Abstract
Background
The aim of this study was to elucidate the molecular epidemiology of carbapenem non-susceptible Enterobacteriaceae(CNSE) isolated in the Eastern region of Heilongjiang Province, China, and the mechanism of carbapenem resistance.
Methods
A total of 53 CNSE isolates were collected in a grade-3 hospital in Heilongjiang province. Sensitivity to antibiotics was determined using the VITEK-2 Compact automatic system. The modified Hodge test (MHT) and modified carbapenem inactivation test (mCIM) were performed for phenotypic identification. Beta-lactamases gene were detected by Polymerase chain reaction(PCR) and DNA sequencing. The transfer of blaNDM and blaKPC was investigated through conjugation experiment. The clinical data of patients were retrospectively reviewed. Homology of Carbapenem-resistant Klebsiella pneumoniae(CRKP) was conducted by multilocus sequence typing (MLST).
Results
CNSE were highly resistant to the majority of antimicrobial agents. The resistance rate was 100% for first, third, fourth generation cephalosporins and enzyme inhibitor compounds. Gentamicin and tobramycin recorded a resistance rate higher than 80%. Less than 30% resistance was detected for amikacin and levofloxacin. Among CNSE 52(98.1%) and 48(90.6%) of CNSE were positive for mCIM and MHT respectively. There were 42 positive blaKPC genes, three blaNDM-1 genes, three blaNDM-5 genes, one blaNDM-7 gene, and six blaIMP-4 genes. Most isolates harbored multiple drug resistance gene, especially as related to extended-spectrum-β-lactamases, blaSHV, blaTEM and blaCTX-M-15 genes.The resistant gene was transferred into recipient Escherichia coli J53 through conjugation in 21.3% (10/47) of the strains. MLST revealed that ST76 (n = 36) was the most predominant clone, followed by ST896, ST323 and ST11. A new one ST 2946 was identity by this study.
Conclusion
The carbapenem resistance phenomenon is alarming and blaKPC-2 is the main resistant gene of CNSE in our hospital. This is the first report of an outbreak caused by blaKPC-2 positive K. pneumoniae ST76 in the Eastern region of Heilongjiang Province, China. Relevant departments should implement infection control and prevention measures to avoid further dissemination of the multi drug-resistant bacteria (MDR).
Keywords: Molecular characterization, Epidemiologic typing, Carbapenem non-susceptible Enterobacteriaceae
Background
The emergence of MDR bacteria is not conducive to infection control. Carbapenems are used as the final treatment agent for infections caused by MDR Gram negative bacteria. Carbapenem resistant Enterobacteriaceae (CRE) infection is responsible for a high mortality rate of 26–44% [1]. It has been reported that the high colonization rate of Klebsiella pneumoniae Carbapenemase(KPC)-producing Klebsiella pneumoniae in patients with ICU is related to the number of comorbidities, administration of carbapenems, β-lactams/lactamase inhibitors, and the time of previous ICU admission [2]. In Asia, the resistance rates of Enterobacteriaceae to carbapenems are increasing [3]. The main mechanisms of carbapenem resistance among Enterobacteriaceae are the production of carbapenemases such as KPC and New Delhi metalloβ-lactamase (NDM), extended spectrum β-lactamase (ESBL) or AmpC β-lactamases(AmpC) enzymes that are accompanied more rarely with loss of outer membrane proteins. These resistant genes are often located on plasmids. Timely detection of carbapenem-producing strains is significant for infection control. Here we investigated the prevalence and resistance characteristics of CNSE in the largest university hospital in the eastern region of Heilongjiang Province, focusing on the resistance mechanism and epidemiologic characteristics.
Methods
Bacterial strains and Antimicrobial susceptibility testing (AST)
This study was conducted from January 2015 to September 2017. All the Enterobacteriaceae isolates were identified and the minimum inhibitory concentrations(MICs) of antibiotics were determined using the Vitek 2 system and the AST-GN card (bioMérieux, France) at the First Affiliated Hospital of Jiamusi University, a 1600 beds hospital. Disk diffusion method was used as a supplementary susceptibility test. As CNSE were considered only those isolates that confirmed as Carbapenems-nonsusceptible (either of ertapenem, imipenem or meropenem) according to Clinical and Laboratory Standard Institutes (CLSI-2016) criteria. Escherichia coli ATCC 25922 and E. coli J53 (sodium azide resistant) were used as the control for antimicrobial susceptibility test and recipient strain for conjugation experiment, respectively.
Phenotype experiment
CNSE isolates were evaluated using mCIM as mentioned before [4]. Use a 1 μL inoculation loop to scrape CNSE strain into 2 mL of MH broth and vortex for 15 s. Meropenem disk (10 μg) were immersed in the 35 °C suspension and removed after 4 h of culture. E. coli ATCC25922 was formulated into a suspension of 0.5McF bacteria and densely coated on MH plates. The meropenem disk soaked in the bacterial suspension was placed on the MH plate and incubated at 35 °C incubator for 6 h. The results showed that the diameter of the inhibition zone of the meropenem disk soaked in the bacterial suspension was 6-15 mm or there are scattered colonies within the bacterium but with a diameter of 16-18 mm, which is positive for the mCIM test. The MHT was used to screen isolates for the production of carbapenemases, according to CLSI 2016 guidelines.
DNA extraction
For each CNSE, five colonies of each CNSE isolate from overnight culture plates, suspended in 1.5 ml Sodium chloride. The suspension was heated at 100 °C for ten minutes, then centrifuged at 4000 rpm for five minutes to remove cellular debris, and after 100 μl of the supernatant was transferred to a new Eppendorf tube. The DNA was stored at − 20 °C.
Molecular detection of resistance genes
PCR and nucleotide sequencing techniques were conducted to detect the presence of carbapenemase genes blaKPC, blaNDM, blaVIM-1, blaVIM-2, blaIMP-4, blaIMP-8, blaOXA-48, blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58, as well as ESBL genes, including blaCTX, blaTEM, blaACC, and blaSHV using primers as described previously [5–7]. Bioedit software was used to analyze test data, and results were compared using online blast software.
Conjugation experiments
The conjugation experiment was carried out using a membrane bonding experiment as previously described [8]. Both the donor (CNSE) and the recipient strains (E.coli J53) were mixed on Luria-Bertani agar at a ratio of 1:3, and the mixtures were incubated for 24 h at 35 °C. Transconjugants were selected in LB broth supplemented with sodium azide (100 μg/ml) and imipenem (1 μg/ml). Colonies which grew on the selective medium were identified by the VITEK-2 Compact system. Strains that harbored carbapenemase and exhibited higher MICs of resistance to carbapenems and cephalosporins than J53 were defined as the transconjugants.
Homology analysis
MLST was performed using seven housekeeping genes of K. pneumoniae which were amplified using primers showed in online databases (http://bigsdb.pasteur.fr/klebsiella/primers_used.html). The products of PCR were sequenced. Sequence types (STs) were determined using online database tools.
Statistical analysis
The SPSS 22.0 program was conducted using Chi-square test for statistical analysis. P<0.05 was statistically significant.
Results
Clinical and epidemiological characteristics
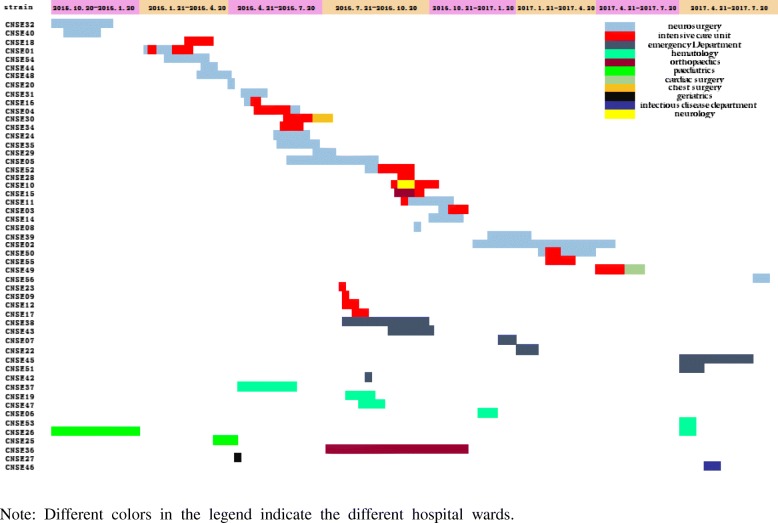
From October 2015 to July 2017 a total of 53 non-duplicated CNSE were isolated from various clinical specimens. The CNSE strains were isolated one from each patient, with age range (5 months - 89 years, median 60.5 years). Among 53 patients 34(64.2%) were male. The mortality of patients with CNSE infections was 22%. Three patients were excluded; one outpatient who had no hospital records and two nosocomial patients who had no longer contact. The majority of the CNSE isolates were K.pneumoniae (75.5%, n = 40) followed by Enterobacter cloacae (9.4%, n = 5), E.coli (7.5%, n = 4), Klebsiella oxytoca (3.8%, n = 2) whereas, Citrobacter freundii and Serratia marcescens included one isolate each (1.9%). Specimen with positive culture for CNSE, included sputum (71.7%, n = 38), blood (15.1%, n = 8), swabs (5.7%, n = 3) and urine and pus (3.8%, n = 2,each). The CNSE isolates emerged from Neurosurgery 35.8% (n = 19), ICU 28.3% (n = 15), emergency room 13.2% (n = 7), hematology 9.4% (n = 5) and other Departments. Department distribution is shown in Fig. 1.
Fig. 1.
A timeline representing the CNSE isolated in relation to the ward and the duration of stay in each ward
Antimicrobial susceptibility
According to the VITEK 2 test results, all CNSE isolates were MDR (Table 1). The CNSE isolates were resistant to ampicillin(100%), ampicillin/sulbactam(100%), piperacillin/tazobactam(100%), cefazolin(100%), ceftazidime(100%), ceftriaxone(100%) and cefepime(100%), to amikacin (7.5%), to ciprofloxacin(39.2%) and levofloxacin (24.5%), to aztreonam (96.2%) and to gentamicin and tobramycin (88.7 and 83% respectively).
Table 1.
Susceptibility to the antimicrobial agents, basic characterization and drug resistance gene of CNSE
| Isolate NO. | MIC(ug/mL) | bla genotype | MHT/mCIM | strain | Culture type | History of antibiotic use | Invasive procedure | outcome | MLST | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMS | TZP | CZO | CAZ | CRO | FEP | ATM | IMP | AMK | GEN | TOB | CIP | LEV | |||||||||
| CNSE01 CNSE02 CNSE15 |
R | R | R | R | R | R | R | R | R | S | R | R | R | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
| CNSE03 CNSE09 CNSE12 CNSE44 CNSE48 |
R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Died | ST76 |
| CNSE04 CNSE05 CNSE07 CNSE14 CNSE16 CNSE29 CNSE30 CNSE31 CNSE32 CNSE34 CNSE35 CNSE54 |
R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
| CNSE06 | R | R | R | R | R | R | R | R | R | S | S | S | S | S | KPC-2, SHV | +/+ | KP | Sputum | YES | YES | Died | ST323 |
| CNSE08 CNSE11 |
R | R | R | R | R | R | R | R | R | S | R | R | R | R | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
| CNSE10 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | NO | NO | Survived | ST76 |
| CNSE17 | R | R | R | R | R | R | R | R | R | S | R | R | S | S | KPC-2, SHV, CTX-M-15 | +/− | KP | Sputum | YES | YES | Died | ST76 |
| CNSE18 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Died | ST76 |
| CNSE19 | R | R | R | R | R | R | R | R | R | S | S | I | R | R | KPC-2, NDM-5, CTX-M-14, TEM | −/+ | EC | Blood | YES | YES | Survived | ND |
| CNSE20 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, NDM-5, SHV, CTX-M-177, TEM | +/+ | KP | Sputum | YES | YES | Died | ST76 |
| CNSE22 | R | R | R | R | R | R | R | R | R | S | R | I | S | S | IMP-4, SHV, TEM | +/+ | KP | Sputum | YES | YES | Died | ST896 |
| CNSE23 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Blood | YES | YES | NR | ST76 |
| CNSE24 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Blood | YES | YES | Survived | ST76 |
| CNSE25 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | NDM-1, TEM, CTX-M-15 | +/+ | EC | Sputum | YES | YES | Survived | ND |
| CNSE26 | R | R | R | R | R | R | R | S | R | S | S | S | S | S | IMP-4 | +/+ | KO | Sputum | YES | NO | Survived | ND |
| CNSE27 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | NDM-7, TEM, CTX-M-15 | −/+ | EC | Urine | YES | YES | Survived | ND |
| CNSE28 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Blood | YES | YES | Died | ST76 |
| CNSE36 | R | R | R | R | R | R | R | R | R | S | R | R | R | I | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Secretion | YES | YES | Survived | ST76 |
| CNSE37 | R | R | R | R | R | R | R | R | I | S | R | I | S | S | IMP-4, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Died | ST2964 |
| CNSE38 | R | R | R | R | R | R | R | R | R | S | R | R | S | S | NDM-1, SHV, CTX-M-15 | −/+ | ECL | Secretion | YES | YES | Survived | ND |
| CNSE39 | R | R | R | R | R | R | R | R | R | S | S | S | S | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
| CNSE40 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KO | Sputum | YES | YES | Survived | ND |
| CNSE41 | R | R | R | R | R | R | R | R | S | S | R | R | R | S | – | −/+ | SM | Secretion | NR | NR | NR | ND |
| CNSE42 | R | R | R | R | R | R | R | R | I | R | R | R | S | S | IMP-4, TEM | +/+ | ECL | Blood | YES | YES | Died | ND |
| CNSE43 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | KPC-2, SHV, TEM, DHA | +/+ | ECL | Sputum | YES | YES | Died | ND |
| CNSE45 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | KPC-2, TEM | +/+ | CF | pus | YES | YES | Survived | ND |
| CNSE46 | R | R | R | R | R | R | R | R | R | S | R | I | R | R | NDM-1 IMP-4, SHV, TEM, CTX-M-15 |
+/+ | ECL | Urine | YES | YES | Survived | ND |
| CNSE47 | R | R | R | R | R | R | R | R | R | S | S | S | R | R | KPC-2, SHV | −/+ | KP | Blood | YES | YES | Survived | ST11 |
| CNSE49 | R | R | R | R | R | R | R | R | R | S | R | R | R | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Blood | YES | YES | Survived | ST76 |
| CNSE50 | R | R | R | R | R | R | R | R | R | S | R | R | R | R | KPC-2, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
| CNSE51 | R | R | R | R | R | R | R | R | R | S | S | S | R | R | KPC-2, IMP-4, TEM, DHA | +/+ | ECL | pus | YES | YES | Survived | ND |
| CNSE52 | R | R | R | R | R | R | R | R | R | S | R | R | I | S | KPC-2, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
| CNSE53 | R | R | R | R | R | R | R | S | R | R | R | R | R | R | NDM-5, SHV | +/+ | EC | Blood | YES | YES | Died | ND |
| CNSE55 | R | R | R | R | R | R | R | R | R | S | R | R | R | S | KPC-2, SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | NR | ST76 |
| CNSE56 | R | R | R | R | R | R | R | R | R | S | R | R | R | S | SHV, TEM, CTX-M-15 | +/+ | KP | Sputum | YES | YES | Survived | ST76 |
AMP ampicillin, AMS ampicillin-sulbactam, TZP piperacillin/tazobactam, CZO cefazolin, CAZ ceftazidime, CRO ceftriaxone, FEP cefepime, ATM aztreonam, IMP imipenem, GEN gentamicin, TOB tobramycin, AMK amikacin, CIP ciprofloxacin and LEV levofloxacin, KP Klebsiella pneumoniae, EC Enterobacter cloacae, EC Escherichia coli, KO Klebsiella oxytoca, CF Citrobacter freundii, SM Serratia marcescens, NR No Record, ND Not Determined
Phenotypic experiment
According to mCIM results, the positive strains were 52 (98.1%). The positive rate of MHT was 90.6%, also shown on Table 1. There was no statistically significant difference between mCIM and MHT in their ability to detect carbapenemase (p = 0.219).
Drug resistant genes
The results from the study of blagenes among CNSE isolates were listed on Table 1. Among 53 CNSE isolates, 42(79. 2%) were KPC-2 producers. Six isolates (11.3%) including three E. cloacae, two K. pneumoniae and one K.oxytoca carried blaIMP-4 genes. Seven isolates (13.2%) produced NDM carbapenemase. One E.coli and two E.cloacae carrying blaNDM-1, two E.coli, one K.pneumoniae carrying blaNDM-5, one E.coli carrying blaNDM-7. ESBL genes were found in 43(81.1%), 43 (81.1%) and 45 (84.9%) isolates carried blaSHV, blaCTX-M and bIaTEM, respectively. Thirty-three K. pneumoniae carried blaSHV, blaCTX-M and blaTEM genes simultaneously. Other drug resistance genes (blaVIM, blaOXA-58, blaOXA-51, blaOXA-48, blaOXA-23, blaOXA-24 and blaACC) were not detected.
Horizontal transfer of blaKPC and blaNDM
Forty seven blaNDM positive or blaKPC positive isolates were selected for conjugation. The conjugation experiments showed that the plasmids with blaNDM and blaKPC from ten CNSE isolates were successfully transferred to recipient E. coli J53. The success rate was 21.3%, and the conjugants exhibited higher resistance to carbapenems compared to J53. The MICs of imipenem and ertapenem for the conjugants ranged from 0.12 to 16 mg/L (Table 2).
Table 2.
The MICs of Cephalosporins and carbapenems for the conjugants
| Isolate NO. | MIC | Drug-resistant gene transfer | ||||
|---|---|---|---|---|---|---|
| CAZ | CRO | FEP | IMP | ETP | ||
| J53 | ≤0.12 | ≤0.25 | ≤0.12 | ≤0.25 | ≤0.12 | – |
| JCNSE09 | 4 | 32 | 0.5 | ≤0.25 | 0.5 | KPC |
| JCNSE15 | 16 | 32 | 4 | 0.5 | 0.5 | KPC |
| JCNSE18 | ≤0.12 | 1 | ≤0.12 | 0.5 | ≤0.12 | KPC |
| JCNSE19 | ≥64 | ≥64 | 8 | 8 | ≥8 | NDM |
| JCNSE24 | 0.25 | ≤0.25 | ≤0.12 | ≤0.25 | ≤0.12 | KPC |
| JCNSE27 | ≥64 | ≥64 | 16 | ≥16 | ≥8 | NDM |
| JCNSE28 | 16 | 16 | ≤0.12 | 0.5 | ≤0.12 | KPC |
| JCNSE35 | 16 | 32 | 16 | ≤0.25 | ≤0.12 | KPC |
| JCNSE38 | ≥64 | ≥64 | 16 | ≥16 | ≥8 | NDM |
| JCNSE40 | 4 | 32 | ≤0.12 | ≤0.25 | ≤0.12 | KPC |
Homology analysis
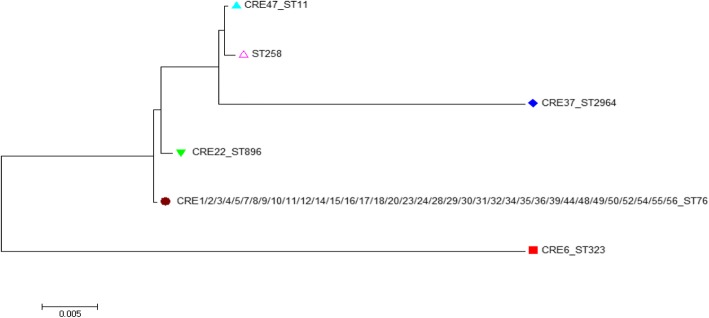
All of five distinct MLST sequence types were observed among the forty CRKP isolates, including ST76 (n = 36), ST11 (n = 1), ST323 (n = 1), ST896 (n = 1) and ST2964 (n = 1). CRKP ST76 isolates were isolated, mainly from neurosurgery (50.0%) and intensive care unit (41.7%). The ST2964 isolate was a novel type found in this study and its DNA sequences were submitted into the Institute Pasteur MLST database for K. pneumoniae (available at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). ST76 has earlier evolution than ST896, ST2964 and ST11. Their evolutionary relationships are shown in Fig. 2.
Fig. 2.
Phylogenetic tree of 40 Klebsiella pneumoniae isolates
Discussion
The increase in ESBL-producing bacteria indicates a rise in the use of carbapenems and this may be the reason for the increasing number of carbapenem resistant enterobacteriaceae in recent years [9]. As the mortality in present study was found as high as 22% (11/50), it was necessary to evaluate the carbapenem resistance trend in our hospital and the gene characteristics responsible for the resistance. Clinical treatment using carbapenem may vary depending on the mechanism of resistance [10].
As it is well known, the production of KPC-type enzyme is the most important mechanism of carbapenem resistance in Enterobacteriaceae. KPC-producing Enterobacteriaceae spp. have emerged globally. BlaKPC is now endemic all over the world. In Europe, especially Austria, Germany, Greece, Russia and United Kingdom; In Latin America, including Brazil and Mexico;The Asia-Pacific, China, Japan, Taiwan; Also found in Israel and the United States [11]. It has disseminated dramatically throughout China, causing serious infectious diseases. In China, the first case of blaKPC was found in Hangzhou, which was isolated from one elderly patient’s sputum in 2004 [12]. Currently, blaKPC has been reported in many regions of China, including Beijing, Zhejiang, Taiwan, and Sichuan [13–16]. To the best of our knowledge, this article is the first report on the prevalence of CNSE in the eastern part of Heilongjiang province.
Metallo-β-lactams, including IMP and NDM, contributed to carbapenem resistance in Enterobacteriaceae, particularly NDM-1, which has caused a global health threat. NDM isolates are highly resistant to many classes of antibiotics and have the potential to spread rapidly to many members of Enterobacteriaceae which can cause nosocomial outbreaks [17]. BlaNDM-1 was first reported in K.pneumoniae from a Swedish patient in 2009 [18]. It exists in many variant forms and has been reported in Africa, Europe, Australia, America and Asia, specifically Turkey, Algeria, France, Italy, Greece, New Zealand, Mexico and China [18–26]. Chen et al. first reported the emergence of blaNDM-1-positive strains in four different provinces in China [27] . BlaNDM-1 widely disseminated in China, the epidemic dissemination of NDM-1-producing Enterobacteriaceae spp. has been confirmed in Henan, Zhejiang, Yunnan, Hunan and other provinces [28–31]. One major reason for this is the rapid dissemination of the blaNDM-1 gene is plasmid location [29]. In our study four E. coli, two E. cloacae and one K. pneumoniae were the seven isolates carrying blaNDM.
In present study CNSE have been found to carry at least two different types of beta-lactamase resistance genes, most isolates (58.5%) carried simultaneously the blaKPC-2, blaCTX-M-15, blaSHV, blaTEM gene, this is consistent with Netikul’s report [32]. The strains that produce KPC can caused the mediation of carbapenem resistance in K. pneumoniae [33]. Most of the strains were resistant to a variety of antimicrobial agents, including cephalosporins, carbapenems, enzyme inhibitors and so on. Indicates that the phenomenon of drug resistance was serious.This may be related to clinicians’ experience of medication. CTX-M can hydrolyze cephalosporin and mono-amide antibiotics, CTX-M exhibits powerful activity against ceftriaxone [34]. According to amino acid sequence similarities, blaCTX-M-15 is a member of the blaCTX-M-1 subgroup. The hydrolysis of ceftazidime by CTX-M-15 was strongest. As is well known, TEM can hydrolyze penicillin and first generation cephalosporin. SHV is resistant to cephalosporin and aztreonam, in particular, it shows drug resistance to third-generation cephalosporins, and some were resistant to beta-lactamase inhibitors. The antimicrobial resistance may be produced by a variety of beta-lactamase genes which enhances antimicrobial resistance and broadens the resistant spectrum and may be the reason why drug resistance of these strains was very strong [35]. Horizontal genes transfer tend to be associated with the high prevalence of ESBLs and the increasing presence of carbapenemases [36]. Conjugation was the reason for carbapenem resistance acquisition in carbapenem resistant enterobacteriaceae [37]. Acquired carbapenemases are encoded by genes located in mobile elements such as transposons and plasmids, which may transfered to different strains and species [38]. Interestingly, not all carbapenemase-producing isolates are carbapenem-resistant [39], this is because, while production of carbapenemase always elevates the MICs of carbapenems, they may not be high enough to be classified as resistance or intermediate resistance [40].
The MHT and mCIM results for phenotypic testing were recommended by CLSI for carbapenemase detection and early screening of carbapenemases to avoid the further spread of resistant bacteria. We evaluated the effectiveness of mCIM and our results indicated that mCIM had high sensitivity and specificity for detecting carbapenemase-producing Enterobacteriaceae. Although one strain gave false positive result in mCIM. False positive results may be due to different resistance mechanisms in carbapenem resistance. Our results showed that MHT had powerful ability to detect carbapenemase positive isolates 48 (90.6%). Among them only four isolates were blaNDM positive and six others were blaIMP. Many studies have showed that MHT often has low sensitivity and specificity for detection of class B carbapenemases [41], and this is consistent with our results. There was no statistical significance in the detection of carbapenems between MHT and mCIM. They are all the particularly useful tool for the detection of carbapenemase producers.
Isolates with the blaKPC gene are spread in many regions such as North America, Latin America and Asia with ST11 and ST258 being the common types [42–44]. The epidemiological information on CRKP in the eastern region of Heilongjiang Province is unknown. Interestingly, blaKPC gene has been detected in our CRKP isolates but none of the isolates were ST11 which were reported in other parts of China. In our study, we demonstrated that the sequence type ST76 was the predominant type among CRKP isolates which are blaKPC-2 positive in Heilongjiang Province. To the best of our knowledge, the ST76 clone has been sporadically reported in Taiwan and America [45, 46]. In Japan, ST76 has been isolated from meat [47]. Zhu reported in 2014 an outbreak caused by NDM-1-producing K. pneumoniae ST76 in neonates in Shanghai [48]. Figure 1 shows the outbreak of CNSE in the department of neurosurgery and ICU of our hospital exhibiting sporadic phenomena in other departments. During the study period, the first case of type ST76 CRKP was detected from a sample coming from the department of neurosurgery.
In addition, several other ST types were found among CRKP isolates, including ST323, ST11, ST896 and the novel ST2964 clone. There are not many reports about ST323 and ST896. ST76 type evolution before ST2964. The common ST258 in the United States differs from the common ST11 in China only by a tonB housekeeping gene, with only a difference of four bases, so the two are closely related. MLST studies indicated that clones of CRKP were widespread. Here, we emphasize the need to actively monitor the spread of CRKP. Our findings supplied the novel epidemiologic data of CRKP in China.
At the beginning of the epidemic, most of the strains were isolated from respiratory tract specimens, which suggested that respiratory tract colonization had occurred. Almost all the patients had one or more severe conditions, of which 19 (35.8%) patients suffered from sudden cerebral hemorrhages, six (11.3%) patients severe head injury caused by trauma, 11 (20.7%) patients patients with myelodysplastic syndrome and others with cancer of the stomach, abdominal closed injury, acute cerebral infraction among other diseases. In our study, 50 (94.3%) patients were treated with invasive procedures, such as the arteriovenous catheter, tracheotomy, ventilator-assisted breathing, among other procedures. Fifty-one patients (96.2%) had a history of antibiotic exposure, which contributed to the production of CNSE. In addition, receiving a transplant was an independent factor in CNSE infection [49]. The prevention and control of CNSE infection or colonization mainly includes the following aspects: necessary interventions for the hand hygiene of medical workers and patients; routine ongoing active monitoring;prescribing medications strictly in accordance with the regulations for the rational use of antimicrobial agents; and isolated infection or colonization of CNSE patients. Currently, a collaboration is needed between the pharmacists, microbiologists, infection control practitioners, and infectious disease clinicians in working together to defeat CNSE [50].
As it is well known, the economy is underdeveloped in the eastern part of Heilongjiang Province and the prevalence of CNSE in such area must be treated with the needed urgency. Therefore, there is an urgent need to establish more and better surveillance in our hospital to prevent further spread of the resistant bacteria.
Conclusions
We report for the first time, the endemic spread of CNSE in the largest university hospital in the eastern region of Heilongjiang Province, China. Even if the prevalence of CNSE were low, we should still be monitored closely. The main drug resistance genes are blaKPC-2, blaNDM, blaTEM, blaSHV and blaCTX-M-15. The spread of CRKP was caused by the selection of blaKPC-2 positive K. pneumoniae ST76 epidemic clones. This will help prevent the widespread spread of CNSE in this hospital and provide a basis for the epidemiology of CNSE in Heilongjiang Province. More research should be done to better understand the causes of CNSE.
Acknowledgments
We thank Dr. Wang Minggui from Huashan Hospital Affiliated to Fudan University for the isolates of J53. We thank Dr. Yu YunSong from Sir Run Run Shaw Hospital affiliated to Zhejiang University for the isolates of EC600. We thank the team of the curators of the Institute Pasteur MLST system (Paris, France) for importing novel alleles, profiles and/or isolates at http://bigsdb.web.pasteur.fr.
Funding
This work was supported by Natural Science Foundation of Heilongjiang Province (D201224), and Heilongjiang Provincial Health and Family Planning Commission on Scientific Research Project (2017–405).
Availability of data and materials
The datasets analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AmpC
AmpC β-lactamases
- CLSI
Clinical and Laboratory Standard Institutes
- CNSE
Carbapenem non-susceptible Enterobacteriaceae
- CRKP
Carbapenem-resistant Klebsiella pneumoniae
- ESBL
Extended spectrum β-lactamase
- KPC
Klebsiella pneumoniae Carbapenemase
- mCIM
Modified carbapenem inactivation method
- MHT
Modified hodge test
- MICs
Minimum inhibitory concentrations
- MLST
Multilocus sequence typing
- NDM-1
New Delhi metalloβ-lactamase-1
- PCR
Polymerase chain reaction
- STs
Sequence types
Authors’ contributions
All authors contributed to this work. XLZ and YW conceived and designed the experiments; XG, JSZ and YJF performed the experiments; XG and JSZ analyzed the data and SSS wrote the paper. XG and JSZ are the first authors. All authors reviewed and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of Jiamusi University Clinical Medical College for research. The committee’s reference number is 0326. Individual informed consent was waived by the ethics committee listed above because this study used currently existing sample collected during the course of routine medical care and did not pose any additional risks to the patients.
Consent for publication
Not applicable.
Competing interests
The authors have declared that no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xue Gong, Email: 291135155@qq.com.
Jisheng Zhang, Email: zhangjisheng215@126.com.
Shanshan Su, Email: 827691363@qq.com.
Yanjun Fu, Email: fuyanjun1981@163.com.
Mingjia Bao, Email: cmubmj@163.com.
Yong Wang, Email: 15331878255@163.com.
Xiaoli Zhang, Email: jmszxl123@163.com.
References
- 1.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis. 2014;20(7):1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papadimitriou-Olivgeris M, Marangos M, Fligou F, Christofidou M, Bartzavali C, Anastassiou ED, Filos KS. Risk factors for KPC-producing Klebsiella pneumoniae enteric colonization upon ICU admission. J Antimicrob Chemother. 2012;67(12):2976–2981. doi: 10.1093/jac/dks316. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Gu B, Huang M, Liu H, Xu T, Xia W, Wang T. Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000-2012 in Asia. Thoracic disease. 2015;7(3):376–385. doi: 10.3978/j.issn.2072-1439.2014.12.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing, 27th ed CLSI supplement M100. Clinical and laboratory standards institute. Wayne: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 5.Zhang JX. Study on bacterial resistance and transmission mechanism of carbapenem non-susceptible Enterobacteriaceae[D] Beijing: Peking Union Medical College Hospital; 2012. [Google Scholar]
- 6.Yang QW. Epidemiology and drug resistance mechanism of carbapenem resistant Enterobacteriaceae in China [D] Beijing: Peking Union Medical College Hospital; 2015. [Google Scholar]
- 7.Pu SL. Resistance Mechanism and Molecular Epidemiology of Carbapenem - resistant Klebsiell pneumoniae.[D] Chongqing: Chongqing Medical University; 2015. [Google Scholar]
- 8.Wang X, Chen G, Wu X, et al. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the blaNDM-1 element and clonal spread of progenitor resistant strains. Front Microbiol. 2015;6:595. [DOI] [PMC free article] [PubMed]
- 9.Torres-Gonzalez P, Cervera-Hernandez M, Niembro-Ortega M, Leal-Vega F, Cruz-Hervert L, García-García L, Galindo-Fraga A, Martinez-Gamboa A, Bobadilla-Del Valle M, Sifuentes-Osornio J, Ponce-de-Leon A. Factors associated to prevalence and incidence of Carbapenem-resistant Enterobacteriaceae fecal carriage: a cohort study in a Mexican tertiary care hospital. PLoS One. 2015;10(10):e0139883. doi: 10.1371/journal.pone.0139883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez F, Van Duin D. Carbapenem-resistant Enterobacteriaceae: a menace to our mostvulnerable patients. Cleve Clin J Med. 2013;80(4):225–233. doi: 10.3949/ccjm.80a.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazmierczak KM, Biedenbach DJ, Hackel M, Rabine S, de Jonge BL, Bouchillon SK, Sahm DF, Bradford PA. Global dissemination of blaKPC into bacterial species beyond Klebsiella pneumoniae and in vitro susceptibility to ceftazidime-avibactam and Aztreonam-avibactam. Antimicrob Agents Chemother. 2016;60(8):4490–4500. doi: 10.1128/AAC.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2006;51(2):763–765. doi: 10.1128/AAC.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J, Yin Z, Zhao Q, Zhao Y, Zhang D, Jiang X, Wu W, Chen W, Wang H, Song Y, Tong Y, Wang J, Li Y, Zhou D. Genomic characterization of novel IncFII-type multidrug resistant plasmids p0716-KPC and p12181-KPC from Klebsiella pneumoniae. Sci Rep. 2017;7(1):5830. doi: 10.1038/s41598-017-06283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai JC, Zhang R, Hu YY, Zhou HW, Chen GX. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob Agents Chemother. 2014;58(2):1146–1152. doi: 10.1128/AAC.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng IL, Liu YM, Wang SJ, Yeh HY, Hsieh CL, Lu HL, Tseng YC, Mu JJ. Emergence of Carbapenemase producing Klebsiella pneumonia and spread of KPC-2 and KPC-17 in Taiwan: a Nationwide study from 2011 to 2013. PLoS One. 2015;10(9):e0138471. doi: 10.1371/journal.pone.0138471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu W, Feng Y, Carattoli A, Zong Z. Characterization of an Enterobacter cloacae strain producing both KPC and NDM Carbapenemases by whole-genome sequencing. Antimicrob Agents Chemother. 2015;59(10):6625–6628. doi: 10.1128/AAC.01275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baran I, Aksu N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clin Microbiol Antimicrob. 2016;15:20. doi: 10.1186/s12941-016-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, Bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L, Yilmaz M, Istanbullu A, Arslan F, Mert A, Bernabeu S, Nordmann P. Spread of NDM-1-producing Enterobacteriaceae in a neonatal intensive care unit in Istanbul, Turkey. Antimicrob Agents Chemother. 2014;58(5):2929–2933. doi: 10.1128/AAC.02047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yousfi M, Mairi A, Bakour S, Touati A, Hassissen L, Hadjadj L, Rolain J. First report of NDM-5-producing Escherichia coli ST1284 isolated from dog in Bejaia, Algeria. New Microbes New Infect. 2015;8:17–18. doi: 10.1016/j.nmni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Mee-Marquet N, Diene SM, Chopin F, Goudeau A, Francois P. Enigmatic occurrence of NDM-7 enzyme in the community. Int J Antimicrob Agents. 2016;47(6):505–507. doi: 10.1016/j.ijantimicag.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Bitar I, Piazza A, Gaiarsa S, Villa L, Pedroni P, Oliva E, Nucleo E, Pagani L, Carattoli A, Migliavacca R. ST405 NDM-5 producing Escherichia coli in northern Italy: the first two clinical cases. Clin Microbiol Infect. 2017;23(7):489–490. doi: 10.1016/j.cmi.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 23.Spyropoulou A, Bartzavali C, Vamvakopoulou S, Marangos M, Anastassiou E, Spiliopoulou I, Christofidou M. The first NDM metallo-β-lactamase producing Klebsiella pneumoniae isolate in a University Hospital of Southwestern Greece. J Chemother. 2016;28(4):350–351. doi: 10.1179/1973947815Y.0000000003. [DOI] [PubMed] [Google Scholar]
- 24.Howard JC, Creighton J, Heffernan H, Werno A. Evidence of transmission of an NDM-5-producing Klebsiella pneumoniae in a healthcare facility in New Zealand. Antimicrob Chemother. 2017;72(3):949–951. doi: 10.1093/jac/dkw498. [DOI] [PubMed] [Google Scholar]
- 25.Torres-Gonzalez P, Bobadilla-Del Valle M, Tovar-Calderon E, Leal-Vega F, Hernandez-Cruz A, Martinez-Gamboa A, Niembro-Ortega MD, Sifuentes-Osornio J, Ponce-de-Leon A. Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-beta-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob Agents Chemother. 2015;59(11):7080–7083. doi: 10.1128/AAC.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Que TL, Chow KH. Identification and characterization of a novel incompatibility group X3 plasmid carrying Bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect. 2012;1(11):e39. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Zhou Z, Jiang Y, Yu Y. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother. 2011;66(6):1255–1259. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Qin S, Xu H, Xu L, Zhao D, Liu X, Lang S, Feng X, Liu HM. New Delhi Metallo-beta-Lactamase 1(NDM-1), the Dominant Carbapenemase Detected in Carbapenem-Resistant Enterobacter cloacae from Henan Province, China. PloS one. 2015;10(8):e0135044. doi: 10.1371/journal.pone.0135044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One. 2015;10(6):e0129454. doi: 10.1371/journal.pone.0129454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng R, Zhang Q, Guo Y, Feng Y, Liu L, Zhang A, Zhao Y, Yang X, Xia X. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan. China Ann Clin Microbiol Antimicrob. 2016;15:10. doi: 10.1186/s12941-016-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Li X, Wang M, Yue H, Li P, Liu Y, Cao W, Yao D, Liu L, Zhou X, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae causing neonatal infection in a teaching hospital in mainland China. Antimicrob Agents Chemother. 2015;59(7):4349–4351. doi: 10.1128/AAC.03868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Netikul T, Kiratisin P. Genetic characterization of Carbapenem-resistant Enterobacteriaceae and the spread of Carbapenem-resistant Klebsiella pneumonia ST340 at a University Hospital in Thailand. PLoS One. 2015;10(9):e0139116. doi: 10.1371/journal.pone.0139116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53(10):4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossolini GM, D'Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect. 2008;14(Suppl 1):33–41. doi: 10.1111/j.1469-0691.2007.01867.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhao XJ, Deng LH, Shi DS, Kang HQ, Jiang F, Ma P. Mechanism of drug resistance of Klebsiella pneumoniae resistant carbapenem pneumonia. Chin J Infect Sci. 2015;25(17):3851–3853. [Google Scholar]
- 36.Datta S, Roy S, Chatterjee S, Saha A, Sen B, Pal T, Som T, Basu S. A five-year experience of carbapenem resistance in Enterobacteriaceae causing neonatal septicaemia: predominance of NDM-1. PLoS One. 2014;9(11):e112101. doi: 10.1371/journal.pone.0112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Franco M, Paone L, Novati R, Giacomazzi CG, Bagattini M, Galotto C, Montanera PG, Triassi M, Zarrilli R. Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789) BMC Microbiol. 2015;15(1):260. doi: 10.1186/s12866-015-0597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daikos GL, Petrikkos P, Psichogiou M, Kosmidis C, Vryonis E, Skoutelis A, Georgousi K, Tzouvelekis LS, Tassios PT, Bamia C, et al. Prospective observational study of the impact of VIM-1 metallo-beta-lactamase on the outcome of patients with Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother. 2009;53(5):1868–1873. doi: 10.1128/AAC.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doi Y, Paterson DL. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med. 2015;36(1):74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girlich D, Poirel L, Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50(2):477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bengoechea JA, Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One. 2015;10(7):e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasteran FG, Otaegui L, Guerriero L, Radice G, Maggiora R, Rapoport M, Faccone D, Di Martino A, Galas M. Klebsiella pneumoniae Carbapenemase–2, Buenos Aires. Argentina Emerg Infect Dis. 2008;14(7):1178–1118. doi: 10.3201/eid1407.070826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Zou MX, Wang HC, Dou QY, Hu YM, Yan Q, Liu WE. An outbreak of infections caused by a Klebsiella pneumoniae ST11 clone coproducing Klebsiella pneumoniae Carbapenemase-2 and RmtB in a Chinese teaching hospital. Chin Med J. 2016;129(17):2033–2039. doi: 10.4103/0366-6999.189049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan JJ, Zheng PX, Wang MC, Tsai SH, Wang LR, Wu JJ. Allocation of Klebsiella pneumoniae bloodstream isolates into four distinct groups by ompK36 typing in a Taiwanese University hospital. J Clin Microbiol. 2015;53(10):3256–3263. doi: 10.1128/JCM.01152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis GS, Waits K, Nordstrom L, Weaver B, Aziz M, Gauld L, Grande H, Bigler R, Horwinski J, Porter S, et al. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin Infect Dis. 2015;61(6):892–899. doi: 10.1093/cid/civ428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano R, Nakano A, Hikosaka K, Kawakami S, Matsunaga N, Asahara M, Ishigaki S, Furukawa T, Suzuki M, Shibayama K, et al. First report of metallo-beta-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother. 2014;58(12):7611–7612. doi: 10.1128/AAC.04265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J, Sun L, Ding B, Yang Y, Xu X, Liu W, Zhu D, Yang F, Zhang H, Hu F. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis. 2016;35(4):611–618. doi: 10.1007/s10096-016-2578-z. [DOI] [PubMed] [Google Scholar]
- 49.Sahin K, Tekin A, Ozdas S, Akin D, Yapislar H, Dilek AR, Sonmez E. Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann Clin Microbiol Antimicrob. 2015;14:44. doi: 10.1186/s12941-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez F, Van Duin D. Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable ptients. Cleve Clin J Med. 2013;80(4):225–233. doi: 10.3949/ccjm.80a.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed during the current study are available from the corresponding author on reasonable request.