Abstract
Antimicrobial resistance is an emerging problem in both humans and animals due to misuse and excessive use of drugs. Resistance in commensal E. coli isolates can be used to predict emergence of resistance in other gut microflora. The aim of this study is to determine the phylogenetic groups and antimicrobial resistance patterns of E. coli from healthy chickens in Uganda. The phylogenetic grouping of 120 fecal E. coli isolates from eastern and central Uganda was derived using the triplex PCR assay and their susceptibility patterns determined by agar disc diffusion method to 5 antimicrobial drugs. Most E. coli is segregated into phylogenetic group A comprising 84%, while 12% and 4% were in groups D and B1, respectively. Similarly most E. coli from central (87%) and eastern Uganda (82%) belonged to group A. Overall, 85 (70%) of E. coli were resistant to antimicrobial drugs, of which 72/101 (70%) are in PG A, 10 of 14 (71.4%) in PG D, and 3 of 5 (60%) in PG B1. Significantly, most of the isolates in PG A from both central (66.7%) and (60.6%) eastern Uganda were resistant to one antimicrobial. Resistance to tetracycline alone or in combination with other drugs for central and eastern Uganda in PG A is 51% and 55%, respectively. Multidrug resistance to tetracycline and ciprofloxacin or nalidixic acid was 10% and 18% in isolates from central and 10% and 12% in isolates from eastern region, respectively. Phylogenetic group A accounts for most of the E. coli in chicken from Uganda. No difference in the resistance rates between the phylogenetic groups of E. coli has been observed. The high prevalence of resistant E. coli strains from different phylogenetic groups in healthy chickens suggests antimicrobial drug selection pressure due to excessive drug in the rearing layer chickens.
1. Background
E. coli is a commensal organism within the gastrointestinal tract of warm blooded animals. In the recent past, strains known to cause illness in animals and humans have emerged [1, 2]. In chicken, pathogenic strains cause respiratory infections, pericarditis, septicemia [3], and colibacillosis [4, 5]. E. coli is a ubiquitous organism. Its adaptation to the diverse ecological niches including the intestinal and extraintestinal sites, as well as sites outside the host [6], is aided by the flexibility of the genome; exchange, retention, and/or loss of accessory genetic elements takes place [6, 7].
Antimicrobial drugs are commonly used in Uganda and other countries to prevent and treat diseases and as growth promoters in poultry proudction [8]. Equivocally, indiscriminate drug use exerts high antibiotic selection pressure on chicken gut coliforms which leads to emergence of antibiotic-resistant E. coli phenotypes [9, 10] and shed in faeces [11]. The presence of resistant E. coli is a strong predictive indicator for emergence of resistance in other organisms (pathogenic and nonpathogenic) within gastrointestinal tract of the chicken [11].
The genetic background of E. coli reflects its evolutionary lineage [12] and strains that evolved along distinct lineages carry specific genetic backgrounds [13]. According to Clermont et al. [14], E. coli segregate into the four major phylogenetic groups, namely, A, B1, B2, and D. The commensal strains belong mainly to phylogenetic groups A and B1 [15]. Strains with phylogenetic groups B2 and D carry virulence determinants [15, 16]. Phylogenetic studies are important to improve the understanding of the lineages of E. coli population; however, such information is not available for E. coli strains in chicken from Uganda. Therefore the study investigates the genetic background and the occurrence of antimicrobial resistance of E. coli from healthy chickens in Uganda.
2. Materials and Methods
2.1. Bacterial Isolates
Previously archived E.coli strains from healthy chicken in central and eastern Uganda collected from May 2010 to September 2011 were used. These had been stored in microbiology laboratory of College of Veterinary Medicine, Animal Resources and Biosecurity, Makerere University. One hundred twenty isolates were identified as E. coli by the standard biochemical tests [17].
2.2. Genomic DNA Was Extracted
Bacterial genomic DNA was extracted using the rapid boiling method described by Wang et al. (2010). A single colony of E. coli was grown overnight on Brain Heart Agar (Oxoid™) for 24 hours at 37°C. A loop-full of colonies was suspended in 0.5 ml of double distilled sterile water; cells were lysed at 95°C for 10 minutes. After cooling to room temperature, the suspension was centrifuged at 12,000 rpm for 3 minutes to remove cell debris. The supernatant containing template DNA was used for PCR.
2.3. Phylogenetic Typing
Triplex PCR-based method as described by Clermont et al. [14] was used. All strains were assigned to 1 of the 4 major E. coli phylogenetic groups (A, B1, B2, and D). The E. coli K-12 (phylogroup A), STEC O111 (phylogroup B1), and O157:H7 (phylogroup D) were used as positive controls.
2.4. Antimicrobial Susceptibility Test
Antimicrobial susceptibility testing was performed on E. coli isolates using Kirby-Bauer disc diffusion method [18] as recommended by CLSI [19]. It was carried out on Mueller-Hinton agar to chloramphenicol 30 μg, nalidixic acid 30 μg, ciprofloxacin 5 μg, gentamicin 10 μg, and tetracycline 30 μg (Oxoid), which are frequently used in poultry production. The mean zone of inhibition of three replicates was used to determine the susceptibility of the isolates [19]. Escherichia coli ATCC 25922 was used as control strain.
3. Results
3.1. Phylogenetic Distribution of E. coli from Chicken in Eastern and Central Uganda
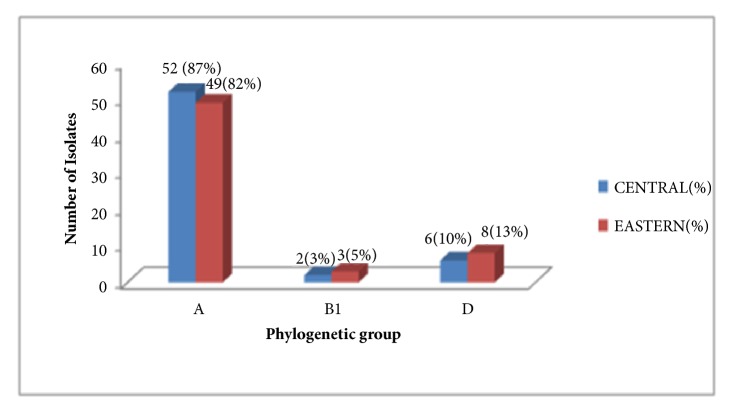
Most E. coli is segregated into phylogenetic group A comprising 84% (101 of 120), while 12% (14) and 45(5) of the isolates were in groups D and B1, respectively. Similarly a majority of E. coli from central Uganda, 52 (87%), was segregated into phylogenetic group A, 6 (10%) in D, and 2 (3.3%) in B1; whereas, for eastern Uganda, 49 (82%) were segregated in A, 3 (5%) in B1, and 8 (13%) in D. None of the E. coli in phylogenetic group B2 was detected (Figures 1 and 2).
Figure 1.
Phylogenetic distribution of E.coli from chicken in Eastern and Central Uganda.
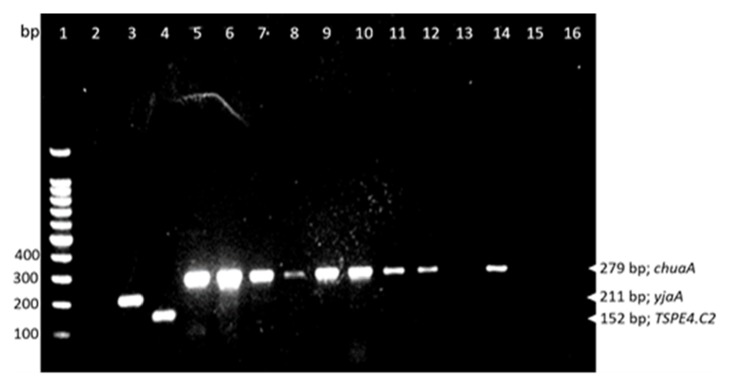
Figure 2.
PGs of selected E.coli isolates from chicken in Uganda. Lane 1, hyper ladder (100bp DNA ladder, Promega Madison, USA); Lane 2, negative control, no DNA template); Lane 3, E. coli K-12 (PG, A); Lane 4, STEC 0111 (PG, B1); Lane 5, 0157:H7 (PG, D); Lanes 6-12 and 14, E.coli with PG-D; Lanes 13,15, and 16, E. coli with PG-A.
3.2. Antibiotic Resistance Profiles of E.coli Isolates in relation to Phylogenetic Groups
Overall, 85 of 120 (70%) E. coli isolates were resistant to antimicrobial drugs. With respect to phylogenetic grouping, 72/101 (70%) in PG A, 10 of 14 (71.4%) in PG D, and 3 of 5 (60%) in PG B1 were resistant. Most of the isolates in PG A from both central (66.7%) and (60.6%) eastern Uganda were resistant to one antimicrobial drug compared to being resistant to two or more antimicrobials (Figure 3).
Figure 3.
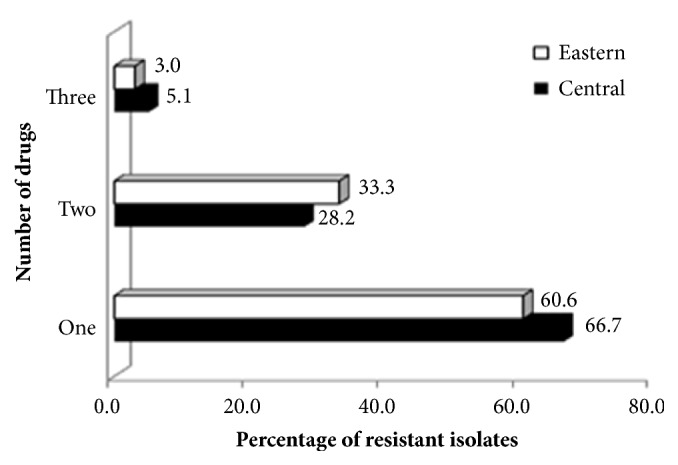
Resistant E. coli isolates to one or multiple antimicrobial drugs.
As regards phylogenetic group A, of the 72 E. coli that are resistant, 86% (62/72) were resistant to tetracycline, 22%, 21%, and 8% were resistant to ciprofloxacin, nalidixic acid, and chloramphenicol, respectively. A total of 39 of 49 (80%) and 33 of 52 (63%) E. coli isolates from central and eastern Uganda, respectively, were resistant. Of these, 20 of 39 (51%) and 18 of 33 (55%) from central and eastern Uganda, respectively, showed resistance to tetracycline alone or in combination with other drugs (Table 1). Multidrug resistance to tetracycline and ciprofloxacin or nalidixic acid was 10% and 18% in isolates from central and 10% and 12% in isolates from eastern region, respectively. Five and 3% of the isolates from central and eastern regions, respectively, were resistant to a combination of tetracycline, ciprofloxacin, and nalidixic acid (Table 1).
Table 1.
Antibiotic resistance profiles of E. coli isolates in phylogenetic groups A and D from central and eastern Uganda.
| Number and percentage [ ] of resistant E. coli isolates in phylogenetic groups A and D | ||||
|---|---|---|---|---|
| Antimicrobial resistance profile | Phylogenetic group A | Phylogenetic group D | ||
|
| ||||
| Central | Eastern | Central | Eastern | |
| N= 49 | N=52 | N=6 | N=8 | |
| T | 20 [51.3] | 18 [55] | 2 [50] | 1 [16.7] |
| C | 3 [7.7] | 0.0 | 0 | 1 [16.7] |
| CIP | 2 [5] | 1 [3] | 0 | 1 [16.7] |
| NA | 1 [2.6] | 2 [6] | 0 | 0 |
| T+NA | 4 [10] | 4 [12] | 1 [25] | 1 [16.7] |
| T+CIP | 4 [10] | 6 [18] | 0 | 0 |
| T+C | 3 [7.7] | 0.0 | 1 [25] | 0 |
| NA+C | 0.0 | 1 [3] | 0 | 0 |
| T+NA+CIP | 2 [5] | 1 [3] | 0 | |
| T+NA+C | 0 | 0 | 0 | 1[16.7] |
| T+CIP+C | 0 | 0 | 0 | 1[16.7] |
| Total number and %age of resistance | 39 [65] | 33 [55] | 4 [66.7] | 6 [75] |
T, tetracycline; C, chloramphenicol; CIP, ciprofloxacin; NA, nalidixic acid
Resistance was second highest for ciprofloxacin in the central region with 20% resistant to ciprofloxacin alone (5%), in combination with tetracycline (10%), or with other two drugs (5%). Similarly, 24% from eastern region were resistant to ciprofloxacin, of which majority (18%) were also resistant to tetracycline. Resistance to nalidixic acid alone or in combination with other drugs was observed in 24% and 17.6% of isolates from eastern and central regions, respectively. Significantly, 15.5% of isolates from central regions were resistant to chloramphenicol and none from the eastern region (Table 1).
Of the 10 resistant isolates in PG D, 4 (40%) from central Uganda were all resistant to tetracycline, while two were in addition resistant to chloramphenicol or nalidixic acid. Similarly all 6 (60%) of isolates from eastern region were resistant to tetracycline and two of these were also resistant to chloramphenicol and ciprofloxacin or nalidixic acid, while in PG 3 of 5 isolates in PG B1 were also resistant to ciprofloxacin (1 isolate) from central and tetracycline alone or in combination with chloramphenicol for each isolate from eastern Uganda (Table 1).
4. Discussion
Phylogenetic grouping was determined using a simple, quick, reproducible assay [14] that is most suitable for resources limited laboratories. The different strains of E. coli were predominantly separated into phylogenetic group A and then D, but none in B2. This comparable phylogenetic distribution of E. coli in chicken in both central and eastern regions of Uganda shows that E. coli within the chicken population evolved from a recent common ancestry and have established mutual coexistence over millions of years. Phylogenetic group A isolates were predominate in our study which considered healthy chicken. More often, phylogenetic group A E. coli isolates are commensal organisms, associated with healthy chicken, and rarely cause disease [20, 21]. Our findings were in agreement with other workers [21, 22]. Conversely, in birds with colibacillosis a majority of E. coli belongs to phylogenetic groups B2 [23]. Similarly, the proportion of phylogenetic group B1 E. coli isolates was the least in strains from chicken in this study. The majority of E. coli from poultry in this group are usually Enteropathogenic E. coli isolates [20, 21]. Phylogenetic group B2 was absent as expected because E. coli in this group are virulent strains of E. coli that usually cause infections in chicken [23]. Phylogenetic group D was observed as the second most isolated group of E. coli from chicken in Uganda. However, it must be noted that the phylotyping method used in [14] could not distinguish between E. coli isolates in groups D and E [24], the latter being associated with severe illness in humans [24]. Thus, there is a need for future studies to confirm the absence or presence of E.coli in group E from chicken in central and eastern Uganda.
In this study we examined antimicrobial resistance in commensal E. coli isolates from healthy layer chickens from central and eastern Uganda. More than 70% of the isolates were resistant, showing that high prevalence of antimicrobial resistant bacteria in chicken in Uganda is widespread due to misuse of antimicrobials during rearing. Evidently, antibiotic drugs are readily available and administered by farmers without prescription [8, 25], factors that promote the emergence of resistance antibiotic in Uganda. The high level of E. coli resistance in chicken is a public health concern, as this organism has a high propensity to disseminate antimicrobial resistance genes to intestinal bacteria in the humans [26, 27]. This may also be an indicator of emerging resistance in other gut microflora within the chicken population.
The isolates were susceptible to gentamicin but more resistant to tetracycline, ciprofloxacin, nalidixic acid, and chloramphenicol. The highest resistance to tetracycline was likely due to extensive use of this drug during chicken rearing for preventive and curative purposes. Tetracycline resistance is easily promoted within E. coli population and among other gut microflora because tetracycline resistance genes are located on mobile genetic elements [27, 28]. No resistance was observed to gentamycin, since only injectable formulations are available and rarely used in chicken in Uganda. Similarly, formulations of chloramphenicol are no longer available for use in chicken and hence minimal resistance to this drug. However, the observed resistance can be due to coselection of chloramphenicol resistance by sulphonamides and streptomycin use [29], which are extensively used. Also horizontal transfer of genes from sources like water contaminated with human sewage may be another contributing factor [30]. We detected isolates resistant to more than two drugs; however, this was less frequent compared to other findings where most isolates were resistant to tetracycline, ciprofloxacin, and chloramphenicol [31].
Since the genome of E. coli is known to frequently exchange genetic elements including resistance genes [7] we attempted to link the antibiotic resistance pattern to the phylogenetic background. Our results, however, do not show any differences in antibiotic resistance of the strains in the different phylogenetic groups which is in agreement with other workers [24]. Conversely, other studies reported association between phylogenetic group B2 and quinolone-susceptible isolates [32, 33] whereas quinolone-resistant isolates were associated with group A in human isolates [32].
5. Conclusion
Phylogenetic group A accounts for most of the E. coli in chicken from Uganda. No difference in the resistance rates between the phylogenetic groups of E. coli has been observed. The high prevalence of resistant E. coli strains from different phylogenetic groups in healthy chickens suggests antimicrobial drug selection pressure due to excessive use in the rearing layer chickens. Rational use of antibiotics may reduce the chances of developing antibiotic-resistant E. coli in chickens from Uganda.
Acknowledgments
The authors acknowledge Mr. Musisi Lubowa, Microbiology Laboratory Manager, Makerere University College of Veterinary Medicine, Animal resources and Biosecurity, for providing the reference bacterial strains.
Data Availability
Data is available from the corresponding author on reasonable request.
Ethical Approval
Permission to carry out the research was granted by research and ethics committee of the College of Veterinary Medicine, Animal Resources and Biosecurity of Makerere University.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
Authors' Contributions
Winston Kabiswa, Ann Nanteza, Gabriel Tumwine, and Samuel Majalija were involved in the conception and design of the study. Ann Nanteza, Gabriel Tumwine, and Samuel Majalija supervised field work and laboratory studies. Winston Kabiswa was involved in data collection and writing manuscript. Winston Kabiswa, Gabriel Tumwine, and Samuel Majalija made substantial contribution in data analysis. Winston Kabiswa, Ann Nanteza Gabriel Tumwine, and Samuel Majalija assisted in writing and revising the manuscript
References
- 1.Mitchell N. M., Johnson J. R., Johnston B., Curtiss R., Mellata M. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Applied and Environmental Microbiology. 2015;81(3):1177–1187. doi: 10.1128/AEM.03524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu S., Jin D., Wu S., et al. Insights into the evolution of pathogenicity of Escherichia coli from genomic analysis of intestinal E. coli of Marmota himalayana in Qinghai–Tibet plateau of China. Emerging Microbes & Infections. 2016;5:p. e122. doi: 10.1038/emi.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matin M. A., Islam M. A., Khatun M. M. Prevalence of colibacillosis in chickens in greater Mymensingh district of Bangladesh. Veterinary World. 2017;10(1):29–33. doi: 10.14202/vetworld.2017.29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Carli S., Ikuta N., Lehmann F. K. M., et al. Virulence gene content in Escherichia coli isolates from poultry flocks with clinical signs of colibacillosis in Brazil. Poultry Science. 2015;94(11):2635–2640. doi: 10.3382/ps/pev256. [DOI] [PubMed] [Google Scholar]
- 5.Kabir S. M. L. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. International Journal of Environmental Research and Public Health. 2010;7(1):89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang J., Hur H.-G., Sadowsky M. J., Byappanahalli M. N., Yan T., Ishii S. Environmental Escherichia coli: ecology and public health implications—a review. Journal of Applied Microbiology. 2017;123(3):570–581. doi: 10.1111/jam.13468. [DOI] [PubMed] [Google Scholar]
- 7.Escobar-Páramo P., Clermont O., Blanc-Potard A.-B., Bui H., Le Bouguénec C., Denamur E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Molecular Biology and Evolution. 2004;21(6):1085–1094. doi: 10.1093/molbev/msh118. [DOI] [PubMed] [Google Scholar]
- 8.Sosa A. D. J., Amábile-Cuevas C. F., Byarugaba D. K., Hsueh P.-R., Kariuki S., Okeke I. N. Antimicrobial Resistance in Developing Countries. Springer; 2010. [DOI] [Google Scholar]
- 9.Oz T., Guvenek A., Yildiz S., et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Molecular Biology and Evolution. 2014;31(9):2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahmoudi-Aznaveh A., Bakhshi B., Najar-Peerayeh S., et al. Commensal E. coli as an Important Reservoir of Resistance Encoding Genetic Elements. International Journal of Enteric Pathogens. 2013;1(2):43–7. doi: 10.17795/ijep13516. [DOI] [Google Scholar]
- 11.Tello A., Austin B., Telfer T. C. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environmental Health Perspectives. 2012;120(8):1100–1106. doi: 10.1289/ehp.1104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eppinger M., Mammel M. K., Leclerc J. E., Ravel J., Cebula T. A. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proceedings of the National Acadamy of Sciences of the United States of America. 2011;108(50):20142–20147. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallman T. J., Ashton P. M., Byrne L., Perry N. T., et al. Applying phylogenomics to understand the emergence of Shiga-toxin-producing Escherichia coli O157:H7 strains causing severe human disease in the UK. Journal of Microbial Genomics. 2015 doi: 10.1099/mgen.0.000029. http://mgen.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clermont O., Bonacorsi S., Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Applied and Environmental Microbiology. 2000;66(10):4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poey M. E., Laviña M. Integrons in uropathogenic Escherichia coli and their relationship with phylogeny and virulence. Microbial Pathogenesis. 2014;77:73–77. doi: 10.1016/j.micpath.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Lee J. H., Subhadra B., Son Y. J., Kim D. H., et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic. Letters in Applied Microbiology. 2016;62(1):84–90. doi: 10.1111/lam.12517. [DOI] [PubMed] [Google Scholar]
- 17.Nolan L. K., Barnes H. J., Vaillancourt J. P., et al. Colibacillosis. In: Swayne D. E., Gilsson J. R., McDougald L. R., Nolan L. K., Suarez D. L., Nair V. L., editors. Diseases of Poultry. 13th. Ames, Iowa: Wiley-Blackwell; 2013. pp. 751–805. [Google Scholar]
- 18.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test. 2009. Accessed on 7th May 2018, http://www.asmscience.org/content/education/protocol/protocol.3189.at. [Google Scholar]
- 19.CLSI. Performance standards for antimicrobial susceptibility testing. Proceedings of the 17th Informational Supplement; 2007; Wayne, Ill, USA. Clinical Laboratory standards Institute; [Google Scholar]
- 20.Duriez P., Clermont O., Bonacorsi S., et al. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001;147(6):1671–1676. doi: 10.1099/00221287-147-6-1671. [DOI] [PubMed] [Google Scholar]
- 21.Coura F. M., Diniz S. A., Silva M. X., et al. Phylogenetic group of escherichia coli isolates from broilers in Brazilian Poultry Slaughterhouse. The Scientific World Journal. 2017;2017:7. doi: 10.1155/2017/5898701.5898701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiki M., Usui M., Akiyama T., et al. Phylogenetic grouping, epidemiological typing, analysis of virulence genes, and antimicrobial susceptibility of Escherichia coli isolated from healthy broilers in Japan. International Journal of Environmental Research and Public Health. 2014;7:89–114. doi: 10.1186/2046-0481-67-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akhtar F., Rabbani M., Muhammad K., et al. Phylogenetic grouping of the pathogenic E. coli isolated from commercial broiler chicken in Pakistan. Journal of Animal and Plant Sciences. 2016;26(5):1242–1246. [Google Scholar]
- 24.Clermont O., Christenson J. K., Denamur E., Gordon D. M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental Microbiology Reports. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 25.Ampaire L., Muhindo A., Orikiriza P., Mwanga-Amumpaire J., Bebell L., Boum Y. A review of antimicrobial resistance in East Africa. African Journal of Laboratory Medicine. 2016;5(1):p. 432. doi: 10.4102/ajlm.v5i1.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landers T. F., Cohen B., Wittum T. E., Larson E. L. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Reports. 2012;127(1):4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts M. C., Schwarz S. Tetracycline and phenicol resistance genes and mechanisms: Importance for agriculture, the environment, and humans. Journal of Environmental Quality. 2016;45(2):576–592. doi: 10.2134/jeq2015.04.0207. [DOI] [PubMed] [Google Scholar]
- 28.Kang Y., Li Q., Yin Z., et al. High diversity and abundance of cultivable tetracycline-resistant bacteria in soil following pig manure application. Scientific Reports. 2018;8(1489) doi: 10.1038/s41598-018-20050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Österberg J., Wingstrand A., Nygaard Jensen A., et al. Antibiotic Resistance in Escherichia coli from Pigs in Organic and Conventional Farming in Four European Countries. PLoS ONE. 2016;11(6):p. e0157049. doi: 10.1371/journal.pone.0157049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitema E. S., Kikuvi G. M., Wegener H. C., et al. An assessment of antimicrobial consumption in food producing animals in Kenya. Journal of Veterinary and Therapeutics. 2001;24(6):385–390. doi: 10.1046/j.1365-2885.2001.00360.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang X.-M., Liao X.-P., Zhang W.-J., et al. Prevalence of serogroups, virulence genotypes, antimicrobial resistance, and phylogenetic background of avian pathogenic escherichia coli in south of China. Foodborne Pathogens and Disease. 2010;7(9):1099–1106. doi: 10.1089/fpd.2010.0542. [DOI] [PubMed] [Google Scholar]
- 32.Röderova M., Halova D., Papousek I., et al. Characteristics of Quinolone Resistance in Escherichia coli Isolates from Humans, Animals, and the Environment in the Czech Republic. Frontiers in Microbiology. 2017;7 doi: 10.3389/fmicb.2016.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skurnik D., Lacheeb S., Bernede C., et al. Integrons and antibiotic resistance in phylogenetic group B2 escherichia coli. Microbial Drug Resistance. 2009;15(3):173–178. doi: 10.1089/mdr.2009.0918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available from the corresponding author on reasonable request.