCC.
Given the male-dominant nature of HCC and clinical success of castration and AR inhibitors in prostate cancer, anti-androgen and -AR as a therapeutic strategy were explored in HCC. However, the results from these clinical trials have been disappointing (16, 44). We found that only a third of HCC tumors significantly express AR and our evidence suggests that only these tumors respond to AR inhibition. Therefore, lack of biomarker guided patient selection is likely an explanation for the previous poor trial results. Moreover, AR inhibition leads to feedback activation of AKT-mTOR signaling, a mechanism also seen in prostate cancer (36, 37, 45), which should also limit the effectiveness of anti-AR therapy. We found that FKBP5 is an AR target gene in HCC and inhibition of AR leads to down-regulation of FKBP5 expression, which leads to feedback activation of AKT-mTOR signaling.
Inhibition of AR leads to feedback activation of AKT-mTOR signaling. Because mTOR is an oncogenic driver and mTOR positively regulates AR, we explored the utility of co-targeting AR and mTOR as a therapeutic strategy in preclinical HCC models. Indeed, enalutamide and rapamycin together yielded stronger anti-HCC activity than each drug alone in vitro and in vivo. Interestingly, the combination of enzalutamide and rapamycin exhibits more potent antitumor activity in the xenograft tumor model than cultured cancer cells, causing elevated apoptotic cell death and tumor regression, a phenomenon not seen in vitro. This observation suggests that AR and mTOR pathways together are essential for HCC cell survival under in vivo tumor microenvironment. Both enzalutamide and rapamycin are FDA-approved oncology drugs, and their combination is well tolerated in mice. This drug combination has the potential to improve HCC therapy for this difficult-to-treat cancer.
Supplementary Material
Acknowledgments
We thank Dr. Bin He for providing AR expressing lentiviral plasmid, Dr. Xianzi Yang for assistance with IHC and illustration.
Funding: This work was supported by the National Institutes of Health (R01CA173519), the National Natural Science Foundation of China (No: 81572440, 81372564, 81730081 and 81372600), the Recruitment Program of Global Experts, the Leading Talent of Guangdong Province, the Natural Science Foundation of Guangdong Province for Distinguished Young Scholar (No: 2015A030306047) and the Research Fund of State Key Laboratory of Oncology in South China.
Footnotes
Competing Interests: None declared.
Patient Consent: Obtained.
Contributions: XFZ conceived the overall project; HZ, XFZ, HYW, XXL and YJZ designed the studies; XFZ, HYW and XXL guided the studies; HZ and YJZ performed the experiments; YY provided supporting work; XFZ and HYW provided laboratory infrastructures; HZ and XFZ wrote the manuscript; HZ prepared the figures and tables; HYW and XXL proofread the manuscript.
References
- 1.Marquardt JU, Thorgeirsson SS. SnapShot: Hepatocellular carcinoma. Cancer Cell. 2014;25:550, e551. doi: 10.1016/j.ccr.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 5.Mokdad AH, Dwyer-Lindgren L, Fitzmaurice C, et al. Trends and patterns of disparities in cancer mortality among us counties, 1980–2014. JAMA. 2017;317:388–406. doi: 10.1001/jama.2016.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verslype C, Van Cutsem E, Dicato M, Arber N, Berlin JD, Cunningham D, De Gramont A, et al. The management of hepatocellular carcinoma. Ann Oncol; Current expert opinion and recommendations derived from the 10th World Congress on Gastrointestinal Cancer; Barcelona. 2008; 2009. pp. vii1–vii6. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 9.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 10.Ma W-L, Lai H-C, Yeh S, Cai X, Chang C. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocrine-Related Cancer. 2014;21:R165–R182. doi: 10.1530/ERC-13-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S-H, Chen P-J, Yeh S-H. Gender disparity in chronic hepatitis B: Mechanisms of sex hormones. Journal of Gastroenterology and Hepatology. 2015;30:1237–1245. doi: 10.1111/jgh.12934. [DOI] [PubMed] [Google Scholar]
- 12.Wang SH, Yeh SH, Lin WH, Wang HY, Chen DS, Chen PJ. Identification of androgen response elements in the enhancer I of hepatitis B virus: a mechanism for sex disparity in chronic hepatitis B. Hepatology. 2009;50:1392–1402. doi: 10.1002/hep.23163. [DOI] [PubMed] [Google Scholar]
- 13.Wu MH, Ma WL, Hsu CL, Chen YL, Ou JH, Ryan CK, Hung YC, et al. Androgen receptor promotes hepatitis B virus-induced hepatocarcinogenesis through modulation of hepatitis B virus RNA transcription. Sci Transl Med. 2010;2:32ra35. doi: 10.1126/scitranslmed.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma WL, Hsu CL, Wu MH, Wu CT, Wu CC, Lai JJ, Jou YS, et al. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology. 2008;135:947–955. 955.e941–945. doi: 10.1053/j.gastro.2008.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 16.Groupe d’Etude et de Traitement du Carcinome H. Randomized trial of leuprorelin and flutamide in male patients with hepatocellular carcinoma treated with tamoxifen. Hepatology. 2004;40:1361–1369. doi: 10.1002/hep.20474. [DOI] [PubMed] [Google Scholar]
- 17.Manesis EK, Giannoulis G, Zoumboulis P, Vafiadou I, Hadziyannis SJ. Treatment of hepatocellular carcinoma with combined suppression and inhibition of sex hormones: a randomized, controlled trial. Hepatology. 1995;21:1535–1542. [PubMed] [Google Scholar]
- 18.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 19.Tsang C, Qi H, Liu L, Zheng X. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112–124. doi: 10.1016/j.drudis.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Tsang CK, Wang S, Li XX, Yang Y, Fu L, Huang W, et al. MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology. 2016;63:1928–1942. doi: 10.1002/hep.28507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matter M, Decaens T, Andersen J, Thorgeirsson S. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. Journal of Hepatology. 2014;60:855–865. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho C, Wang C, Mattu S, Destefanis G, Ladu S, Delogu S, Armbruster J, et al. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology. 2012;55:833–845. doi: 10.1002/hep.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Pilo GM, Li X, Cigliano A, Latte G, Che L, Joseph C, et al. Inactivation of fatty acid synthase impairs hepatocarcinogenesis driven by AKT in mice and humans. J Hepatol. 2016;64:333–341. doi: 10.1016/j.jhep.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P, Ge M, Hu J, Li X, Che L, Sun K, Cheng L, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology. 2017;66:167–181. doi: 10.1002/hep.29183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samarin J, Laketa V, Malz M, Roessler S, Stein I, Horwitz E, Singer S, et al. PI3K/AKT/mTOR-dependent stabilization of oncogenic far-upstream element binding proteins in hepatocellular carcinoma cells. Hepatology. 2016;63:813–826. doi: 10.1002/hep.28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buitrago-Molina LE, Pothiraju D, Lamle J, Marhenke S, Kossatz U, Breuhahn K, Manns MP, et al. Rapamycin delays tumor development in murine livers by inhibiting proliferation of hepatocytes with DNA damage. Hepatology. 2009;50:500–509. doi: 10.1002/hep.23014. [DOI] [PubMed] [Google Scholar]
- 27.Zhu AX, Kudo M, Assenat E, Cattan S, Kang YK, Lim HY, Poon RT, et al. Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib: the EVOLVE-1 randomized clinical trial. Jama. 2014;312:57–67. doi: 10.1001/jama.2014.7189. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, Yang J-Y, et al. TCPA: a resource for cancer functional proteomics data. Nat Meth. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas Janice D, Zhang Y-J, Wei Y-H, Cho J-H, Morris Laura E, Wang H-Y, Zheng XFS. Rab1A Is an mTORC1 Activator and a Colorectal Oncogene. Cancer Cell. 2014;26:754–769. doi: 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagasue N, Yu L, Yukaya H, Kohno H, Nakamura T. Androgen and oestrogen receptors in hepatocellular carcinoma and surrounding liver parenchyma: impact on intrahepatic recurrence after hepatic resection. Br J Surg. 1995;82:542–547. doi: 10.1002/bjs.1800820435. [DOI] [PubMed] [Google Scholar]
- 32.Tavian D, De Petro G, Pitozzi A, Portolani N, Giulini SM, Barlati S. Androgen receptor mRNA under-expression in poorly differentiated human hepatocellular carcinoma. Histol Histopathol. 2002;17:1113–1119. doi: 10.14670/HH-17.1113. [DOI] [PubMed] [Google Scholar]
- 33.Yoon G, Kim JY, Choi YK, Won YS, Lim IK. Direct activation of TGF-beta1 transcription by androgen and androgen receptor complex in Huh7 human hepatoma cells and its tumor in nude mice. J Cell Biochem. 2006;97:393–411. doi: 10.1002/jcb.20638. [DOI] [PubMed] [Google Scholar]
- 34.Yu Z, Gao YQ, Feng H, Lee YY, Li MS, Tian Y, Go MY, et al. Cell cycle-related kinase mediates viral-host signalling to promote hepatitis B virus-associated hepatocarcinogenesis. Gut. 2014;63:1793–1804. doi: 10.1136/gutjnl-2013-305584. [DOI] [PubMed] [Google Scholar]
- 35.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 36.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, et al. FKBP51 Affects Cancer Cell Response to Chemotherapy by Negatively Regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Mikhailova M, Bose S, Pan CX, deVere White RW, Ghosh PM. Regulation of androgen receptor transcriptional activity by rapamycin in prostate cancer cell proliferation and survival. Oncogene. 2008;27:7106–7117. doi: 10.1038/onc.2008.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Duan Y, Zheng X. Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discov Today. 2011;16:325–331. doi: 10.1016/j.drudis.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vizoso FJ, Rodriguez M, Altadill A, Gonzalez-Dieguez ML, Linares A, Gonzalez LO, Junquera S, et al. Liver expression of steroid hormones and Apolipoprotein D receptors in hepatocellular carcinoma. World J Gastroenterol. 2007;13:3221–3227. doi: 10.3748/wjg.v13.i23.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalra M, Mayes J, Assefa S, Kaul AK, Kaul R. Role of sex steroid receptors in pathobiology of hepatocellular carcinoma. World J Gastroenterol. 2008;14:5945–5961. doi: 10.3748/wjg.14.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mora GR, Prins GS, Mahesh VB. Autoregulation of androgen receptor protein and messenger RNA in rat ventral prostate is protein synthesis dependent. J Steroid Biochem Mol Biol. 1996;58:539–549. doi: 10.1016/0960-0760(96)00079-9. [DOI] [PubMed] [Google Scholar]
- 44.Di Maio M, Daniele B, Pignata S, Gallo C, De Maio E, Morabito A, Piccirillo MC, et al. Is human hepatocellular carcinoma a hormone-responsive tumor? World J Gastroenterol. 2008;14:1682–1689. doi: 10.3748/wjg.14.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, Will M, et al. Feedback suppression of PI3Kalpha signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kbeta. Cancer Cell. 2015;27:109–122. doi: 10.1016/j.ccell.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.