Abstract
We report a family from Tabuk, Saudi Arabia, previously screened for Acrodermatitis Enteropathica (AE), in which two siblings presented with typical features of acral dermatitis and a pustular eruption but differing severity. Affected members of our family carry a rare genetic variant, p.Gly512Trp in the SLC39A4 gene which encodes a zinc transporter; disease is thought to result from zinc deficiency. Similar mutations have been reported previously; however, the variable severity within cases carrying the p.Gly512Trp variant and in AE overall led us to hypothesise that additional genetic modifiers may be contributing to the disease phenotype. Therefore whole genome sequencing was carried out in five family members, for whom material was available to search for additional modifiers of AE; this included one individual with clinically diagnosed AE. We confirmed that the p.Gly512Trp change in SLC39A4 was the only candidate homozygous change which was sufficiently rare (ExAC allele frequency 1.178e-05) and predicted deleterious (CADD score 35) to be attributable as a fully penetrant cause of AE. To identify other genes which may carry relevant genetic variation, we reviewed the relevant literature and databases including Gene Ontology Consortium, GeneMANIA, GeneCards, and MalaCards to identify zinc transporter genes and possible interacting partners. The affected individual carried variants in RECQL4 and GPAA1 genes with ExAC allele frequency <0.01 and CADD score >10. p.Gly512Trp is highly likely to be the pathogenic variant in this family. This variant was previously detected in a Tunisian proband with perfect genotype-phenotype segregation suggestive of pathogenicity. Further research is required in this area due to small sample size, but attention should be given to RECQL4 and GPAA1 to understand their role in the skin disease.
1. Introduction
Acrodermatitis Enteropathica (AE) is a rare inherited metabolic condition that affects zinc absorption and inheritance is often seen in an autosomal recessive pattern [1]. The frequency of inherited cases is estimated at 1:500,000 individuals with no obvious correlation in race or gender [2]. The manifestations of AE include alopecia, diarrhoea, dermatitis, growth retardation, and behavioural changes. Although AE has been attributed to fully penetrant homozygous mutations in the SLC39A4 gene, severity is variable even within individual families [3] suggesting that additional genetic modifiers may play a role.
Measurement of plasma zinc is the commonest method of identifying patients with AE, and reduced levels <60 ug/dL were noted in patients B03 and B06 in the original study which were correlated with their typical AE symptoms. However, limitations of zinc testing have been highlighted by Garza-Rodriguez et al. (2015) [4] who reported a rare case of AE presenting with two novel missense mutations of the SLC39A4 gene associated with periorificial and acral dermatitis only and with normal plasma levels of zinc.
The family in this study was previously screened and analysed by Abu-Duhier et al. 2017 [3] to identify the pathogenic mutation. In this analysis DNA samples obtained from eleven individuals were amplified by polymerase chain reaction and Sanger sequenced. This comprised both parents and all the siblings including the affected individuals with their respective partners. A homozygous alternate allele in chromosome 8 at position 145638714 was identified in the gene SLC39A4 (solute carrier 39 member 4) which is a zinc transporter. This mutation was present only in the two affected members B03 and B06 presenting with the AE phenotype (Figure 1). The mutation gave rise to a change from the neutral aliphatic amino acid glycine to the highly hydrophobic aromatic tryptophan, with genotype c.1534G>T at position p.Gly512Trp. Although there was strong evidence that this mutation is the cause of AE in this family, severity was variable suggesting that other modifiers may be present. Moreover, because sequencing was limited to a small number of genes, it is possible that alternative homozygous pathogenic change in a zinc transporter gene may have been missed. Others have reported the p.Gly512Trp change in a patient with AE [1], but the role of genetic modifiers has not previously been examined.
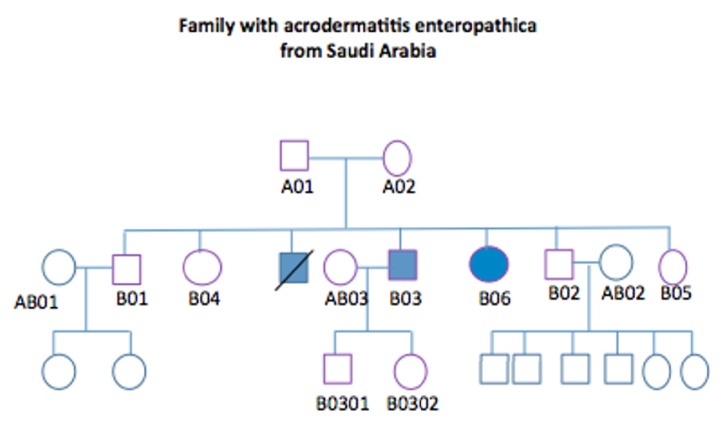
Figure 1.
Pedigree analysis to demonstrate the family of AE and the sequenced individuals for this study. Genome sequence of the following samples: A01, A02, B03, AB01, and AB02. Only B03 and B06 are affected (one male sibling was also affected, but he died). Note. Shading in blue displays the individuals affected by the disease.
To exhaustively define the genetic basis of AE in our patients, the whole genome of available family members has now been evaluated. In this study, we aimed to identify pathogenic and relevant modifier genes associated with familial AE. On this occasion the proband B03, his parents A01 and A02, and unaffected family members AB01 and AB02 were examined by whole genome sequencing. Two additional family members were utilised as population controls in order to eliminate shared, but nonrelevant alleles.
2. Materials and Methods
Ethical approval was obtained from both University of Tabuk and University of Sheffield, and written informed consent was obtained to use genetic data for research purposes. Blood samples were available from 5 members of the family with hereditary AE and were initially sequenced (as discussed above), followed by whole genome sequencing. The individuals studied were the affected child B03, his parents A01 and A02, and two unrelated family members AB01 and AB02 who were included as individuals from the same population background (Figure 1). Both lanes of three HiSeq 2500 Rapid Run were used to multiplex and sequence the five samples. To produce fastq files, bcl2fastq version 1.8.4 was used with adapter trimming further modified to accept a single mismatch in the index sequence. BWA ALN version 0.7.5a was used to map reads by lane to the human reference genome hg19. Picard version 1.101 was used to mark duplicate reads followed by realignment around InDels using GATK version 2.6-5-gba531bd. Picard was then used to merge lane-level bam files, with additional marking of duplicates and realignment around InDels on the complete bam file. GATK HaplotypeCaller was used to call variants. The minimal calling quality was accepted at 1, allowing 10 alternative haplotypes. GATK was further used to calculate coverage statistics.
Variant calling files of the five individuals were automatically annotated using wANNOVAR to provide information including genomic annotation, frequency of variant observed in controls, and CADD scores. “R” software was used to analyse the annotated variants. Relevant literature was reviewed and GO consortium data used to identify all zinc transporter genes including SLC39A4. Variants in exonic regions of zinc transport genes were filtered by ExAC frequency and CADD score using “R” to identify rare, predicted deleterious variants present in a homozygous form (see sup materials). An ExAC frequency <0.01 and CADD score >10 were employed to suggest a deleterious variant. (ExAC allele frequency suggests the relative frequency of an allele at a genetic locus in a population [5]; CADD score helps measure the level of toxicity of a genetic variant such that a value of 10 defines the variant to be within 10% of a damaging variant in the human genome.) The analysis was repeated in the other sequenced individuals to determine whether the toxic mutation was present in relatives.
To extend the analysis beyond zinc transporter genes, GeneMANIA was used to identify genetic interactions of SLC39A4. Interacting partners were screened for potential rare deleterious variants although the requirement for homozygosity was relaxed to include heterozygous variants.
3. Results
3.1. Only the Homozygous p.Gly512Trp SLC39A4 Variant Is Sufficiently Rare and Deleterious to Cause AE in Our Pedigree
The identified deleterious SLC39A4 variant p.Gly512Trp (with ExAC frequency of 1.178e-05 and CADD score of 35) was present in homozygous form in B03 (Table 1). As would be expected the same p.Gly512Trp change was found to be present in an heterozygous form in both parents, A01 and A02, but was not found in the unaffected relatives. The remainder of identified variants in SLC39A4 and all other screened zinc transport genes demonstrated either a low CADD score or a high ExAC frequency implying a benign or uncertain significance (Sup Materials 2).
Table 1.
“R” software was used to identify the deleterious zinc transporter gene in B03. An EXAC frequency of <0.01 and CADD score of >10 were accepted. Excluding synonymous variants, at position Chr8:145638714 of the SLC39A4 gene, c.G1534T and p.G512W, portrayed as a rare mutation in comparison to other identified genes and genetic variants. Note. Please see Sup Materials 2 for complete list of zinc transporter genes derived by “R” software for B03.
| Gene | Chromosome | Start-End | Exon | Nucleotide substitution | Protein substitution | EXAC Frequency | CADD Score |
|---|---|---|---|---|---|---|---|
| SLC39A4 | Chr8 | 145638714 | 10 | c.G1534T | p.G512W | 1.178e-05 | 35 |
3.2. Screening SLC39A4 Interacting Partners Identified Rare Deleterious Variants in Candidate Modifier Genes
The patient B03 suffered a particularly severe form of AE and therefore is a good candidate to identify additional deleterious genetic modifiers of AE. To identify candidate genes which are known to interact with SLC39A4, we used GeneMANIA (https://genemania.org) to identify relevant protein-protein interactions, coexpressed genes, and genes of related function based on transcription and phenotypic screening profiles. The interaction databases used by GeneMANIA included BioGRID and Pathway Commons, both including primary research studies. Databases GeneCards and MalaCards were then used to obtain information on the identified genes (Sup Materials 3). The interacting genes were screened for rare deleterious variants which may have negatively impacted upon the function of SLC39A4 and thus the AE phenotype in our patient. With specific focus on exonic regions of the genes and nonsynonymous mutations, “R” analysis demonstrated two genes that were within acceptable ExAC allele frequency and CADD score range (<0.01, >10): GPAA1 and RECQL4, suggestive of significant interaction with SLC39A4 in developing the disease phenotype in B03.
4. Discussion
AE appears to occur mainly in France and Tunisia. The p.Gly512Trp variant was previously identified in a Tunisian family [1]. All published mutations in the human SLC39A4 gene have been collated (Table 2). In the majority of cases, pathogenic variants were present in a homozygous or compound heterozygous state in AE. Families identified across a number of countries with AE have been shown to carry over 30 different mutations in SLC39A4, including deletions, nonsense, missense, and splice-site alterations [6]. Schmitt et al. [1] reported mutations including p.Gly512Trp and p.549delLeu affecting amino acids that were highly conserved between a number of species. Variants p.Gly512Trp and p.549delLeu were detected in homozygous state in Tunisian and Swedish probands, respectively, and the toxic mutation was not identified in any of 164 control chromosomes from North Africa included in their study. Moreover, there was perfect segregation between the genotype and phenotype in the pedigree suggesting a high likelihood for AE to be associated with SLC39A4 gene. As previously described by Abu-Duhier et al. [3], the mutation was located in a putative transmembrane domain and likely to alter zinc absorption by reducing transcription/ translation of SLC39A4. Heterozygous individuals tend to be asymptomatic carriers rather than manifesting the disease phenotype [7]. A significant feature of AE overall and our family in particular is variable phenotype even between individuals with the same genetic change in SLC39A4 [3]. It is anticipated the AE phenotype could be dependent on either modifier genes or an unknown putative AE gene. Hence we have screened for additional genetic modifiers of SLC39A4 function within an individual with a particularly severe AE phenotype [3].
Table 2.
Modified table from Schmitt et al. (2009) on genetic mutations identified up to date with AE associated SLC39A4 gene. Outlined in bold is the mutation of interest that has been identified in a Tunisian family. Note. Some reports failed to provide adequate information to complete the table.
| Exon | Mutation (Nucleotide) | Influence on Amino Acid | Clinical Significance | Other findings | Study |
|---|---|---|---|---|---|
| Exon 6 | c.1191insC | p.Gln398fsX18 | Pathogenic | (i) Homozygous (ii) Frameshift |
Coromilas et al. 2011 |
|
| |||||
| Exon 5 | c.850G>A | p.Glu284Lys | Likely/benign | (i) Missense (ii) France (iii) Homozygous |
Kury et al., 2002 |
|
| |||||
| Exon 1 | c.184T>C | p.Cys62Arg | Uncertain Significance | (i) Missense (ii) May affect protein conformation due to loss of disulfide bond (iii) Tunisia (iv) Homozygous |
Kury, et al. 2003 |
|
| |||||
| Exon 1 | c.143T>G | p.Leu48X | Likely pathogenic | (i) Nonsense (ii) Homozygous (iii) Tunisia (iv) Lacks putative zinc binding site |
Kury et al. 2003 Kharfi et al. 2005 Nakano et al. 2003 |
|
| |||||
| Intron 1 | c.192+19G>A | Donor splice site error (possibly) |
Likely /Pathogenic |
(i) France (ii) Compound heterozygous/homozygous (iii) Possibly altering transcripts through mis-splicing |
Kury et al. 2002 |
|
| |||||
| Exon 2 | c.283C>T | p.Arg95Cys | Pathogenic | (i) Missense (ii) Japanese (iii) Compound heterozygous (iv) Abolishes restriction enzyme site for Faul |
Nakano et al. 2003 |
|
| |||||
| Exon 2 | c.318C>A | p.Asn106Lys | Pathogenic | (i) Missense (ii) France (iii) Compound Heterozygous (iv) Deletion in one allele – failed expression of gene |
Wang et al. 2002 |
|
| |||||
| Intron 2 | c.475-2A>G | Acceptor splice site error (possibly) | Uncertain Significance | (i) Nonsense (ii) Homozygous (iii) France (iv) Appearance of premature stop codon |
Kury et al. 2003 |
|
| |||||
| Exon 2 | c.251C>T | p.Pro84Leu | Likely Benign | (i) Missense (ii) Various countries (iii) Possibly compound heterozygous (iv) Various amino acid change |
Wang et al. 2002 |
|
| |||||
| Exon 3 | c.511G>T | p.Val171Leu | Uncertain Significance | (i) Missense (ii) Heterozygous (iii) Caucasian |
Schmitt et al. 2009 |
|
| |||||
| Exon 3 | c.599C>T | p.Pro200Leu | Pathogenic | (i) Missense (ii) reduces Vmax/alter protein folding (iii) France + Austria (iv) Compound heterozygous/homozygous |
Kury et al. 2002 |
|
| |||||
| Exon 3 | c.631C>T | p.Gln211X | Likely Pathogenic | (i) Nonsense (ii) Truncated protein (iii) Tunisia (iv) Homozygous |
Meftah et al. 2006 |
|
| |||||
| Exon 3 | c.641_642ins10 | p.Ser214ArgfsX30 | Unknown | (i) Frameshift | Santiago et al. 2011 |
|
| |||||
| Exon 4 | c.766delC | p.Leu256SerfsX16 | Likely Pathogenic | (i) Deletion (ii) Spanish (iii) Truncated protein (iv) Heterozygous |
Schmitt et al. 2009 |
|
| |||||
| Exon 4 | c.751C>T | p.Arg251Trp | Benign | (i) Missense (ii) Homozygous (iii) France |
Wang et al. 2002 |
|
| |||||
| Exon 5 | c.850G>A | p.Glu284Lys | Likely/benign | (i) Missense (ii) Homozygous |
Kury et al. 2003 |
|
| |||||
| Exon 5 | c.909G>C | p.Gln303His | Pathogenic | (i) Missense (ii) Homozygous (iii) Substitution of highly conserved amino acid (iv) Japan |
Nakano et al. 2003 |
|
| |||||
| Exon 5 | c.926G>A | p.Cys309Tyr | Unknown | (i) Missense | Wang et al. 2002 |
|
| |||||
| Exon 5 |
c.968_971del AGTC |
p.Ser324ArgfsX24 | Likely/ Pathogenic |
(i) Frameshift (ii) France (iii) Compound Heterozygous (iv) Alter protein function |
Kury et al. 2002 |
|
| |||||
| Exon 6 | c.989G>A | p.Gly330Asp | Likely Pathogenic | (i) Cellular mislocalization (ii) Missense (iii) Egypt (iv) Homozygous |
Wang et al. 2002 |
|
| |||||
| Exon 6 | c.1016_1017ins53 | p.Thr357AlafsX10 | Unknown | (i) Frameshift (ii) Premature termination codon (iii) Heterozygous (iv) Japan |
Nakano et al. 2003 |
|
| |||||
| Exon 6 | c.1115T>C | p.Leu372Pro | Likely Pathogenic | (i) Reduced protein levels (ii) Missense (iv) Egypt (iv) Homozygous |
Wang et al. 2002 |
|
| |||||
| Exon 6 | c.1120G>A | p.Gly374Arg | Pathogenic | (i) Reduced protein levels (ii) Missense (iii) France (iv) Homozygous |
Kury et al. 2002 |
|
| |||||
| Exon 6 | c.1141A>G | p.Thr381Ala | Uncertain Significance | (i) Missense (ii) Heterozygous (iii) Caucasian |
Schmitt et al. 2009 |
|
| |||||
| Exon 6 | c.1115T>G | p.Leu372Arg | Unknown | (i) Missense | Li et al. 2010 |
|
| |||||
| Intron 6 | c.1150-2A>G | Acceptor splice site error (possibly) | Uncertain significance | (i) Homozygous (ii) France |
Wang et al. 2002 |
|
| |||||
| Exon 7 | c.1203G>A | p.Trp401X | Likely/ pathogenic |
(i) Nonsense (ii) Compound Heterozygote (iii) Austria (iv) Absence of zinc binding site |
Kury et al. 2003 |
|
| |||||
| Exon 7 | c.1223delC | p.Ala408fsX481 | Unknown | (i) Frameshift | Vardi et al. 2009 |
|
| |||||
| Exon 7 | c.1223_1227delCCGGG | p.Trp411ArgfsX7 | Uncertain significance | (i) Frameshift (ii) Founder Effect (iii) Tunisian (iv) Homozygous |
Kury et al. 2002 |
|
| |||||
| Exon 7 | c.1229T>C | p.Leu410Pro | Uncertain Significance | (i) Missense | Wang et al. 2002 |
|
| |||||
| Intron 7 | c.1287+2T>C | Acceptor splice site error (possibly) | Uncertain Significance | Park et al. 2010 | |
|
| |||||
| Exon 9 | c.1438G>T | p.Glu480Sto | Unknown | Stop | Nakano et al. 2009 |
|
| |||||
| Exon 9 | c.1462_147411+delAGACTGAGCCCAGG | p.Arg488SerfsX2 | Unknown | (i) Frameshift | Wang et al. 2008 |
|
| |||||
| Exon 10 | c.1534G>T | p.Gly512Trp | Pathogenic |
(i) Missense (ii) Tunisia (iii) Homozygous (iv) Affect amino acids |
Schmitt et al. 2009 |
|
| |||||
| Exon 10 | c.1576G>A | p.Gly526Arg | Pathogenic | (i) Reduces Vmax (ii) Missense (iii) France (iv) Homozygous |
Kury et al. 2002 |
|
| |||||
| Exon 11 | c.1784G>T | p.Gly595Val | Uncertain significance | (i) Missense (ii) Tunisia |
Kharfi et al. 2010 |
|
| |||||
| Exon 11 | c.1646_1648delTGC | p.549delLeu | Pathogenic | (i) Deletion (ii) Sweden (iii) Homozygous (iv) Affect amino acids |
Schmitt et al. 2009 |
|
| |||||
| Exon 12 | c.1888G>C | p.Gly630Arg | Pathogenic for mental retardation/X-Linked | (i) Reduced protein levels (ii) Missense (iii) Homozygous (iv) Jordan |
Wang et al. 2002 |
Our data suggests that the p.Gly512Trp change is the likely cause of disease in our family because it is the only homozygous, rare, and predicted deleterious change within a zinc transporter gene which is present within the affected patient and absent (or heterozygous) in unaffected family members. In addition we identified two rare and predicted deleterious genetic variants in SLC39A4 interacting partners which may have a role in the development of the particularly severe AE phenotype in our patient: RECQL4 and GPAA1 (Table 4). Interestingly Nistor et al. [8] reported that the complete clinical triad of features in AE was only documented in 20% of patients, questioning if only one gene is responsible and if modifier genes could be involved in the variable clinical characteristics. Patients with mutation in the RECQL4 gene have several characteristic features similar to AE; for example, Bernstein et al. [9] reported a RECQ disorder: Bloom syndrome, a rare autosomal recessive condition, which presents with mental retardation, immunodeficiency, male infertility, and increased chance of cancer. Similar features occur in some patients with AE, in the form of immunodeficiency, mental retardation, and infertility. In addition, Mann et al. [10] identified distinctive skin abnormalities in a mutant RECQL4 mouse model. Although the GPAA1 gene is known to be expressed in skin (Table 3), no disorder has been identified to date that has been directly affected by GPAA1 genetic mutation, although rare cases have been reported in which GPAA1 gene amplification and RNA and protein overexpression occurred in hepatocellular carcinoma [11]. Further research is required to explore a putative link between GPAA1 and RECQL4 mutations and AE.
Table 4.
GPAA1 and RECQL4 genes both demonstrated a likelihood of being modifier genes of SLC39A4 in developing AE in B03. Note. Please see Sup Materials 3 for “R” software version of complete list of interacting partners with SLC39A4 gene for B03.
| Gene | Chromosome | Start-End | Exon | Nucleotide substitution | Protein substitution | EXAC Frequency | CADD Score |
|---|---|---|---|---|---|---|---|
| GPAA1 | Chr8 | 145140564 | 11 | c.G1540A | p.A514T | 0.0053 | 15.04 |
| RECQL4 | Chr8 | 145740364 | 9 | c.C1576T | p.L526F | 0.0007 | 16.37 |
Table 3.
GeneMANIA presented the interacting partners with SLC39A4 gene. The databases GeneCards and MalaCards and the literature were reviewed to identify the function, disorder, and tissue expression. Note. The documented information only includes the main signs, symptoms, and tissue expression.
| Gene | Description | Disease | Function | Signs/Symptoms | Tissue Expression |
|---|---|---|---|---|---|
| SLC39A4 | Solute Carrier Family 39 Member 4 | Acrodermatitis enteropathica | Encodes for a ZIP family. Required for zinc uptake in the intestine. | Growth retardation, immune-system dysfunction, alopecia, diarrhea, dermatitis. | Mostly Lungs and intestine. Overexpressed in small intestine, fetal gut and CD8 Tcells. |
|
| |||||
| TSTA3 | Tissue Specific Transplantation Antigen P35B |
Leukocyte adhesion deficiency, type II | Catalyzes the reactions epimerase and reductase in GDP-D-mannose. | Infections, persistent leukocytosis, mental and growth retardation. | Esophagus, stomach and pancreas. Overexpressed in oral epithelium and breast |
|
| |||||
| SCRIB | Scribbled Planar Cell Polarity Protein | Neural tube defects. Tick-Borne Encephalitis. | Regulates epithelial and neuronal morphogenesis. | Cleft lip, myelocystocele, urinary incontinence, hydrocephalus and lipomas. | Intestine, Ovary and Testis. Overexpressed in pancreas. |
|
| |||||
| GPAA1 | Glycosylphosphatidylinositol Anchor Attachment | N/A | Links proteins to cell surface membrane. | N/A | Nervous system, skin, lungs. Overexpressed in Nasal epithelium. |
|
| |||||
| CYC1 | Cytochrome C1 | Mitochondrial complex III deficiency | Mediates the transfer of electron from Rieske iron sulphur protein to cytochrome. | Lactic acidosis, infection, insulin-responsive hyperglycemia and ketoacidosis. | Lungs, Skin, Nervous system. Overexpressed in heart. |
|
| |||||
| RECQL4 | RecQ Like Helicase 4 | Baller-Gerold Syndrome. RAPADILINO Syndrome. | DNA helicases unwind double stranded DNAs and may modulate chromosome segregation. | Fusion of bones (neonates), slow growth, missing/malformed kneecaps. | Ubiquitously expressed. Overexpressed in Testis and Thymus. |
|
| |||||
| EXOSC4 | Exosome Component 4 | N/A | Participates in cellular RNA processing and degradation. | N/A | Skin. Overexpressed in whole blood, testis and breast. |
Our data supports pathogenicity of the p.Gly512Trp change in SLC39A4, but in addition we have identified potentially deleterious variants in SLC39A4 interacting partners which may be important to disease pathogenesis and are potential therapeutic targets.
Acknowledgments
This work was funded by University of Tabuk Prince Fahd Bin Sultan Research Chair in collaboration with University of Sheffield. Research contact grant number is R/130649-11-1. The authors are grateful to the family from Tabuk for participating in this study. The authors also thank Sheffield Diagnostic Genetics Service, Sheffield Children's NHS Trust, for genome sequencing.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
This work complies with ethical standards. The study was approved by University of Tabuk and University of Sheffield ethical committees.
Consent
The participants gave their consent after they received verbal and written explanation including patient information. Individual consent has been signed by each individual.
Disclosure
Faisal Abu-Duhier is the Dean of the Faculty of Applied Medical Science; Andrew Messenger and Andrew McDonagh are Consultant Dermatologists at Sheffield Teaching Hospitals; Youssef Bakri is the Head of the Biology Department, Faculty of Sciences, Rabat; and Vivetha Pooranachandran, Paul Heath, Johnathon Cooper-Knock, and Rachid Tazi-Ahnini are Research Scientists at the University of Sheffield. The authors authorised the publication of this manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Rachid Tazi-Ahnini and Faisel Abu-Duhier were the principal investigators. Faisel Abu-Duhier was responsible for the acquisition of clinical materials. Faisel Abu-Duhier, Paul R. Heath, and Rachid Tazi-Ahnini were responsible for the concept and design of the study. Vivetha Pooranachandran, Rachid Tazi-Ahnini, and Paul R. Heath were responsible for the analysis and interpretation of data. Vivetha Pooranachandran and Johnathan Cooper-Knock were involved in designing software for genetic analysis. Andrew J. G. McDonagh, Andrew G. Messenger, Youssef Bakri, and Rachid Tazi-Ahnini drafted the manuscript. All authors participated in the study design, critical revision, and final approval of the manuscript. Paul R. Heath and Rachid Tazi-Ahnini contributed equally to this work.
Supplementary Materials
Supplementary materials include codes derived for “R” software analysis of the genome data, code used to identify the mutated zinc transporter genes in B03 sample, and code used to identify interacting partners with SLC39A4 gene in causing the disease in B03.
References
- 1.Schmitt S., Küry S., Giraud M., Dréno B., Kharfi M., Bézieau S. An update on mutations of the SLC39A4 gene in acrodermatitis enteropathica. Human Mutation. 2009;30(6):926–933. doi: 10.1002/humu.20988. [DOI] [PubMed] [Google Scholar]
- 2.Maverakis E., Fung M. A., Lynch P. J., et al. Acrodermatitis enteropathica and an overview of zinc metabolism. Journal of the American Academy of Dermatology. 2007;56(1):116–124. doi: 10.1016/j.jaad.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Abu-Duhier F., Lovewell T., McDonagh A., Messenger A., Ibrahimi A., Tazi-Ahnini R. First report of SLC39A4 mutation in acrodermatitis enteropathica family from the Middle East. International Journal of Dermatology. 2017;56(5):e97–e100. doi: 10.1111/ijd.13463. [DOI] [PubMed] [Google Scholar]
- 4.Garza-Rodríguez V., De La Fuente-García A., Liy-Wong C., et al. Acrodermatitis enteropathica: A novel SLC39A4 gene mutation in a patient with normal zinc levels. Pediatric Dermatology. 2015;32(3):e124–e125. doi: 10.1111/pde.12555. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi Y., Yang S., Nykamp K., Garcia J., Lincoln S. E., Topper S. E. Pathogenic variant burden in the ExAC database: An empirical approach to evaluating population data for clinical variant interpretation. Genome Medicine. 2017;9(1, article no. 13) doi: 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Küry S., Kharfi M., Kamoun R., et al. Mutation spectrum of human SLC39A4 in a panel of patients with acrodermatitis enteropathica. Human Mutation. 2003;22(4):337–338. doi: 10.1002/humu.9178. [DOI] [PubMed] [Google Scholar]
- 7.Nakano A., Nakano H., Nomura K., Toyomaki Y., Hanada K. Novel SLC39A4 mutations in acrodermatitis enteropathica. Journal of Investigative Dermatology. 2003;120(6):963–966. doi: 10.1046/j.1523-1747.2003.12243.x. [DOI] [PubMed] [Google Scholar]
- 8.Nistor N., Ciontu L., Frasinariu O.-E., Lupu V. V., Ignat A., Streanga V. Acrodermatitis Enteropathica. Medicine (United States) 2016;95(20) doi: 10.1097/MD.0000000000003553.e3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein K. A., Gangloff S., Rothstein R. The RecQ DNA helicases in DNA repair. Annual Review of Genetics. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mann M. B., Hodges C. A., Barnes E., Vogel H., Hassold T. J., Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Human Molecular Genetics. 2005;14(6):813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 11.McKillop I. H., Moran D. M., Jin X., Koniaris L. G. Molecular pathogenesis of hepatocellular carcinoma. Journal of Surgical Research. 2006;136(1):125–135. doi: 10.1016/j.jss.2006.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials include codes derived for “R” software analysis of the genome data, code used to identify the mutated zinc transporter genes in B03 sample, and code used to identify interacting partners with SLC39A4 gene in causing the disease in B03.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.