Abstract
Homeobox genes are key toolkit genes that regulate the development of metazoans and changes in their regulation and copy number have contributed to the evolution of phenotypic diversity. We recently identified a whole genome duplication (WGD) event that occurred in an ancestor of spiders and scorpions (Arachnopulmonata), and that many homeobox genes, including two Hox clusters, appear to have been retained in arachnopulmonates. To better understand the consequences of this ancient WGD and the evolution of arachnid homeobox genes, we have characterized and compared the homeobox repertoires in a range of arachnids. We found that many families and clusters of these genes are duplicated in all studied arachnopulmonates (Parasteatoda tepidariorum, Pholcus phalangioides, Centruroides sculpturatus, and Mesobuthus martensii) compared with nonarachnopulmonate arachnids (Phalangium opilio, Neobisium carcinoides, Hesperochernes sp., and Ixodes scapularis). To assess divergence in the roles of homeobox ohnologs, we analyzed the expression of P. tepidariorum homeobox genes during embryogenesis and found pervasive changes in the level and timing of their expression. Furthermore, we compared the spatial expression of a subset of P. tepidariorum ohnologs with their single copy orthologs in P. opilio embryos. We found evidence for likely subfunctionlization and neofunctionalization of these genes in the spider. Overall our results show a high level of retention of homeobox genes in spiders and scorpions post-WGD, which is likely to have made a major contribution to their developmental evolution and diversification through pervasive subfunctionlization and neofunctionalization, and paralleling the outcomes of WGD in vertebrates.
Keywords: homeobox genes, development, gene duplication
Introduction
Developmental programs precisely orchestrate proliferation and differentiation to build multicellular organisms. Many of the key regulatory factors and pathways utilized in development are conserved between species like the Wnt and Delta/Notch signaling pathways and transcription factors (TF) such as those encoded by the homeobox genes (Reviewed by Carroll et al. 2005; Rokas 2008). Many studies in recent decades have shown that changes in the expression and copy number of these tool kit genes can lead to the evolution of phenotypic differences among species (Averof and Patel 1997; Stern 1998; Ronshaugen et al. 2002; Carroll et al. 2005; Liubicich et al. 2009; Werner et al. 2010; Guerreiro et al. 2013; Koshikawa et al. 2015; Kvon et al. 2016). Therefore, understanding the evolution of these genes can provide important insights into the development and evolution of metazoans.
The homeobox genes encode a large superclass of TFs (Garcia-Fernandez 2005; Hoegg and Meyer 2005; Pascual-Anaya et al. 2012; Holland 2015; Ferrier 2016). They are characterized by encoding a homeodomain, which is usually 60 amino acids in length and folds to form a structure with three α-helices and an N-terminal domain (Ortiz-Lombardia et al. 2017). The third α-helix and N-terminal domain confer the specificity to the binding of the homeodomain to the major and minor groove of the DNA double helix, respectively (Hanes and Brent 1991; Chu et al. 2012; Ortiz-Lombardia et al. 2017). This conservation of sequence facilitates the characterization of many homeobox genes based solely on their homeodomain sequence (Holland et al. 2007), although there are also a variety of other DNA binding domains found in metazoan homeobox genes, which provide additional identification characteristics and biological functions (Burglin and Affolter 2016).
During the evolution of metazoans the expansion of homeobox gene number via duplication has been associated with multicellularity and the increase in morphological complexity (Garcia-Fernandez 2005; Hoegg and Meyer 2005; Pascual-Anaya et al. 2012; Holland 2015). The initial multiplication and divergence of proto-homeobox genes started early in evolution and created several classes of homeobox genes (Pascual-Anaya et al. 2012; Ferrier 2016). In the urbilaterian, the homeobox genes are hypothesized to have formed a large “Giga-homeobox” cluster, containing several homeobox families (Ferrier 2016). In metazoans, this Giga-cluster also included the metazoan specific ANTP class of homeobox genes (Ferrier 2016). Subsequent tandem duplications of each of the different classes generated clusters of similar homeobox class genes such as the ParaHox, SuperHox, SINE/Six, TALE/Irx, PRD/HRO clusters (Ferrier 2016). These clusters were then fragmented in the genome of the bilaterian ancestor, and have been subject to lineage specific retention, loss, and further duplication during bilaterian evolution (Ferrier 2016).
We recently found that in arachnids there had been a whole genome duplication (WGD) in a common ancestor of arachnopulmonates (spiders, scorpions, and Pedipalpi [Uropygi and Amblypygi]; Sharma, Kaluziak, et al. 2014; Schwager et al. 2017). Like the independent WGDs in vertebrates, after this event many duplicated homeobox genes have been retained in spiders and scorpions, including two clusters of Hox genes (Lynch et al. 2006; Putnam et al. 2008; Cao et al. 2013; Sharma, Schwager, et al. 2014; Di et al. 2015; Qu et al. 2015; Sharma et al. 2015; Schwager et al. 2017). Furthermore, divergence in the expression of ohnologs in spiders, including the Hox genes, suggests there has been neofunctionalization and subfunctionalization of many of these genes since the WGD (Pechmann et al. 2015; Turetzek et al. 2016, 2017; Schwager et al. 2017).
Here, we systematically compare the repertoires of homeobox genes between the arachnopulmonates with an ancestral WGD, the spiders Parasteatoda tepidariorum and Pholcus phalangioides, and the scorpions Centruroides sculpturatus and Mesobuthus martensii (Di et al. 2015), with arachnids that have no evidence for an ancestral WGD, the harvestman Phalangium opilio, the pseudoscorpions Neobisium carcinoides and Hesperochernes sp., and the tick Ixodes scapularis, as well as several mandibulate arthropods. We find pervasive duplication and retention of homeobox genes in arachnopulmonates, and synteny analysis of homeobox genes in P. tepidariorum also revealed several more duplicated ancient homeobox clusters (Ferrier 2016), in addition to the Hox clusters. To explore the fate and role of these duplicated genes further we compared the expression profiles of ohnologs during spider embryogenesis and found striking differences in their levels and temporal expression. Furthermore, comparison of the spatial expression of duplicated homeobox genes between P. tepidariorum and their single copy homologues in P. opilio suggests that there has been extensive neofunctionalization and subfunctionalization in embryogenesis during arachnopulmonate evolution. Taken together, our work shows that WGD greatly expanded the repertoire of homeobox genes in arachnopulmonates and that this contributed to diversification in their developmental gene regulatory networks and may have contributed to evolutionary innovations in these animals as has been postulated in other animal lineages (Van de Peer et al. 2009; Huminiecki and Conant 2012).
Results
Comparison of Homeobox Gene Families in Arachnids and Other Arthropods
To systematically identify homeobox repertoires we searched for the characteristic homeodomain sequence in a range of available and new arachnid transcriptomes. In a transcriptome of the spider P. phalangioides (Turetzek et al., in preparation), we identified 78 homeobox families (fig. 1 and supplementary file 1, Supplementary Material online), which is similar to the 80 families identified previously in the spider P. tepidariorum (Schwager et al. 2017) and to the 82 families found in the scorpions C. sculpturatus and M. martensii (Di et al. 2015; Schwager et al. 2017).
Fig. 1.
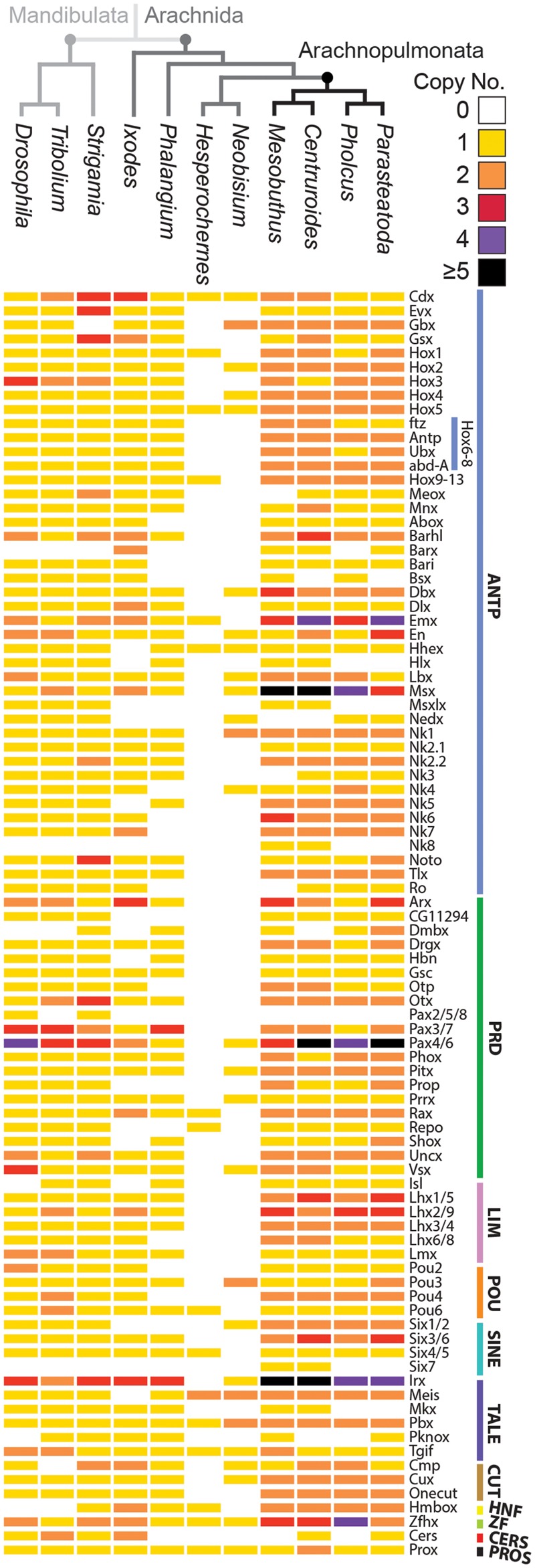
Comparison of homeobox repertoires in arthropods reveals pervasive duplication in arachnopulmonates. The copy number of homeobox families is generally greater in arachnopulmonates compared with other arthropods across all classes, except Cers and Pros. Homeobox genes are classified based on Holland et al. (2007) and the number of paralogs in each family is color coded. The Hox6-8 family has been broken down further to show the specific copy numbers of ftz, Antp, Ubx, and abdA.
For lineages that were thought not to have an ancestral WGD, we surveyed existing transcriptomes from the tick I. scapularis, the harvestman P. opilio, and the pseudoscorpion Hesperochernes sp., as well as sequencing a transcriptome for another pseudoscorpion N. carcinoides. The number of homeobox families found in I. scapularis (70) (fig. 1 and supplementary file 1, Supplementary Material online) was comparable to arachnopulmonates and mandibulates (S. maritima—83; A. mellifera—77; T. castaneum—80; D. melanogaster—80) (Zhong et al. 2008; Zhong and Holland 2011; Chipman et al. 2014). However, we only managed to recover genes from 65 families in P. opilio and just 27 and 16 families in N. carcinoides and Hesperochernes sp., respectively, which likely represent only a subset of families present in these arachnids (fig. 1 and supplementary file 1, Supplementary Material online).
The assignment of homeobox genes into families was verified using a maximum likelihood tree constructed using the homeodomain sequences (supplementary fig. 1, Supplementary Material online). This analysis provided good support for the annotation of each homeodomain to a homeobox gene family, as families were generally monophyletic and had >70% bootstrap support. The general topology of the tree also grouped the homeobox classes together consistent with Holland et al. (2007).
Comparisons of the repertoires of homeobox families between these species suggest particular patterns of retention and loss of homeobox families in arthropod lineages (fig. 1). Overall, excluding the harvestman and pseudoscorpion data due to incompleteness, 60 of the known 87 homeobox families were present in all species surveyed, indicating a reasonable retention of most families.
Families that were present in vertebrates, arachnids and the myriapod, but absent in insects were the HNF and Dmbx families (Zhong et al. 2008; Zhong and Holland 2011; Chipman et al. 2014). Another family that was present in vertebrates and arachnids but missing from the mandibulates surveyed was the Barx family (Zhong et al. 2008; Zhong and Holland 2011; Chipman et al. 2014). The only family not present in arachnids but present in mandibulates and vertebrates was the Pax2/5/8 family.
There were also some retention/loss differences among arachnid species. While Nedx is present in spiders, it appears to have been lost in the scorpions and I. scapularis, although there is a single copy in the pseudoscorpion N. carcinoides (fig. 1 and supplementary file 1, Supplementary Material online). The Hlx, Msxlx, and Mkx families also appear to be missing from spiders but present in the scorpions and the mandibulates surveyed (fig. 1).
Pervasive Duplication of Homeobox Genes in Arachnopulmonates
Although the number of homeobox families is fairly similar between arthropod species surveyed, except the harvestman and pseudoscorpions, the actual number of genes varied considerably between arachnopulmonates and nonarachnopulmonate arthropods. The spider P. phalangioides had a total of 132 homeobox genes (supplementary file 1, Supplementary Material online), which is comparable to the 145 in P. tepidariorum and the 156 found in the scorpions C. sculpturatus and M. martensii (Di et al. 2015; Schwager et al. 2017). In contrast, the nonarachnopulmonate species I. scapularis, P. opilio, N. carcinoides, and Hesperochernes sp. had 96, 69, 32, and 17 homeobox genes, respectively (supplementary file 1, Supplementary Material online). The most complete nonarachnopulmonate data set represented by I. scapularis compared well to the number of homeobox genes previously identified in S. maritima (113), T. castaneum (105), and D. melanogaster (104) (Zhong et al. 2008; Zhong and Holland 2011; Chipman et al. 2014).
We found that 58%, 50%, 59%, 57% of homeobox families in P. tepidariorum, P. phalangioides, C. sculpturatus, and M. martensii are duplicated, compared with 24% in the tick, 3% in the harvestman, 19% in the centipede, beetle, and fly. This shows that many more of the arachnopulmonate homeobox families are comprised of multiple genes copies compared with other arthropods. In total, 34 families are duplicated in all four arachnopulmonate species (fig. 1), which may indicate that these were duplicated in a single event and subsequently retained in the ancestor of the Araneae and Scorpiones lineages. 18 of these 34 families are not duplicated in any of the nonarachnopulmonate species surveyed. Furthermore, 38 families are duplicated in both spiders, whereas 46 families are duplicated in both scorpions (fig. 1).
The families in arachnopulmonates that contain more than two copies, such as Pax4/6 and Irx, are also duplicated in the mandibulate species surveyed. This perhaps suggests that these were duplicated in the arthropod ancestor and that further paralogs were generated in arachnopulmonates due to the WGD (fig. 1).
Homeobox Gene Ohnologs and Tandem Duplicates in P. tepidariorum
It has already been shown that duplicated Hox clusters were retained after the ancestral WGD in arachnopulmonates (Schwager et al. 2017). Therefore, we next investigated if other homeobox gene clusters have also been retained. Of the 45 homeobox gene families that are duplicated in P. tepidariorum, 40 families are represented by paralogs that are located on different scaffolds, hereafter called dispersed paralogs. Some of these dispersed paralogs are present as duplicated clusters in the genome.
One homeobox cluster that is present across protostomes and deuterostomes is the NK cluster (Garcia-Fernandez 2005; Ferrier 2016). In P. tepidariorum, we identified scaffolds that contained duplicated remnants of this cluster. There were two clusters that contained Nk7 and Tlx/C15 paralogs, which have the same transcriptional orientation on each scaffold (fig. 2A). Each of these clusters also contained other ANTP class genes that are usually found in the NK cluster (Lbx, Bap, tin, Hhex, and Msx). However, of these five genes only Msx is duplicated, though the other two Msx paralogs are not located in the NK clusters. This indicates differential retention/loss between these duplicate NK clusters in P. tepidariorum.
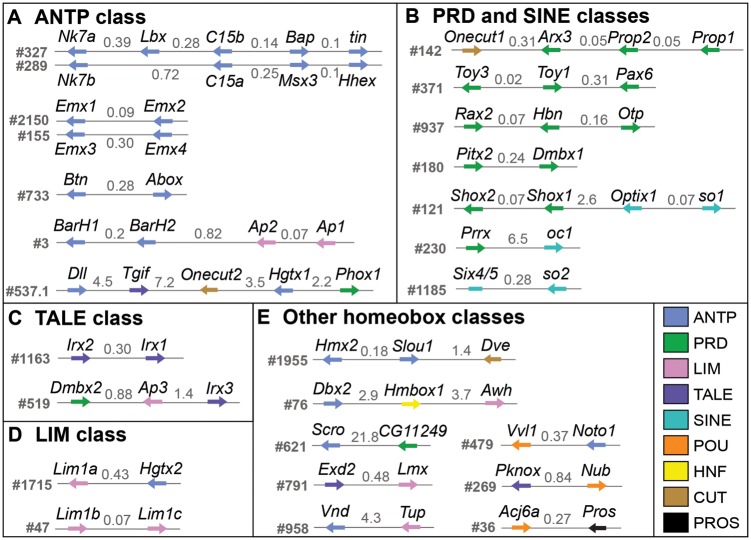
Fig. 2.
Homeobox gene clustering in the Parasteatoda tepidariorum genome. (A) Scaffolds containing at least two ANTP class genes. (B) Scaffolds containing PRD and SINE class gene clusters. (C) Scaffolds containing the Irx family of the TALE class. (D) Scaffolds with Lhx1/5 family of the LIM class. (E) Other scaffolds with at least two homeobox genes. All other homeobox genes were localized to individual scaffolds. The intergenic distances are indicated in Mb. Parasteatoda tepidariorum DoveTail assembly scaffold numbers are to the left of each cluster. Arrows depict the direction of transcription. Nonhomeobox genes are not shown.
We also identified other clusters of homeobox genes that are duplicated and retained to various extent in P. tepidariorum. There is evidence for a duplication of the SINE/Six cluster on scaffolds #121 and #1185 (fig. 2B). This cluster, found in both protostomes and deuterostomes, is usually composed of three genes commonly arranged in the order Optix, sine oculis (so), and Six4/5 (Ferrier 2016). On both scaffolds there are so genes followed by one paralog of Optix on scaffold #121 and the single Six4/5 gene on scaffold #1185. There are also other paralogs of Optix in P. tepidariorum but they are dispersed in the genome. We also identified clusters of ANTP, TALE, and LIM class genes. There are two scaffolds that each contained two tandem paralogs of Emx genes, and these clusters have maintained the same transcriptional orientation (fig. 2A). For the TALE class, two Irx/mirr paralogs were identified on one scaffold and a single copy of Irx was present on another scaffold along with Dmbx2 and Ap3 (fig. 2C). We also identified a scaffold containing two Lhx1/5 paralogs and another with a single copy of Lhx1/5 and one of the Hgtx paralogs (fig. 2D).
We also found eight homeobox families with tandemly duplicated paralogs: the BarH, Lhx5/9, Pax4/6, Prop, and Shox families as well as the aforementioned mentioned Emx, Irx, and Lhx1/5 families (fig. 2). These tandem duplicates were all found in the same transcriptional orientation apart from the Pax4/6 cluster. This means that of the retained duplicate homeobox families, 50% were found as dispersed paralogs, whereas only 6% have conclusively resulted from tandem duplications. Collectively this implies that there has been a greater contribution of WGD than tandem duplication to the expansion of arachnopulmonate homeobox repertoires.
Expression of Homeobox Genes in a P. tepidariorum Embryogenesis
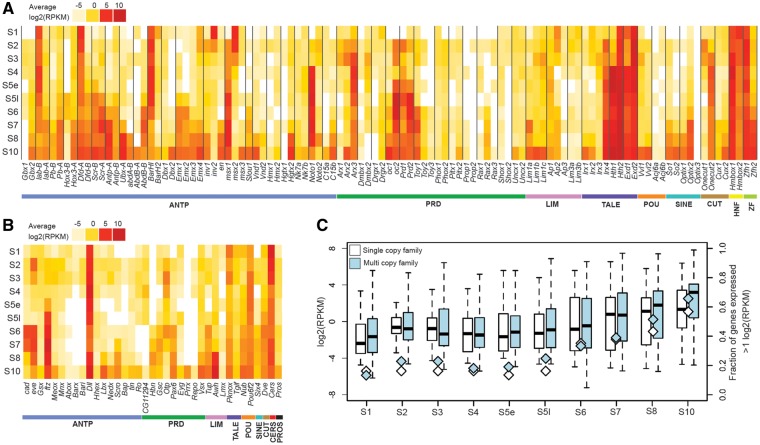
We next investigated the expression of homeobox genes in P. tepidariorum by quantifying their levels in RNA-Seq data covering the first ten stages of embryogenesis of this spider. All 145 annotated homeobox genes were found to be expressed in at least one of the ten embryonic stages assayed, with the exception of Slou2 (fig. 3A and B).
Fig. 3.
Expression of homeobox genes in Parasteatoda tepidariorum from S1 to S10. The transcriptome profile of P. tepidariorum AUGUSTUS gene models for (A) duplicated and (B) single copy Homeobox genes. (C) The average expression of all homeobox genes increases from S1 to S2, likely corresponding to onset of zygotic transcription (Pechmann et al. 2017). The numbers of families expressed >1 log2(RPKM) also increase from S1 to S2. The mean expression level is lower and fewer families are expressed around S4/S5e. After which the mean expression level and number of families continues to increase.
There is an increase in the average expression of single copy and duplicated homeobox genes from S1 to S2 (fig. 3C). The number of homeobox genes expressed >1 log2(RPKM) also increases between these first two stages, especially in the case of the multicopy genes. This observation is likely to be explained by the onset of zygotic transcription at S2 (Pechmann et al. 2017). After S2 both the average expression level and number of genes expressed decreases to the lowest levels around early S5 after which the number of genes and the average expression also increases (fig. 3C).
Interestingly, one homeobox gene that is highly expressed at S1 was Distal-less (Dll) (fig. 3B). This is much earlier than previously reported at S5 (detected by ISH) and its roles in segment specification and limb development (Pechmann et al. 2011). Furthermore, expression of Pt-cad and Pt-eve was also earlier detected at S1 and then increased at S2, again earlier than previously detected using ISH (fig. 3B) (Schönauer et al. 2016). Therefore, it is possible that Pt-Dll, Pt-cad, and Pt-eve are maternally deposited in this spider and are involved in presently unknown functions during early embryogenesis.
Expression Divergence of Duplicated P. tepidariorum Homeobox Genes in the Embryonic Transcriptome
To assess the divergence in the expression of duplicated P. tepidariorum homeobox genes, the RNA-Seq profiling was then analyzed to compare the expression levels of dispersed and tandem paralogs during embryogenesis in this spider (fig. 3A).
The spatial and temporal expression of Hox paralogs in P. tepidariorum was previously analyzed using ISH and showed that Hox genes from both clusters are expressed in the classical collinear fashion across the AP axis (Schwager et al. 2017). Interestingly, both the previous ISHs and our RNA-Seq profiling reveal that one paralog of each Hox gene is always expressed earlier than the other, except for the Pt-abdA paralogs (Schwager et al. 2017). Overall, the timing of Hox expression in the RNA-Seq data matches well with onset of expression detected by ISH (fig. 3A). However, both Pt-lab-A and Pt-Dfd-A were highly expressed from S1 onward, indicating earlier expression than detected by ISH (Pechmann et al. 2015; Schwager et al. 2017). These results are consistent with previous findings that P. tepidariorum Hox paralogs have probably been subject to subfunctionalization and/or neofunctionalization (Pechmann et al. 2015; Schwager et al. 2017).
Other dispersed paralogs that were present in clusters were the NK class families Nk7 and Tlx/C15 (fig. 2A). The Pt-Nk7 paralogs are both expressed at very low levels throughout most of embryogenesis apart from S10 when they both increase in expression (fig. 3A). The Pt-C15 paralogs, however, exhibit divergence in their timing and level of expression, with Pt-C15b showing increased expression around S7 to S10, compared with Pt-C15a, which is barely expressed at any of the ten stages (fig. 3A).
There were also several cases of dispersed (nonclustered) paralogs, which have diverged in the level and timing of their expression (fig. 3A). For example, Pt-Hth2 is expressed throughout all ten stages, whereas Pt-Hth1 is only expressed from S4 to S10 and these genes have demonstrably different expression patterning during limb development in this spider (Turetzek et al. 2017). Other dispersed paralogs that show aspects of divergence including Pt-Gbx, Pt-Msx, Pt-Noto, Pt-Arx, Pt-Onecut, Pt-Hmbox, and Pt-Zfh (fig. 3A), as well as the en/Inv family. Pt-en is expressed at S7 in the RNA-Seq data (fig. 3A), which is consistent with ISHs that show expression of en starts at early S8 in forming segments in P. tepidariorum (Schwager 2008). The Pt-Inv1 paralog shows similar expression, however Pt-Inv2 appears to be maternally loaded and down regulated at S2 when zygotic transcription starts (fig. 3A). Therefore, the timing of expression between Pt-en/Pt-Inv paralogs suggests that they have diverged in function.
A few dispersed paralogs exhibited very similar expression profiles such as Pt-Pitx, Pt-Phox, and Pt-Vvl (fig. 3A). However, it is possible that expression difference may occur later in development or during adult stages and this analysis does not account for any differences in the spatial expression pattern of these genes that may have occurred. This suggests that overall there has been evolutionary changes in the cis-regulation of most dispersed paralogs resulting in divergence in expression levels and transcriptional timing between paralogs.
Divergence of Tandem Paralog Expression during P. tepidariorum Embryogenesis
Tandem duplicates, like dispersed duplicates, also exhibit both conserved and divergent expression profiles. The Emx family contains four paralogs, of which pairs of paralogs are found on two different scaffolds (fig. 2A). Paralogs Pt-Emx1 and Pt-Emx2 have similar expression, which increases from S6 to S10 (fig. 3A). In contrast the other two paralogs, Pt-Emx3 and Pt-Emx4, are both expressed later from S7/S8 to S10 (fig. 3A). There is some early expression of Pt-Emx4, however, overall it appears that Pt-Emx paralogs that are on the same scaffold have more similar expression profiles.
The Irx family is also represented by four paralogs, two found in tandem (Pt-Irx1 and Pt-Irx2) and two dispersed (Pt-Irx3 and Pt-Irx4) (fig. 2C). The tandem duplicates are both expressed only at S10 (fig. 3A), while Pt-Irx3 is expressed only at S3 and the Pt-Irx4 paralog is expressed from S2 to S10 at fairly consistent levels (fig. 3A).
The Lim1/5 family is represented by two paralogs on one scaffold and a third paralog on a separate scaffold (fig. 2D). The two Pt-Lim1/5 paralogs on the same scaffold had very similar expression, with low levels at S3 but stronger expression at S10 (fig. 3A). In contrast the single Pt-Lim1/5 paralog on the other scaffold was expressed from S7 to S10 (fig. 3A).
The remaining tandem duplicates, Pt-BarH, Pt-Prop, and Pt-Shox, all showed divergent expression between paralogs (figs. 2A and B and 3A). For example, the Pt-BarH1 paralog is strongly expressed from S1 to S6, whereas the other paralog appears to be expressed only in S1 and then again at S10 (fig. 3A).
Comparison of Duplicated P. tepidariorum Homeobox Gene Expression with Single Copy Orthologs in P. opilio
To polarize the expression patterns of duplicated homeobox genes in a phylogenetic context, we analyzed the embryonic expression patterns of a subset of duplicated homeobox gene families in P. tepidariorum and compared the expression of selected spider genes to their single copy orthologs in P. opilio.
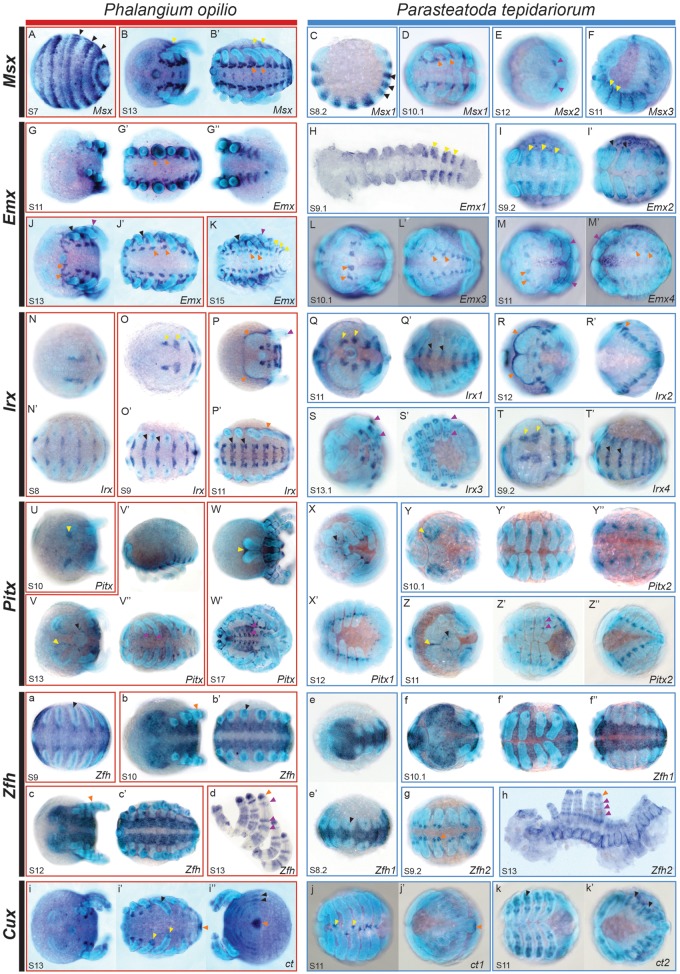
The Msx family provides a likely example of neofunctionalization in the spider (fig. 4A–F). The likely ancestral expression pattern of this gene, possibly represented by Po-Msx, is mostly maintained in Pt-Msx1 (fig. 4A–D). Pt-Msx2 has probably gained a new expression domain in the chelicerae (fig. 4E). Pt-Msx3 is also expressed in a conserved pattern at the base of the prosomal appendages (fig. 4F).
Fig. 4.
Expression of Parasteatoda tepidariorum paralogs compared with single copy orthologs in Phalangium opilio. Expression patterns of Msx (A–F), Emx (G–M′), Irx (N–T′), Pitx (U–Z″), Zfh (a–h), and Cux (i–k′) genes in P. tepidariorum (blue boxes) and P. opilio (red boxes). The early striped expression of Po-Msx (A) matches that of Pt-Msx1 (C), indicated by black arrows. The patches of Po-Msx expression (B′) in each segment along the ventral midline are similar to Pt-Msx1 (D), shown with orange arrows. Expression of Po-Msx and Pt-Msx3 (B, B′ and F) are similar in the region around the base of the appendages (yellow arrows). Pt-Msx2 has undergone possible neofunctionalization (E, purple arrows), with expression in the chelicerae that is not seen for Po-Msx. There is similar expression of Po-Emx (K) in the lateral parts of the opisthosoma compared with Pt-Emx1 (H) and Pt-Emx2 (I), shown with yellow arrows. The expression of Po-Emx around the base of the appendages is only seen for Pt-Emx2 (I′), black arrows. The other two P. tepidariorum paralogs, Pt-Emx3 and Pt-Emx4, both have expression in the precheliceral region and in patches along the ventral midline, which is also present in P. opilio (G′ and J–M′), indicated by orange arrows. The Po-Emx expression in the limbs (J and K) is similar to Pt-Emx4 (M and M′), purple arrows. The expression of Po-Irx in the precheliceral region (N, O, and P) is seen for Pt-Irx1 (Q) and Pt-Irx4 (T), shown by yellow arrows. These two paralogs also have expression in the opisthosoma (Q′ and T′), which matches Po-Irx (N′, O′ and P′), black arrows. The expression of Po-Irx around the germ band (P and P′) can be seen for Pt-Irx2 (R and R′), indicated by orange arrows. There is possibly more elaborate expression of Pt-Irx3 (S and S′) in the limbs compared with Po-Irx (P), shown by purple arrows. Pt-Pitx1 and Pt-Pitx2 expression along the ventral midline (X′, Y′ and Z′), which in combination are similar to that seen for Po-Pitx (V″ and W′) (purple arrows). Expression of Po-Pitx in the precheliceral anterior furrows (U, V, and W, yellow arrows) is seen for Pt-Pitx2 (Y and Z, yellow arrows). However the small dots of Po-Pitx expression around the stomodeum (V, black arrows) is shared between the two Pt-Pitx paralogs (X and Z, black arrows). Expression of Po-Zfh along the ventral midline (a–c′) is seen for the Pt-Zfh1 paralog (e–f″). The later expression of Po-Zfh in the distal tips of limbs (b and d, orange arrows), in bands along the limbs (d, purple arrows) and faint expression throughout the embryo is mirrored by Pt-Zfh2 (g and h). The expression of Po-ct (i–i″) has clearly subfunctionalized in P. tepidariorum with Pt-ct1 having expression in distal tips of limbs (j, yellow arrows) and in the posterior of the germ band (j′, orange arrows). The expression of Pt-ct2 (k and k′) resembles the striped expression of Po-ct in the opisthosoma (i″) and in the mesoderm of the appendages (i′), indicated by black arrows. All embryos are orientated with the anterior to the left. Images within a box are different views of the same embryo. Images H and h are flat mounted embryos. One side of the prosoma has been removed in (h) to aid flat mounting. Opisthosomal limbs have been dissected from P. opilio in (d).
While we observed an apparent case of neofunctionalization in the Msx family there were several families (Emx, Irx, Pitx, Zfh, and Cux) that appear to have undergone subfunctionalization. In the Emx family, the expression pattern of the single copy of Po-Emx is subdivided between the four paralogs found in P. tepidariorum (fig. 4G–M′). Expression of the tandem paralogs Pt-Emx1 and Pt-Emx2 was observed in stripes in the anterior of each opisthosomal segment and Pt-Emx2 also has expression at the base of prosomal appendages. In contrast, both Pt-Emx3 and Pt-Emx4 are expressed in the precheliceral segment and in patches in each segment along the ventral midline, which collectively form a similar expression seen for Po-Emx (fig. 4G′ and J–M′). Therefore, expression of Pt-Emx paralogs is most similar between the tandem paralogs (fig. 3A) consistent with the RNA-Seq profiles of these genes in P. tepidariorum. Nevertheless, some differences are still present between tandem duplicates, mostly in their prosomal appendage domain.
Another likely case of subfunctionalization occurs in the Irx family (fig. 4N–T′). In this family, Pt-Irx1, Pt-Irx2, and Pt-Irx4 appear to have subdivided the expression pattern between them compared with Po-Irx (fig. 4N–P′). Pt-Irx1 and Pt-Irx4 have very similar expression domains, with expression in patches in the precheliceral segment and along the anterior boarder of prosomal and opisthosomal segments (fig. 4Q, Q′, T, and T′). However, Pt-Irx4 expression extends more laterally in the opisthosomal segments, compared with Pt-Irx1. Furthermore, the onset of Pt-Irx4 expression is earlier and continues until later in embryogenesis compared with Pt-Irx1. The other expression domain of Po-Irx around the dorsal boundary edge of the germ band is shared with the Pt-Irx2 paralog. Finally, Pt-Irx3 has possibly gained a completely new domain in the prosomal appendages of later stages and therefore possibly represents another case of neofunctionalization in P. tepidariorum (fig. 4S).
For the single copy of Po-Pitx (fig. 4U–W′), expression during embryogenesis was observed in the precheliceral region, which closely resembled the expression of Pt-Pitx2 in precheliceral region of this spider (fig. 4Y and Z). Other expression domains of Po-Pitx around the stomodeum and the patches along the ventral midline were shared between the Pt-Pitx paralogs (fig. 4X–Z″), indicating possible subfunctionalization of Pt-Pitx paralogs.
The Pt-Zfh paralogs have also undergone expression divergence (fig. 4e–h) such that they represent subfunctionalization compared with Po-Zfh expression (fig. 4a–d). Po-Zfh expression initially starts along the ventral midline of the germ band with emerging expression within the opisthosomal appendages (fig. 4a–b′). This expression is mirrored by the Pt-Zfh1 paralog (fig. 4e′), whereas the expression of Pt-Zfh2 matches that of later Po-Zfh expression, with bands of expression in the limbs and faint expression surrounding the coxa and opisthosomal organs (fig. 4d and h).
Po-ct expression has also probably been subfunctionalized between Pt-ct paralogs (fig. 4i–k′). Po-ct is expressed in the tips of the prosomal appendages and at the very posterior of the germ band matching the expression of Pt-ct1, while the expression of Po-ct in the mesoderm of prosomal appendages, and opisthosoma matches Pt-ct2 expression (fig. 4i–k′).
We also found that expression of Hmx paralogs in P. tepidariorum were highly divergent and again these paralogs perhaps represents an additional example of subfunctionalization (supplementary fig. 2I–K, Supplementary Material online). Pt-Hmx1 is mainly expressed in the prosomal appendages while Pt-Hmx2 is expressed in a pair of cell clusters in the precheliceral region (supplementary fig. 2I–K, Supplementary Material online).
Loss of embryonic expression was found in three of the ten families analyzed (Gbx, Dbx, and Vnd), where one paralog has retained the likely ancestral pattern as compared with P. opilio, while the expression of the other could not be detected during P. tepidariorum embryogenesis by ISH (supplementary fig. 2A–H′, Supplementary Material online). Additionally, in the case of Pt-Gbx2, only the prosomal appendage expression observed in Po-Gbx is conserved (supplementary fig. 2C–E, Supplementary Material online), while this gene has also possibly gained a novel expression domain in the opisthosomal limb buds (supplementary fig. 2F, Supplementary Material online). It remains possible that the paralogs for which we did not detect expression during embryogenesis are expressed later during juvenile or adult stages.
Discussion
Homeobox Gene Repertoires in Chelicerates
Homeobox genes encode an important group of transcription factors that regulate a wide range of developmental processes (Zagozewski et al. 2014; Bataille et al. 2015; Du and Taylor 2015; Zuniga 2015; Krumlauf 2016). Consequently they have received substantial attention and are often characterized and compared within and between animal genomes to better understand their evolution and development. Among arthropods, the insects have been sampled the most extensively and robustly, but there has been limited characterization of these genes in other arthropod groups. For example, among the chelicerates, systematic analysis of the homeobox gene repertoires has only been carried out previously for horseshoe crabs and the scorpion M. martensii (Di et al. 2015; Kenny et al. 2015). Therefore, in order to better understand the homeobox repertoires in chelicerates, we surveyed the two spiders P. tepidariorum and P. phalangioides; another scorpion, C. sculpturatus; the pseudoscorpions N. carcinoides, Hesperochernes sp.; the harvestman P. opilio, and the tick I. scapularis.
Overall we found a similar complement of homeobox classes and families verifying that chelicerates share and have retained similar homeobox repertoires to other arthropods (fig. 1). However several families were observed that are possibly specific to scorpions (Nk8 and Six7), and the Nedx family in spiders was not found in other arachnids except one of the pseudoscorpions. These particular families may therefore regulate lineage specific features during scorpion and spider development. Furthermore, the Barx family, which is found in chelicerates but not in other arthropods, may coordinate specific aspects of chelicerate development.
Aside from the incomplete data set from the pseudoscorpions and the harvestman, we found the fewest homeobox families in the tick I. scapularis indicating that they have either been lost in this arachnid or there is incomplete sequence information for all families. However, the lineage of parasitiforms, and their putative sister group, the acariforms, also exhibit a greater loss of conserved miRNA families compared with other arachnid lineages (Leite et al. 2016). Mite genomes in particular can exhibit marked genome compaction, dynamic rearrangements of homeobox clusters, and associated loss of many transcription factors (Hoy 2009; Grbic et al. 2011). Therefore it is likely that there is actual loss of homeobox genes in I. scapularis. Interestingly, we also observed long-branch lengths for several tick homeodomains, but it is not known if these functional changes are related to the loss of genes, to rapid evolution of gene function, or to the underlying accelerated rate of evolution inherent to this order (Sharma, Schwager, et al. 2014). Note that while we found only a few families in the two pseudoscorpion species, this likely reflects their representation in the transcriptomes analyzed rather than true losses in this lineage.
Expansion of Homeobox Genes after WGD in the Ancestor of Arachnopulmonates
Previous work identified duplicated homeobox genes in chelicerates (Nossa et al. 2014; Di et al. 2015; Kenny et al. 2015), such as Hox genes in spiders and scorpions (Schwager et al. 2007; Sharma, Schwager, et al. 2014; Sharma et al. 2015), as well as other homeobox genes involved in spider eye development (Samadi et al. 2015; Schomburg et al. 2015). However, apart from a scorpion and horseshoe crabs there was no previous systematic analysis of homeobox duplication in chelicerates and in particular how these repertoires have been shaped by WGD in the ancestor of arachnopulmonates.
We found many more duplicated homeobox families in arachnopulmonate species (51–59%) compared with other arthropods surveyed, including I. scapularis (24%), P. opilio (3%), pseudoscorpions (19% and 6%), and several mandibulates (19%) (fig. 1). Indeed, the proportion of duplicated homeobox families found in P. tepidariorum or C. sculpturatus is greater than found in either the BUSCO (41%) or OMA (20.5%) data sets (Schwager et al. 2017). In fact 18 homeobox families were represented by two paralogs in all four arachnopulmonates but were only single copy in all other arthropods surveyed. This makes up a considerable proportion of the 63–78 duplicates identified in P. tepidariorum and C. sculpturatus compared with mandibulates and ticks with respect to the BUSCO-Ar database.
It was previously shown that two clusters of Hox genes have been retained in arachnopulmonates following WGD, whereas only one Hox cluster with single copies of most Hox genes is found in P. opilio, I. scapularis, and T. urticae (Sharma et al. 2012; Pace et al. 2016). Indeed this appears to be a general consequence of WGD: there are two complete and two partial clusters of Hox genes in horseshoe crabs (Nossa et al. 2014). In addition, in vertebrate lineages multiple clusters of Hox genes have been produced by several WGD events (Hoegg and Meyer 2005; Mungpakdee et al. 2008; Pascual-Anaya et al. 2012).
We also found evidence for the duplication of clusters of other homeobox genes in arachnopulmonates in the form of duplicated ANTP (NK cluster), SINE, TALE, and LIM class genes (fig. 2A, C, and D). The inferred ancestral order of arachnopulmonate NK cluster genes (Nk7, Lbx, Tlx, bap, tin, Msx) is consistent with their predicted order in the protostome–deuterostome ancestor (Garcia-Fernandez 2005; Ferrier 2016), requiring just an inversion containing Lbx and Tlx (fig. 2A). Other ANTP class genes in P. tepidariorum are also clustered, which is suggestive of remnants of the mega cluster, however these were not retained as duplicates (fig. 2). A HRO cluster containing Hbn, Rax2, and Otp was also present, and provides further evidence, along with data from S. maritima, that this cluster is a feature of arthropods and other protostomes (fig. 2B) (Mazza et al. 2010; Chipman et al. 2014; Ferrier 2016). However, the order of the three genes in P. tepidariorum is different to other arthropods, suggesting that there has been an inversion in the lineage leading to this spider (Mazza et al. 2010).
In insects and myriapods, the SINE/Six cluster has degraded and all three genes are dispersed in the genome (Chipman et al. 2014; Ferrier 2016). This suggests that the SINE/Six cluster was present in the arthropod ancestor and then has subsequently been degraded in mandibulates but retained in chelicerates. The clusters of ANTP, PRD, SINE, TALE, and LIM class genes in P. tepidariorum suggests that spiders have retained many features of the hypothetical clustering of homeobox genes in the bilaterian ancestor (Ferrier 2016). Furthermore, several of these clusters are duplicated and there are different patterns of gene loss/retention and rearrangements, for example, fewer genes have been lost in the Hox cluster compared with the NK cluster.
Retention of gene duplicates in arachnopulmonates has also been observed for other important developmental genes including Wnts and frizzled4, and dachshund (dac), as well as venom and silk genes (Schwager et al. 2007, 2017; Janssen et al. 2010, 2015; Haney et al. 2014, 2016; Clarke et al. 2015; Pechmann et al. 2015; Samadi et al. 2015; Schomburg et al. 2015; Turetzek et al. 2016, 2017). Furthermore, miRNAs are also pervasively duplicated in arachnopulmonate genomes (Leite et al. 2016). This suggests that the retention of duplicated homeobox genes and other developmental toolbox genes after WGD in arachnopulmonates has played an important role in the evolution of development of these animals. The high rate of retention of duplicated homeobox genes after WGD in arachnopulmonates is similar to that observed after the two rounds of WGD in vertebrates (Dehal and Boore 2005; Maere et al. 2005; Holland et al. 2008; McGrath et al. 2014; Schwager et al. 2017). Indeed most of the homeobox gene families duplicated in arachnopulmonates are also duplicated in vertebrates, but interestingly the Noto, Drgx, Hmbox families are only duplicated in the former (supplementary fig. 3, Supplementary Material online) (Zhong et al. 2008; Zhong and Holland 2011). This indicates that arachnopulmonates and vertebrates have independently retained and utilized duplicated copies of these important transcription factors and this likely contributed to the developmental evolution, novel phenotypes, and adaptation of these two phyla. Furthermore, families that were only present as single copies in vertebrates and arachnopulmonates were Bsx, Hlx, and Mkx, which indicates that these families fail to retain duplicates in both lineages after WGDs. An intriguing counterpoint for future investigation is therefore horseshoe crabs, which have been shown to have likely undergone two rounds of WGD, but exemplify morphological external stasis and evolutionary relictualism (Sharma, Schwager, et al. 2014; Kenny et al. 2015; Schwager et al. 2017).
Divergence in the Expression of Homeobox Paralogs
How has the ancestral WGD in arachnopulmonates contributed to their evolution and the development of lineage specific features? It has already been shown that one paralog of dac in the spider has a distinct and novel role (by comparison to the ancestral function of this gene within Arthropoda), being responsible for patterning the distal boundary of the arachnid-specific podomere, the patella (Turetzek et al. 2016). Furthermore, the arrangement of structures in the opisthosoma of scorpions coincides with the staggered expression of paralogous Hox gene expression (Sharma, Schwager, et al. 2014), suggesting that divergences in Hox paralogs may in part be responsible for innovations of the scorpion body. Moreover, the Hox paralogs of spiders have also divergences in their temporal and spatial expression (Schwager et al. 2007, 2017), while other homeobox paralogs also show differential expression among the developing eyes (Samadi et al. 2015; Schomburg et al. 2015).
In our study we did not identify any homeobox gene paralogs in P. tepidariorum with the same temporal expression profile (fig. 3A), and ISHs on a subset of paralogs also showed divergence in the spatial expression between P. tepidariorum paralogs. Comparisons of gene expression of P. tepidariorum paralogs with their single copy ortholog in P. opilio suggest that most of the surveyed paralogs have likely undergone subfunctionalization, usually in the developing appendages and nervous system. The P. tepidariorum Msx genes have also apparently been subject to possible neofunctionalization in the case of Msx2 in developing chelicerae.
The expression of Pitx and Zfh genes between P. opilio and P. tepidariorum, in conjunction with their known expression in Drosophila, provide particularly strong evidence for subfunctionalization. In Drosophila, Pitx is expressed in several tissues including a subset of ventral somatic muscles and in neural cells (Vorbrüggen et al. 1997). These expression patterns in Drosophila are consistent with Po-Pitx expression (fig. 4), suggesting that this expression pattern is ancestral. In P. tepidariorum, both Pitx paralogs also show metameric patterning along the ventral neuroectoderm, with Pt-Pitx1 most similar to the Drosophila and P. opilio CNS expression and Pt-Pitx2 showing both CNS and mesodermal expression (fig. 4U–Z″).
The expression patterns of Zfh in Drosophila and P. opilio are also very similar, again implying the ancestral expression of this gene (fig. 4a–d). Early expression of the Drosophila Zfh2 ortholog is seen in the brain and ventral CNS (Lai et al. 1991), with later expression seen in leg imaginal discs as an initially broad domain at the centre of the disc that develops into rings of expression in each leg segment and in a domain throughout the tarsus (Guarner et al. 2014). These patterns are similar to that seen for P. opilio (fig. 4a–d). However in P. tepidariorum, early CNS expression and later limb expression has been divided between the Zfh paralogs. Pt-Zfh1 is strongly expressed in the CNS and initial limb buds, while Pt-Zfh2 is observed in the later pattern expression in rings and at the distal tips of the limbs (fig. 4e–h). This expression divergence observed for the Pitx and Zfh paralogs exemplify the pervasive temporal and spatial subfunctionization of genes that has likely occurred in the spider and probably other arachnopulmonates.
Conclusion
Our study has revealed the first comparative genomic picture of the repertoires of homeobox genes in arachnids. This shows that there has been a high level of gene retention of these developmental genes since the WGD in the common ancestor of arachnopulmonates. Furthermore, most of the P. tepidariorum homeobox gene paralogs exhibit differences in their timing and spatial expression, and when compared with their single copy homologues in P. opilio. This suggests there has been pervasive subfunctionalization and/or neofunctionalization of these genes since WGD. It will be interesting to further investigate the roles of these genes in spider development to ascertain their contribution to the evolution of development and diversification of these arachnids, especially with respect to emergence of novel traits including silk glands and book lungs. Furthermore, future comparisons of ohnologs between arachnopulmonates and vertebrates should provide exciting new insights into the general consequences of WGD in animals.
Materials and Methods
Identification of Homeobox Genes in Arachnids
To identify homeobox genes in arachnid species, we analyzed both existing resources and also new transcriptomic data generated in this study. Existing protein predictions were collected for the tick Ixodes scapularis (PRJNA16232), the harvestman Phalangium opilio (PRJNA236471), and the pseudoscorpion Hesperochernes sp. (PRJNA254752).
For further characterization of homeobox genes in arachnids we also generated de novo transcriptomes for the spider Pholcus phalangioides and the pseudoscorpion Neobisium carcinoides. For P. phalangioides RNA isolation, library preparation and sequencing with Illumina HiSeq2000 was previously described (Janssen et al. 2015). A de novo transcriptome assembly (Turetzek N, Torres-Oliva M, Kaufholz F, Prpic NM, Posnien N, in preparation) was performed with Trinity version r20140717 (Haas et al. 2013) with the following settings: –seqType fq –JM 240 G – run_as_paired –CPU 6 and using Trimmomatic for quality trimming and filtering (Bolger et al. 2014). For the pseudoscorpion N. carcinoides, RNA was extracted from the whole body, sequenced with Illumina HiSeqll and de novo assembly of the transcriptome was carried out using Trinity v 2.0.3 (Grabherr et al. 2011) under default parameters and using Trimmomatic for quality control. The raw sequence reads for P. phalangioides and the pseudoscorpion N. carcinoides have been deposited in the SRA with accession numbers PRJNA448805 and PRJNA438779, respectively.
Longest open reading frames (ORFs) were predicted from the transcriptomes of P. phalangioides and the pseudoscorpion N. carcinoides as well as from the existing nucleotide transcriptome of the harvestman P. opilio (PRJNA236471) and the pseudoscorpion Hesperochernes sp. (PRJNA254752) using TransDecoder v3.0.0 (Haas et al. 2013). To retain putative proteins the sequence homology and protein domains of predicted ORFs were then analyzed, respectively, with BLASTP v2.2.28+ (e-value 1e−6) (Altschul et al. 1990) using the UniProt Swiss-Prot database (UniProt 2015), and HMMER v3.1 (Wheeler and Eddy 2013) using the Pfam v30.0 database (Finn et al. 2016).
The protein sequences from P. phalangioides, I. scapularis, P. opilio, and the two pseudoscorpions were then searched for the presence of homeodomain sequences using BLASTP v2.2.28+ (Altschul et al. 1990) with query amino acid homeodomain sequences from all ten species in HomeoDB (Zhong et al. 2008; Zhong and Holland 2011) combined with homeodomain sequences from P. tepidariorum (Schwager et al. 2017), C. sculpturatus (Schwager et al. 2017), M. martensii (Di et al. 2015), Strigamia maritima (Chipman et al. 2014). All the initial BLASTP hits with >30% percentage identity were retained. Next, the full protein sequences of the BLASTP hits were then analyzed using the Conserved Domain Database (CDD) search tool (Marchler-Bauer et al. 2015) to confirm the presence of homeodomains as well as annotate other functional domains. BLASTP hits that did not have homeodomains identified by CDD were removed. Transcripts within a species that had identical protein sequences predicted to encode homeodomains were manually checked and identical nucleotide transcripts or isoforms were removed. Specific BLAST searches for PROS class genes also identified a Pros gene (MMa30254) in M. martensii not reported previously by Di et al. (2015). All identified homeobox genes and their sequences are given in supplementary file 1, Supplementary Material online. By concentrating on the detection of homeobox genes based on the presence of homeodomains some partial transcripts of homeobox genes that lack this domain, or may have highly divergent homeodomains, may be missing in our data set.
Phylogenetic Analysis of Arachnid Homeodomains
The predicted homeobox genes were then classified based on phylogenetic analysis of the homeodomain sequences they encode. Amino acid sequences of homeodomains from two spiders (P. tepidariorum and P. phalangioides), two scorpions (C. sculpturatus and M. martensii) two pseudoscorpions (Hesperochernes sp. and N. carcinoides), the harvestman P. opilio, the tick I. scapularis, the myriapod (centipede) S. maritima and three insects Apis mellifera, Tribolium castaneum, and Drosophila melanogaster were aligned with ClustalW (Larkin et al. 2007), excluding unusual PROS HPD sequences and the Cs-Emx1 homeodomain because it has a large insertion.
Phylogenetic analyses, using only unique homeodomain sequence alignments, were performed in RAxML, with support levels estimated using the rapid bootstrap algorithm (1000 replicates) (Stamatakis et al. 2008), under the PROTGAMMALG model of amino acid substitution—that was identified as best fitting using a custom Perl script from the Exelixis Lab website (https://sco.h-its.org/exelixis/web/software/raxml/hands_on.html). Homeodomain proteins were classified based on the homology of their homeodomains to known homeodomain containing proteins and annotated with nomenclature following that of Holland et al. (2007).
Synteny Analysis of Homeobox Genes in P. tepidariorum
To investigate the arrangement of homeobox genes in P. tepidariorum we used the high quality HiRise/DoveTail genome assembly (Schwager et al. 2017). The scaffold location and coordinates of the previously identified homeobox genes (Schwager et al. 2017) were extracted from the GFF file, which contains coordinates of AUGUSTUS gene models relative to the HiRise/DoveTail genome, and were used to calculate the gaps between genes.
Analysis of Homeobox Gene Expression in P. tepidariorum Embryogenesis
Homeobox gene expression levels were analyzed during P. tepidariorum embryogenesis using RNA sequencing. RNA was extracted using the Dynabeads mRNA DIRECT Kit (Ambion) from 10–100 embryos of each successive developmental stage (stage [S]1–S4, S5 early and S5 late, S6–S8 and S10; Akiyama-Oda and Oda 2003; Mittmann and Wolff 2012). Two replicate sets of mRNAs were independently obtained from two pairs of parents. The mRNAs were fragmented using the NEBNext RNase III RNA Fragmentation Module (New England BioLabs) and then used to construct DNA libraries with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs) and NEBNext Multiplex Oligos for Illumina (Index Primers Set 1, New England Biolabs). The libraries were sequenced using the 150-cycle format of the Illumina MiSeq Reagent Kit v3. The resulting sequence reads were subjected to adaptor trimming using the CLC Genomics Workbench 7.0.3 (Qiagen), and quality of the sequences was confirmed with FastQC v0.11.2 (Andrews 2011). The trimmed raw reads have been deposited in the SRA with accession number PRJNA448775 (Iwasaki-Yokozawa et al. 2018). Replicates for each stage were aligned to the P. tepidariorum reference transcriptome (Schwager et al. 2017) using TopHat v2 (Kim et al. 2013). Outputs files were sorted and indexed with Samtools v1.2 (Li et al. 2009) and RPKM expression levels were quantified using HTSeq-count (Anders et al. 2015) and custom Perl scripts. Heatmaps were generated in R v3.2.3 (R Core Team 2015) using the ComplexHeatmap package (Gu et al. 2016).
Parasteatoda tepidariorum and P. opilio Cultures
An inbred culture of P. tepidariorum (from a strain collected in Göttingen, Germany) was maintained at Oxford Brookes University and fed on a diet of Drosophila vestigial mutants and Gryllodes sigillatus, with a 12:12 h light:dark cycle at 25°C. The culture of P. opilio was maintained at the University of Wisconsin, Madison, WI and fed on a diet of fish flakes supplemented with Acheta domesticus nymphs, with a 14: 10 light: dark cycle at 20°C.
Cloning of Gene Fragments and Probe Synthesis
cDNA was generated using QuantiTech (Qiagen) with RNA extracted (Qiazol) from S1 to S14 P. tepidariorum embryos and from S7 to S17 for P. opilio. Gene fragments were amplified by PCR and cloned into the TOPO-TA vector (ThermoFisher Scientific). Primer sequences are provided in supplementary table 1, Supplementary Material online. RNA probes were transcribed with T3 (11031163001—Roche) or T7 polymerase (10881775001—Roche), with DIG RNA labeling mix (11277073910—Roche), from PCR fragments generated from TOPO-TA clones following standard protocols.
In Situ Hybridization in P. tepidariorum and P. opilio
Colourmetric in situ hybridization (ISH) for P. tepidariorum and P. opilio was performed as previously described (Akiyama-Oda and Oda 2003; Sharma et al. 2012). Embryos were counterstained with DAPI (Roche—10236276001) for ∼20 mins to visualize nuclei. Embryos were imaged using a Zeiss Axio Zoom V.16 and a Nikon SMZ25, and overlays were generated in Photoshop CS6.
Supplementary Material
Acknowledgments
This work was supported by a Leverhulme Trust grant (RPG-2016-234) to A.P.M., Nigel Groome studentships to D.J.L. and L.B.G., a Marie Skłodowska-Curie Fellowship for J.L.-F. (655814), and the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) awards to H.O. (15K07139) and Y.A.-O. (26440130). P.P.S. was supported by National Science Foundation grant IOS-1552610. N.T. and N.M.P. were supported by grants from the Deutsche Forschungsgemeinschaft grant numbers PR 1109/4-1 and PR 1109/6-1.
References
- Akiyama-Oda Y, Oda H.. 2003. Early patterning of the spider embryo: a cluster of mesenchymal cells at the cumulus produces Dpp signals received by germ disc epithelial cells. Development 1309:1735–1747. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 2153:403–410. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 312:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Last accessed January 25, 2015. [Google Scholar]
- Averof M, Patel NH.. 1997. Crustecean appendage evolution associated with changes in Hox gene expression. Nature 3886643:682–686. [DOI] [PubMed] [Google Scholar]
- Bataille L, Frendo JL, Vincent A.. 2015. Hox control of Drosophila larval anatomy; the alary and thoracic alary-related muscles. Mech Dev. 138(Pt 2):170–176. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR, Affolter M.. 2016. Homeodomain proteins: an update. Chromosoma 1253:497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Yu Y, Wu Y, Hao P, Di Z, He Y, Chen Z, Yang W, Shen Z, He X.. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun. 4:2602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD.. 2005. From DNA to diversity .Oxford: Blackwell Publishing. [Google Scholar]
- Chipman AD, Ferrier DE, Brena C, Qu J, Hughes DS, Schroder R, Torres-Oliva M, Znassi N, Jiang H, Almeida FC, et al. 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 1211:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu SW, Noyes MB, Christensen RG, Pierce BG, Zhu LJ, Weng Z, Stormo GD, Wolfe SA.. 2012. Exploring the DNA-recognition potential of homeodomains. Genome Res. 2210:1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TH, Garb JE, Hayashi CY, Arensburger P, Ayoub NA.. 2015. Spider transcriptomes identify ancient large-scale gene duplication event potentially important in silk gland evolution. Genome Biol Evol. 77:1856–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL.. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 310:e314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Z, Yu Y, Wu Y, Hao P, He Y, Zhao H, Li Y, Zhao G, Li X, Li W, et al. 2015. Genome-wide analysis of homeobox genes from Mesobuthus martensii reveals Hox gene duplication in scorpions. Insect Biochem Mol Biol. 61:25–33. [DOI] [PubMed] [Google Scholar]
- Du H, Taylor HS.. 2015. The role of Hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb Perspect Med. 61:a023002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier DEK. 2016. Evolution of homeobox gene clusters in animals: the Giga-cluster and primary vs. secondary clustering. Front Ecol Evol. 4:1–13. [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez J. 2005. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 612:881–892. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 297:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic M, Van Leeuwen T, Clark RM, Rombauts S, Rouze P, Grbic V, Osborne EJ, Dermauw W, Ngoc PC, Ortego F, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 4797374:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 3218:2847–2849. [DOI] [PubMed] [Google Scholar]
- Guarner A, Manjon C, Edwards K, Steller H, Suzanne M, Sanchez-Herrero E.. 2014. The zinc finger homeodomain-2 gene of Drosophila controls Notch targets and regulates apoptosis in the tarsal segments. Dev Biol. 3852:350–365. [DOI] [PubMed] [Google Scholar]
- Guerreiro I, Nunes A, Woltering JM, Casaca A, Nóvoa A, Vinagre T, Hunter ME, Duboule D, Mallo M.. 2013. Role of a polymorphism in a Hox/Pax-responsive enhancer in the evolution of the vertebrate spine. Proc Natl Acad Sci U S A. 11026:10682–10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 88:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD, Brent R.. 1991. A genetic model for interaction of the homeodomain recognition helix with DNA. Science 2514992:426–430. [DOI] [PubMed] [Google Scholar]
- Haney RA, Ayoub NA, Clarke TH, Hayashi CY, Garb JE.. 2014. Dramatic expansion of the black widow toxin arsenal uncovered by multi-tissue transcriptomics and venom proteomics. BMC Genomics 36615:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney RA, Clarke TH, Gadgil R, Fitzpatrick R, Hayashi CY, Ayoub NA, Garb JE.. 2016. Effects of gene duplication, positive selection and shifts in gene expression on the evolution of the venom gland transcriptome in widow spiders. Genome Biol Evol. 81:228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Meyer A.. 2005. Hox clusters as models for vertebrate genome evolution. Trends Genet. 218:421–424. [DOI] [PubMed] [Google Scholar]
- Holland PW. 2015. Did homeobox gene duplications contribute to the Cambrian explosion? Zool Lett. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutierrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. 2008. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 187:1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA.. 2007. Classification and nomenclature of all human homeobox genes. BMC Biol. 5:47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy MA. 2009. The predatory mite Metaseiulus occidentalis: mitey small and mitey large genomes. BioEssays 315:581–590. [DOI] [PubMed] [Google Scholar]
- Huminiecki L, Conant GC.. 2012. Polyploidy and the evolution of complex traits. Int J Evol Biol. 2012:292068.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki-Yokozawa S, Akiyama-Oda Y, Oda H.. 2018. Genome-scale embryonic developmental profile of gene expression in the common house spider Parasteatoda tepidariorum. Data Brief. 19:865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Le Gouar M, Pechmann M, Poulin F, Bolognesi R, Schwager EE, Hopfen C, Colbourne JK, Budd GE, Brown SJ, et al. 2010. Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol Biol. 10372:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Schönauer A, Weber M, Turetzek N, Hogvall M, Goss GE, Patel NH, McGregor AP, Hilbrant M.. 2015. The evolution and expression of panarthropod frizzled genes. Front Ecol Evol. 3:1–14. [Google Scholar]
- Kenny NJ, Chan KW, Nong W, Qu Z, Maeso I, Yip HY, Chan TF, Kwan HS, Holland PW, Chu KH, et al. 2015. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity. 1162:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 144:R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, Carroll SB.. 2015. Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc Natl Acad Sci U S A. 11224:7524–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. 2016. Hox genes and the hindbrain: a study in segments. Curr Top Dev Biol. 116:581–596. [DOI] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissieres V, Pickle CS, Plajzer-Frick I, Lee EA, et al. 2016. Progressive loss of function in a limb enhancer during snake evolution. Cell 1673:633–642 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Fortini ME, Rubin GM.. 1991. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 34(2–3):123–134. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 2321:2947–2948. [DOI] [PubMed] [Google Scholar]
- Leite DJ, Ninova M, Hilbrant M, Arif S, Griffiths-Jones S, Ronshaugen M, McGregor AP.. 2016. Pervasive microRNA duplication in chelicerates: insights from the embryonic microRNA repertoire of the spider Parasteatoda tepidariorum. Genome Biol Evol. 87:2133–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liubicich DM, Serano JM, Pavlopoulos A, Kontarakis Z, Protas ME, Kwan E, Chatterjee S, Tran KD, Averof M, Patel NH.. 2009. Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc Natl Acad Sci U S A. 10633:13892–13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Roth JJ, Wagner GP.. 2006. Adaptive evolution of Hox-gene homeodomains after cluster duplications. BMC Evol Biol. 6:86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y.. 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 10215:5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI.. 2015. CDD: nCBI’s conserved domain database. Nucleic Acids Res. 43(Database issue):D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza ME, Pang K, Reitzel AM, Martindale MQ, Finnerty JR.. 2010. A conserved cluster of three PRD-class homeobox genes (homeobrain, rx and orthopedia) in the Cnidaria and Protostomia. Evodevo 11:3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Gout JF, Johri P, Doak TG, Lynch M.. 2014. Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 2410:1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann B, Wolff C.. 2012. Embryonic development and staging of the cobweb spider Parasteatoda tepidariorum C. L. Koch, 1841 (syn.: achaearanea tepidariorum; Araneomorphae; Theridiidae). Dev Genes Evol. 2224:189–216. [DOI] [PubMed] [Google Scholar]
- Mungpakdee S, Seo HC, Angotzi AR, Dong X, Akalin A, Chourrout D.. 2008. Differential evolution of the 13 Atlantic salmon Hox clusters. Mol Biol Evol. 257:1333–1343. [DOI] [PubMed] [Google Scholar]
- Nossa CW, Havlak P, Yue JX, Vincent KY, Brockmann J, Putman NH.. 2014. Joint assembly and genetic mapping of the Atlantic horshoe crab genome reveals ancient whole genome duplication. Giga Sci. 93:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lombardia M, Foos N, Maurel-Zaffran C, Saurin AJ, Graba Y.. 2017. Hox functional diversity: novel insights from flexible motif folding and plastic protein interaction. Bioessays 394:1600246.. [DOI] [PubMed] [Google Scholar]
- Pace RM, Grbic M, Nagy LM.. 2016. Composition and genomic organization of arthropod Hox clusters. Evodevo 7:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Anaya J, D’Aniello S, Kuratani S, Garcia-Fernàndez J.. 2012. Evolution of Hox gene clusters in deuterostomes. BMC Dev Biol. 1326:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann M, Benton MA, Kenny NJ, Posnien N, Roth S.. 2017. A novel role for Ets4 in axis specification and cell migration in the spider Parasteatoda tepidariorum. Elife 6(e27590):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann M, Khadjeh S, Turetzek N, McGregor AP, Damen WG, Prpic NM.. 2011. Novel function of Distal-less as a gap gene during spider segmentation. PLoS Genet. 710:e1002342.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann M, Schwager EE, Turetzek N, Prpic NM.. 2015. Regressive evolution of the arthropod tritocerebral segment linked to functional divergence of the Hox gene labial. Proc Biol Sci. 2821814:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 4537198:1064–1071. [DOI] [PubMed] [Google Scholar]
- Qu Z, Kenny NJ, Lam HM, Chan TF, Chu KH, Bendena WG, Tobe SS, Hui JH.. 2015. How did arthropod sesquiterpenoids and ecdysteroids arise? Comparison of hormonal pathway genes in noninsect arthropod genomes. Genome Biol Evol. 77:1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Rokas A. 2008. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 42:235–251. [DOI] [PubMed] [Google Scholar]
- Ronshaugen M, McGinnis N, McGinnis W.. 2002. Hox protein mutation and macroevolution of the insect body plan. Nature 4156874:914–917. [DOI] [PubMed] [Google Scholar]
- Samadi L, Schmid A, Eriksson BJ.. 2015. Differential expression of retinal determination genes in the principal and secondary eyes of Cupiennius salei Keyserling (1877). Evodevo 6:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg C, Turetzek N, Schacht MI, Schneider J, Kirfel P, Prpic NM, Posnien N.. 2015. Molecular characterization and embryonic origin of the eyes in the common house spider Parasteatoda tepidariorum. Evodevo 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönauer A, Paese CL, Hilbrant M, Leite DJ, Schwager EE, Feitosa NM, Eibner C, Damen WG, McGregor AP.. 2016. The Wnt and Delta-Notch signalling pathways interact to direct pair-rule gene expression via caudal during segment addition in the spider Parasteatoda tepidariorum. Development 14313:2455–2463. [DOI] [PubMed] [Google Scholar]
- Schwager EE. 2008. Segmentation of the spider Achaearanea tepidariorum investigated by gene expression and functional analysis of the gap gene hunchback. PhD thesis, University of Cologne, Germany.
- Schwager EE, Schoppmeier M, Pechmann M, Damen WG.. 2007. Duplicated Hox genes in the spider Cupiennius salei. Front Zool. 4:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, Akiyama-Oda Y, Esposito L, Bechsgaard J, Bilde T, et al. 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 151:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PP, Kaluziak ST, Perez-Porro AR, Gonzalez VL, Hormiga G, Wheeler WC, Giribet G.. 2014. Phylogenomic interrogation of arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol. 3111:2963–2984. [DOI] [PubMed] [Google Scholar]
- Sharma PP, Santiago MA, Gonzalez-Santillan E, Monod L, Wheeler WC.. 2015. Evidence of duplicated Hox genes in the most recent common ancestor of extant scorpions. Evol Dev. 176:347–355. [DOI] [PubMed] [Google Scholar]
- Sharma PP, Schwager EE, Extavour CG, Giribet G.. 2012. Hox gene expression in the harvestman Phalangium opilio reveals divergent patterning of the chelicerate opisthosoma. Evol Dev. 145:450–463. [DOI] [PubMed] [Google Scholar]
- Sharma PP, Schwager EE, Extavour CG, Wheeler WC.. 2014. Hox gene duplications correlate with posterior heteronomy in scorpions. Proc Biol Sci. 2811792:20140661.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 575:758–771. [DOI] [PubMed] [Google Scholar]
- Stern DL. 1998. A role of Ultrabithorax in morphological differences between Drosophila species. Nature 3966710:463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetzek N, Khadjeh S, Schomburg C, Prpic NM.. 2017. Rapid diversification of homothorax expression patterns after gene duplication in spiders. BMC Evol Biol. 171:168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetzek N, Pechmann M, Schomburg C, Schneider J, Prpic NM.. 2016. Neofunctionalisation of a duplicate dachshund gene underlies the evolution of a novel leg segment in arachnids. Mol Biol Evol. 331:109–121. [DOI] [PubMed] [Google Scholar]
- UniProt C. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43(Database issue):D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 1010:725–732. [DOI] [PubMed] [Google Scholar]
- Vorbrüggen G, Constien R, Zilian O, Wimmer EA, Dowe G, Taubert H, Noll M, Jäckle H.. 1997. Embryonic expression and characterization of a Ptx1 homolog in Drosophila. Mech Dev. 68(1–2):139–147. [DOI] [PubMed] [Google Scholar]
- Werner T, Koshikawa S, Williams TM, Carroll SB.. 2010. Generation of a novel wing colour pattern by the Wingless morphogen. Nature 4647292:1143–1148. [DOI] [PubMed] [Google Scholar]
- Wheeler TJ, Eddy SR.. 2013. nhmmer: dNA homology search with profile HMMs. Bioinformatics 2919:2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagozewski JL, Zhang Q, Pinto VI, Wigle JT, Eisenstat DD.. 2014. The role of homeobox genes in retinal development and disease. Dev Biol. 3932:195–208. [DOI] [PubMed] [Google Scholar]
- Zhong Y-F, Butts T, Holland PWH.. 2008. HomeoDB: a database of homeobox gene diversity. Evol Dev. 105:516–518. [DOI] [PubMed] [Google Scholar]
- Zhong Y-F, Holland PWH.. 2011. HomeoDB2: functional expansion of a comparative homeobox gene database for evolutionary developmental biology. Evol Dev. 136:567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A. 2015. Next generation limb development and evolution: old questions, new perspectives. Development 14222:3810–3820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.