Abstract
Translation of the CRISPR/Cas9 system to human therapeutics holds high promise. Specificity remains a concern, however, especially when modifying stem cell populations. We show that existing rationally-engineered Cas9 high fidelity variants have reduced on-target activity using the therapeutically relevant ribonucleoprotein (RNP) delivery method. Therefore, we devised an unbiased bacterial screen to isolate variants that retain activity in the RNP format. Introduction of a single point mutation, R691A (HiFi Cas9), retained high on-target activity while reducing off-target editing. HiFi Cas9 induces robust AAV6-mediated gene targeting at five therapeutically-relevant loci (HBB, IL2RG, CCR5, HEXB, TRAC) in human CD34+ hematopoietic stem and progenitor cells (HSPCs) as well as primary T-cells. We also show that the HiFi Cas9 mediates high-level correction of the sickle cell disease (SCD)-causing Glu6Val mutation in SCD patient derived HSPCs. We anticipate that HiFi Cas9 will have wide utility for both basic science and therapeutic genome editing applications.
INTRODUCTION
Repurposing bacterial “CRISPR” systems to facilitate RNA-guided cleavage of complementary DNA has revolutionized the field of gene editing in mammalian cells 1–4. S. pyogenes Cas9 has been the most highly used CRISPR system and its use for genome editing only requires the presence of the Cas9 nuclease and a guide RNA (gRNA). In S. pyogenes, the guide RNA is a complex of two species, a universal trans-activating crRNA (tracrRNA) and a target-specific crisprRNA (crRNA). The guide RNAs can be shortened and fused to result in a “single guide” RNA (sgRNA). Cas9/gRNA is directed to search for NGG protospacer adjacent motifs (PAMs) in the genome and if sufficient complementarity exists between the guide and the target, conformational changes occur and the now fully active Cas9 cleaves the DNA, resulting in a targeted double-stranded break (DSB)5,6. The lesion can be healed by the error-prone Non-Homologous End-Joining (NHEJ) pathway, which often results in creation of an insertion/deletion (INDEL) of one to several bases7. If a homologous donor DNA template is provided during the DSB repair, the homologous recombination (HR) repair pathway can generate more precise changes to the target DNA, including single nucleotide changes to insertion of large gene cassettes8,9.
While cleavage at sites with perfect complementarity to the protospacer domain of the gRNA typically occurs with highest efficiency, cleavage at sites having one to several bases of mismatch with the gRNA can occur. These undesired cleavage events are called “off-target effects” (OTEs)10, and their existence should be minimized for therapeutic applications of genome editing technologies. The Cas9 nuclease and gRNA can be delivered as DNA expression cassettes in plasmids or viral vectors, as purified RNAs with Cas9 mRNA, or as a Cas9-gRNA ribonucleoprotein (RNP) complex. As a general rule, methods that result in a higher, more sustained level of Cas9 and gRNA in the cell yield higher OTEs11,12. Methods using RNP complexes, where recombinant Cas9 protein is pre-complexed with gRNA and directly delivered to cells, is preferable to DNA-based and even RNA-based expression methods. RNP delivery results in an initially high level of the genome editing machinery followed by rapid decay, leading to a “fast on, fast off” strategy that results in highly efficient editing while minimizing OTEs13–19. RNP delivery also is a method to avoid the intra-cellular innate immune response that is highly sensitive in sensing the delivery of foreign nucleic acids in human primary cells. Nevertheless, off-target cleavage can and does occur, so methods to improve Cas9 specificity are important to improve therapeutic genome editing applications.
Allogeneic hematopoietic stem and progenitor cell (HSPC) transplantation (allo-HSCT) can provide a one-time and life-long cure for patients with serious genetic hematological diseases such as primary immunodeficiencies20 and hemoglobinopathies21,22, if a suitable immunologically matched donor can be found23. However, the immunologic toxicities of allo-HSCT have prevented it from being applied broadly. Cas9 genome editing of autologous HSPCs has the power to be a safer and more effective therapy for a myriad of genetic diseases. Recent studies have shown that disease-causing genetic errors can be corrected using CRISPR/Cas9 in long-term engrafting HSPCs that are preserved in downstream effector lineages15,16,24,25. However, the risk exists that Cas9-induced off-target cleavage events could lead to loss of some critical stem cell function, activate oncogenes, or inactivate a tumor suppressor and remains a hypothetical concern for translation of this methodology to clinical trials. While several groups have described novel mutant Cas9 enzymes with reduced off-target cleavage activity26–28, these experiments were performed with plasmid-based Cas9 delivery several groups have described novel mutant Cas9 enzymes with reduced off-target cleavage activity26–28, these experiments were performed with plasmid-based Cas9 delivery systems in immortalized cell lines, as opposed to RNP delivery. Therefore, discovery of a Cas9 mutant that does not sacrifice on-target activity while reducing OTEs in the RNP context would have great impact on therapeutic genome editing.
Using an unbiased bacterial screening approach, we identified a single point mutation (R691A) in Cas9 (hereafter referred to as” high fidelity Cas9”, or “HiFi Cas9”) that reduces global OTEs while maintaining high on-target activity when used as an RNP complex. When compared to the R691A HiFi Cas9, we demonstrate that the rationally-designed eSpCas9(1.1)26, SpCas9-HF127, and HypaCas928 high-fidelity Cas9 mutants suffer reduced on-target editing at many sites when used as an RNP. We demonstrate clinical utility of HiFi Cas9 in targeting several important disease-associated loci for HR in clinically-relevant primary human CD34+ HSPCs and T-cells. We also show robust correction of the sickle cell disease (SCD)-causing Glu6Val mutation in HSPCs, while reducing OTEs up to 20 fold compared to wild-type (WT) Cas9.
RESULTS
Existing Cas9 mutants with improved specificity also exhibit reduced on-target activity with RNP delivery
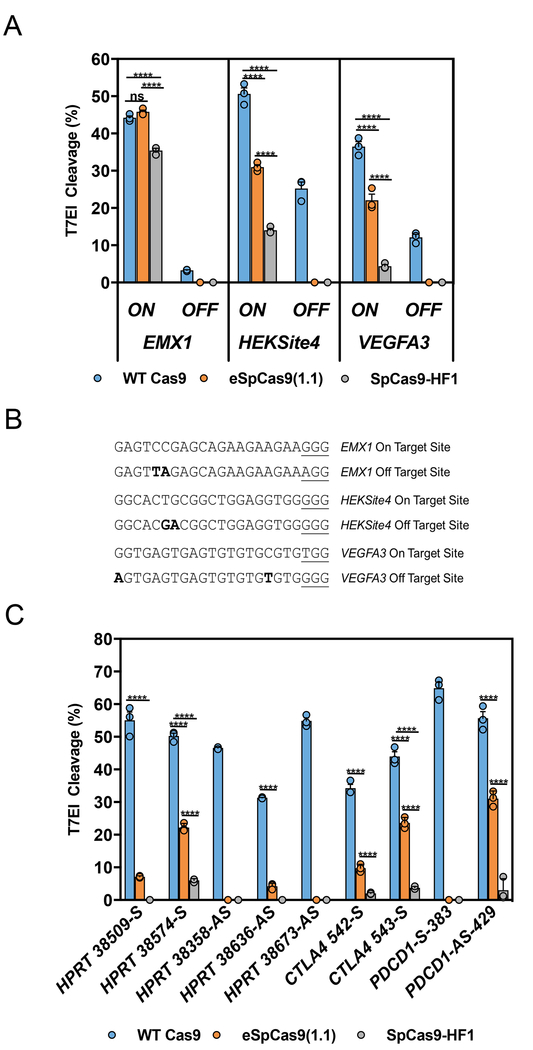
While delivery of the Cas9:gRNA complex as a RNP can dramatically reduce OTEs, certain guide sequences still cleave off-target sites regardless of the delivery mechanism15,16,29. We compared the relative on- and off-target cleavage activities using RNP delivery in HEK293 cells for WT Cas9 and two published mutants, eSpCas9(1.1) (K848A, K1003A, and R1060A) or SpCas9-HF1 (N497A, R661A, Q695A, and Q926A)26,27 at three previously characterized guide sites, VEGFA3, EMX1, and HEKSite4 (Figs. 1a and 1b). The eSpCas9(1.1) mutant showed an on-target editing efficiency that was similar to WT Cas9 with the EMX1 guide; however, it only functioned at 60% of WT when using both the HEKSite4 or VEGFA3 guides. The SpCas9-HF1 protein had even lower activity, showing 28% of WT with the HEKSite4 guide and 12% of WT with the VEGFA3 guide (Fig. 1a,b). A reduction in Cas9 activity for SpCas9-HF1 and eSpCas9(1.1) delivered as a RNP has been previously described, confirming the disadvantage of using these engineered Cas9 variants in the RNP format for high efficiency gene editing16,30. On-target editing activity in RNP format was further tested using 9 guides that target sites within the human HPRT, CTLA4, and PDCD1 genes. The eSpCas9(1.1) mutant on average produced only 23% of the WT Cas9 editing activity, with the best guide in this set showing 56% of the WT activity. The SpCas9-HF1 mutant showed even lower performance and on average produced only 4% of the WT Cas9 editing activity, with the best guide in this set showing 12% of the WT activity (Fig. 1c). Thus these mutants, both of whom bear multiple amino acid changes, show a pronounced reduction in on-target activity when used with short duration RNP delivery.
Figure 1. On-target activity of high-fidelity Cas9 mutants in human cells with ribonucleoprotein (RNP) delivery.
(a) Editing efficiency of the WT (blue), eSpCas9(1.1) (orange), or SpCas9-HF1 (gray) Cas9 proteins with crRNAs that target EMX1, HEKSite4, or VEGFA3 loci in HEK293 cells. The on-target site “ON” and off-target site “OFF” for each guide are indicated and sequence maps are listed in Fig. 1b. (b) Sequence maps corresponding to the on-target and off-target sites analyzed in Fig. 1a. Underlined sequences correspond to the PAM site and bold sequences indicate nucleotide mismatches. (c) On-target editing efficiency of the WT (blue), eSpCas9(1.1) (orange), or SpCas9-HF1 (gray) Cas9 proteins with crRNAs that target the indicated sites within the HPRT, CTLA4, and PDCD1 loci in HEK293 cells. Bars represent mean ± s.e.m., n=3 independent experiments performed at different times. ****P < 0.0001, NS (not significant) = P ≥ 0.05, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.
Bacterial selection of novel Cas9 mutants maintain on-target editing efficiency while reducing off-target editing
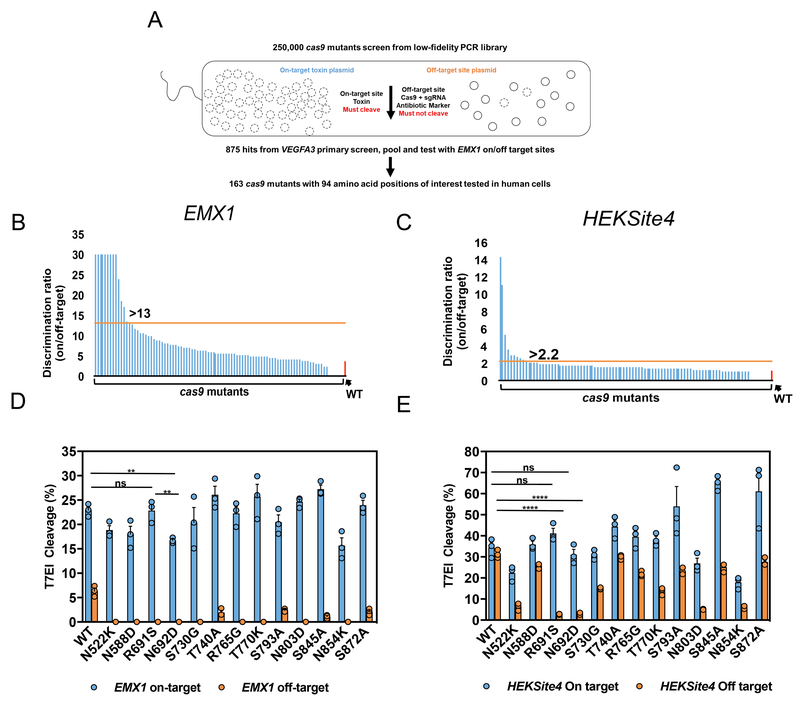
We performed an unbiased and high-throughput bacterial selection scheme to identify Cas9 mutants with an improved combination of on-target potency and specificity. We reasoned that mutants selected with this optimization approach would perform well with RNP delivery, which requires that total editing be achieved with transient expression of Cas9 protein. Positive selection E. coli bacterial screening methods have been adapted to isolate Cas9 mutants with altered PAM recognition specificities31,32. In these studies, if Cas9 fails to cleave the on-target plasmid, toxin expression leads to cell lysis whereas if Cas9 cleaves the on-target plasmid, toxin is not produced and the cells survive and proliferate. We adapted this strategy to select for reduced off-target editing by adding a known off-target site to the Cas9 and sgRNA expression plasmid (that also contained an antibiotic resistance gene) such that surviving colonies represent cells where Cas9 both 1) cleaved the on-target site in the toxin plasmid, and 2) did not cleave the off-target site in the antibiotic resistance selection plasmid (Fig. 2a).
Figure 2. Unbiased bacterial selection for cas9 mutants that reduce off-target editing and maintain on-target potency.
(a) A general schematic for the bacterial selection scheme to isolate cas9 mutants with reduced off-target editing and high on-target activity. Approximately 250,000 clones were screened for cleavage of the on-target VEGFA3 toxin plasmid, and avoidance of cleavage of the off-target plasmid. We isolated 875 surviving colonies from the primary screen, and pooled surviving plasmids were screened a second time using EMX1 guide and target sites. Ultimately 163 cas9 mutant plasmids were sequenced with mutations at 94 positions occurring twice or more. (b,c) Discrimination ratio (on/off-target editing efficiency) of WT Cas9 (red bar) and all 94 cas9 point mutations (blue bars) delivered by plasmid into HEK293 cells with the EXM1 (b) or HEKSite4 (c) gRNAs (2-part). Horizontal lines (orange) indicate the minimum discrimination ratios (indicated numerically above bars) used to select mutants for further testing. Mutants that maintained greater than 50% on-target editing and reduced off-target editing beyond the limits of detection were arbitrarily set at 30 (EMX1 only) to indicate that they were carried forward into subsequent screening steps. (d,e) On- and off-target editing efficiency facilitated by WT and mutant Cas9 plasmids from Fig. 2b,c examined with crRNAs that target the EMX1 (d) and HEKSite4 (e) delivered as by lipofection into HEK293 cells. The on-target site “ON” (blue – left Y-axis) and off-target site “OFF” (orange – right Y-axis) for each guide are indicated and sequence maps are listed in Fig. 1b. Bars represent mean ± s.e.m., n=9 independent experiments performed at different times. **P<0.01, ****P < 0.0001, NS (not significant) = P ≥ 0.05, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.
The primary screen was performed using a random Cas9 open reading frame mutant library of ~250,000 clones (average 5-7 mutations per kb) and on/off-target site pairs using the VEGFA3 guide (Figs. 1b and 2a). We isolated 875 surviving colonies that were pooled and used to perform a secondary screen with on/off-target site pairs using the EMX1 guide (Fig. 2b). We isolated 163 survivors from the second screen and DNA sequencing identified 94 amino acid positions where mutations were found at least twice (Supplementary Fig. 1a). All 94 humanized Cas9 mutant plasmids were tested in HEK293 cells for both on- and off-target editing efficiency targeting EMX1 or the off-target problematic HEKSite4 guide (Supplementary Figs. 1b and 1c)27. In total, 13 mutants were identified that reduced off-target editing with both EMX1 and HEKSite4 guides and exhibited an on-target editing efficiency at least 50% of the level of WT Cas9 (Figs. 2b and 2c). Overall, the R691S and N692D mutants demonstrated the best combination of on/off-target editing efficiencies (Figs. 2d and 2e). In most cases, alternative substitution of these mutants to alanine yielded a similar or improved phenotype, so we focused on alanine variants for subsequent studies (Supplementary Figs. 2a and 2b). When we compared the activity of R691A and N692A mutants to the previously described high-fidelity Cas9 mutants using the HEKSite4 or EMX1 guides, they induced INDELs at similar frequencies, suggesting that these single point mutations did not influence on-target activity (Supplementary Figs 2c and 2d).
The R691A high fidelity (HiFi) Cas9 mutant demonstrates superior on-target editing with RNP delivery
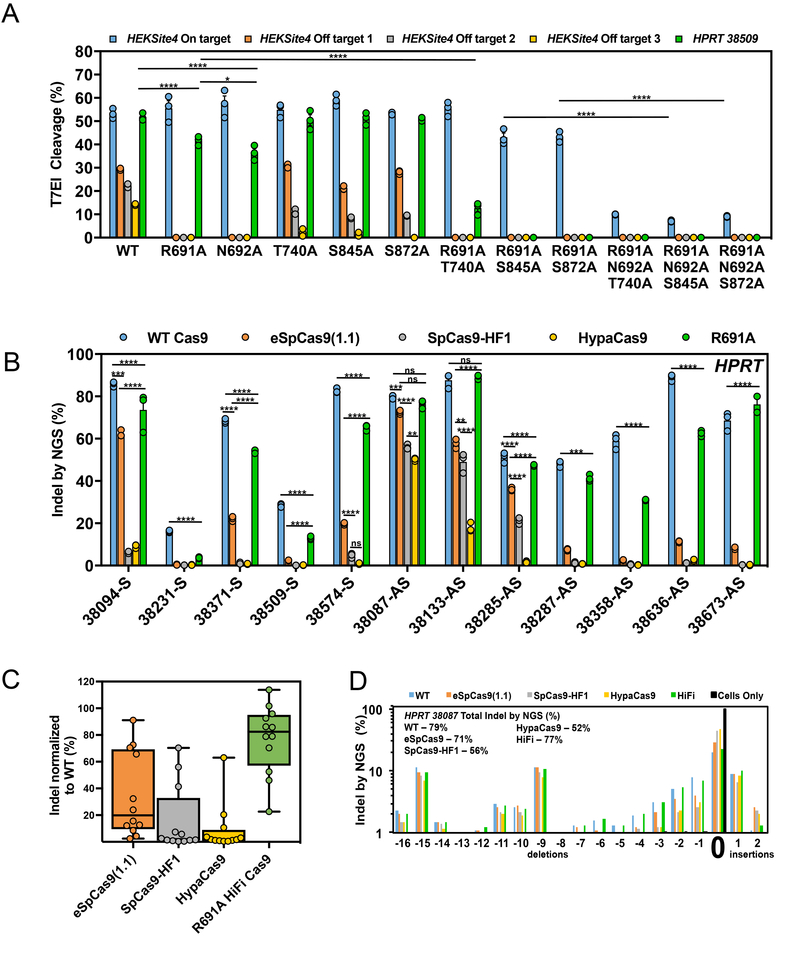
We examined combinations of mutations first using plasmid delivery and then tested the most promising single and multiple mutants in RNP format. While coupling additional mutations to either R691A or N692A eliminated residual off-target editing in every case tested, a majority of these mutant combinations also compromised on-target editing efficiency, including the R691A N692A double mutant (Supplementary Fig. 2e). Of note, plasmid-delivered R691A/T740A, R691A/S845A, and R691A/S872A double mutants eliminated off-target editing without influencing on-target editing efficiency. These results suggest that there can be a tipping point in which optimizing on off-target activity can compromise on-target activity.
We then purified promising recombinant single, double, or triple high-fidelity Cas9 mutants and examined both on- and off-target editing with RNP delivery. On-target editing efficiencies were scrutinized using both a “low activity” guide, HPRT-38509, that is ideally suited for distinguishing subtle differences in on-target editing efficiency, as well as the HEKSite4 guide. Overall, the single R691A mutant exhibited the best overall activity and specificity (Fig. 3a). When delivered as RNP, mutant N692A showed a modest but reproducible reduction in on-target editing efficiency relative to R691A, which is consistent with prior observations using plasmid-based delivery (Supplementary Figs. 2a and 2b). Interestingly, all recombinant triple mutants tested significantly reduced on-target activity compared to WT and HiFi Cas9, suggesting that the previously published mutants may have been over engineered to function as an RNP as all published mutants have 3 or more amino acid substitutions.
Figure 3. The R691A high fidelity (HiFi) Cas9 mutant maintains editing activity when delivered as an RNP.
(a) On-and off-target editing efficiency of the WT and bacteria-selected mutant Cas9 proteins with crRNAs that target the HEKSite4 (blue – on-target, orange – off-target 1, gray – off-target 2, yellow – off-target 3) and HPRT-38509 (green - on-target) loci in HEK293 cells. (b) On-target editing efficiency of WT Cas9 (blue), eSpCas9(1.1) (orange), SpCas9-HF1 (gray), HypaCas9 (yellow), and R691A (green) mutant proteins examined using 12 crRNAs that target different sites within the HPRT locus. Numbers included in the target site name indicate the first nucleotide of the target site with respect to the transcription start site of HPRT. Bars represent mean ± s.e.m., n=3 independent experiments performed at different times. (c) Box and whiskers plot showing aggregated on-target efficiency data of the eSpCas9(1.1) (orange), SpCas9-HF1 (gray), HypaCas9 (yellow), and R691A (green) mutants examined at the HPRT locus in Fig. 3b. The on-target editing efficiencies for each mutant at each site were normalized as a percent of WT Cas9 to account for varying editing efficiencies between guides. Horizontal lines represent median while showing the maximum and minimum values, n=12 from aggregated data in Fig. 3b. (d) Repair profile plot demonstrating the frequency and location of deletions and insertions relative to the unaltered HPRT-38087 target cleavage site (indicated by “0”). Experiments were performed with cells only (black bars), WT Cas9 (blue bars), eSpCas9(1.1) (orange bars), SpCas9-HF1 (gray bars), HypaCas9 (yellow bars), and HiFi (green bars). The overall INDEL frequencies for each experiment are indicated. Bars represent mean ± s.e.m., n=3 independent experiments performed at different times. For statistical comparisons, compare the edge of both sides of the horizontal lines. *P<0.05, **P<0.01, ***P<0.001, ****P < 0.0001, NS (not significant) = P ≥ 0.05, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.
We further interrogated the R691 position by constructing all 19 possible amino acid substitutions and tested these mutants with plasmid delivery to evaluate all possible substitutions at this position. Surprisingly, many of these substitutions (13/19) maintained good on-target editing efficiency while reducing off-target editing below that of the WT R691; only the R691K and R691P mutations failed to improve off-target editing (Supplementary Figs. 3a and 3b). We tested the most promising mutants (R691D, R691G, R691H, R691Y, and R691W) in RNP format but none showed any improvement over the original R691A (Supplementary Figs. 3c and 3d).
A more comprehensive survey was performed to compare the relative on-target activity of the R691A HiFi Cas9 mutant with WT Cas9 and three other improved fidelity mutants eSpCas9(1.1), SpCas9-HF1, and HypaCas9 (R692A/M694A/Q695A/H698A)28. The 5 different Cas9 RNPs were tested in HEK293 cells targeting 12 sites within the human HPRT locus with INDEL detection performed using next-generation sequencing (NGS). The normalized median on-target editing efficiencies for each mutant was: 20% for eSpCas9(1.1), 2% for SpCas9-HF1, 1.7% for HypaCas9, and 82% for the R691A HiFi Cas9 mutant (Figs. 3b and 3c). A more extended comparison of WT Cas9 vs HiFi Cas9 RNP on-target activity was performed with guide RNAs that target an additional 48 sites, and these results showed a similar trend where the HiFi Cas9 mutant on average functioned at 92% the efficiency of WT Cas9 (Supplementary Fig. 3e). NGS analysis of the INDEL spectrum between WT and high-fidelity Cas9 mutants indicates that there is no obvious difference in repair outcomes between enzyme variants (Fig. 3d). Thus the R691A HiFi Cas9 retains editing efficiency comparable to WT Cas9 when used in RNP format while the 3 other mutants show reduced editing efficiencies.
Quantitative reduction in off-target editing using the HiFi mutant
The GUIDE-seq procedure10 can be used to empirically identify real off-target sites in cells. This method relies on DSB capture of short oligos to “mark” cleavage events in living cells. GUIDE-seq, while effective at finding off-target cleavage sites, often yields inconsistent quantitation of low-frequency editing events. Quantification of off-target editing was therefore done using amplicon-based NGS in separate experiments amplifying sites identified by GUIDE-seq.
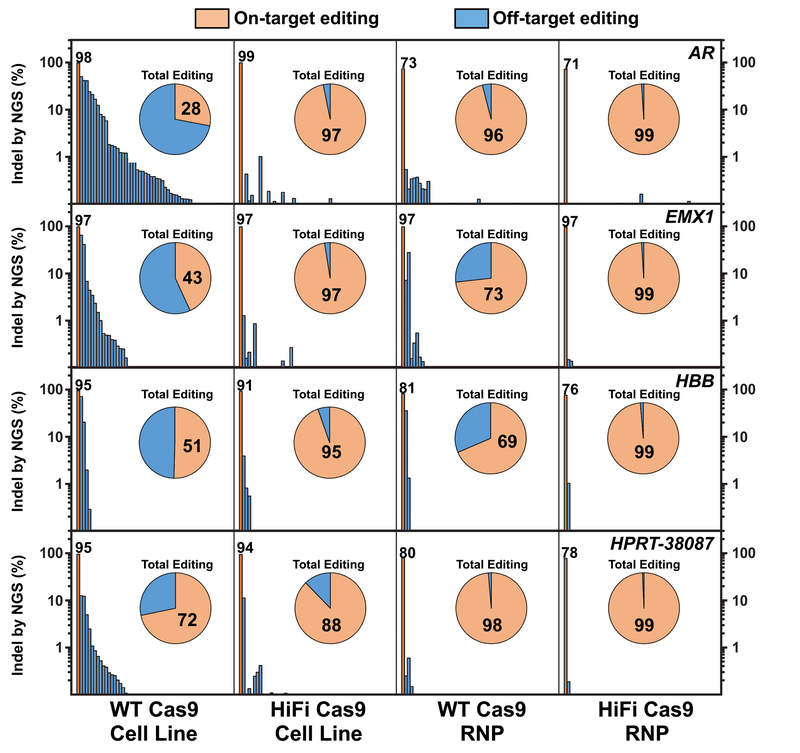
The off-target sites for gRNAs that target genes EMX1, AR, HPRT, and HBB were empirically determined by using the GUIDE-seq procedure with electroporation of gRNA (10 μM) and the GUIDE-seq tag into HEK293 cells that stably express low levels of WT or HiFi Cas9 as a transgene (Supplementary Figs. 4a and 4b). The R691A HiFi Cas9 cell line showed a striking reduction in off-target editing, even with of constitutive Cas9 expression (Supplementary Fig. 4a). Quantitative assessment of INDEL frequency was then carried out using multiplex PCR primer pool-based33 amplicon NGS at top sites predicted using CRISPOR34 as well as GUIDE-seq sites. With constitutive WT Cas9 expression, INDEL formation at the on-target locus occurred at 95-98% frequency, but there were sufficient OTEs that constituted of 28-72% of total editing events (Fig. 4). Constitutive expression of HiFi Cas9, however, maintained 91-99% INDEL formation at the on-target site that resulted in 88-97% of total editing events. While RNP delivery of WT Cas9 reduced OTEs compared to the WT Cas9 cell line, HiFi Cas9 dramatically reduced off-target editing such that on-target events were in every case 99% (or greater) of the total editing events detected. All of the previously reported high fidelity Cas9 mutant proteins reduce off-target editing relative to WT Cas9, but OTE reduction occurred at the expense of on-target editing efficiency (Supplementary Fig. 5).
Figure 4. HiFi Cas9 globally reduces off-target activity with both stable Cas9 expression and RNP delivery.
On-target (orange bars) and off-target editing (blue bars) as determined by NGS for the AR, EMX1, HBB, and HPRT-38087 (top to bottom) gRNAs (4 μM) delivered into HEK293 cells that express WT or HiFi Cas9, or complexed to WT or HiFi Cas9 and delivered as RNP (4 μM) into standard HEK293 cells (left to right). INDEL formation percentages at the on-target loci are indicated directly above the orange on-target bars. All amplicons are rank-ordered (highest to lowest) by INDEL formation percentage as determined for the WT Cas9 stable cell line. Pie charts indicate the fractional percentage of total on-target (orange) and off-target (blue) editing. Fractional on-target editing percentage is indicated in black typeface within the orange on-target portion of the pie chart. The Y-axis is plotted as log-10 scale on both left and right sides of the graph for clarity. The Cas9 protein and delivery method is indicated at the bottom of the graph.
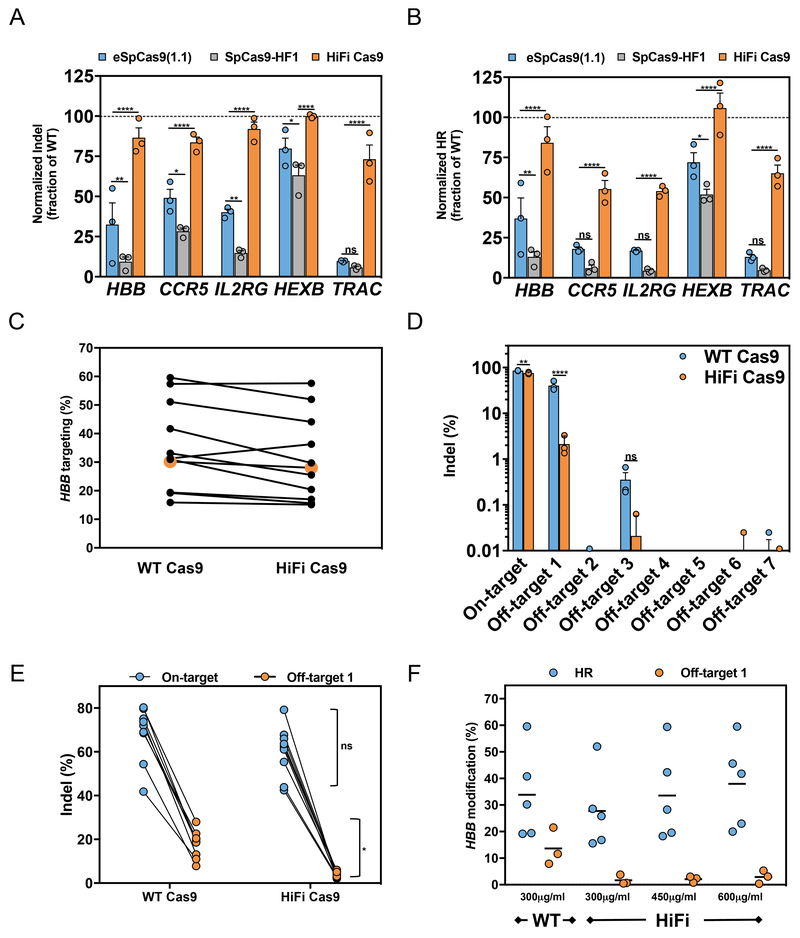
The R691A HiFi Cas9 mutant delivered as RNP is highly active and specific in CD34+ HSPCs
We next examined on-target editing efficiencies of WT, HiFi and other published improved-specificity Cas9 proteins in clinically-relevant human primary cells. RNPs were prepared using chemically-modified sgRNAs12 that target common disease-associated genes (HBB, CCR5, IL2RG, HEXB, and TRAC) and were electroporated into CD34+ HSPCs. The R691A HiFi Cas9 mutant demonstrated near-WT levels of INDEL formation across all sites (~75% of WT Cas9 activity for all five genes) (Fig. 5a and Supplementary Fig. 6a). The previously described improved-specificity Cas9 mutants delivered as RNP showed markedly reduced on-target editing efficiencies, with the eSpCas9(1.1) showing less than ~50% activity of WT in four out of five genes and the SpCas9-HF1 Cas9 showing less than ~25% of WT efficiency.
Figure 5. RNP HiFi Cas9 facilitates near-WT on-target editing potency with large OTE reductions in primary CD34+ HSPCs.
(a) On-target editing efficiencies of WT Cas9 (normalized dotted line), eSpCas9(1.1) (light blue), SpCas9-HF1 (gray), and R691A (orange) mutant proteins examined using chemically-modified sgRNAs that target 5 clinically-relevant loci. Cas9 proteins and sgRNAs were delivered as RNP (1:2.5 – Cas9:sgRNA) by nucleofection into CD34+ HSPCs. INDEL frequencies were calculated by TIDE. Bars represent mean ± s.e.m., n=3 independent experiments performed in three healthy CD34+ cord blood donors. (b) Homologous recombination (HR) frequencies of Cas9 variants were examined as in Fig. 5a, except following the formation of site-specific Cas9 DSBs, electroporated cells were transduced with ssAAV6 homologous donor templates as described in the materials and methods. For HBB and CCR5, HR was measured via GFPhigh expression via FACS; HR at IL2RG and HEXB was measured by assessing allelic-targeted integration via ddPCR; HR at TRAC locus was measured by tNGFR expression. Bars represent mean ± s.e.m., n=3 independent experiments performed in three CD34+ cord blood donors. (c) CD34+ HSPCs were electroporated as described above with either WT HBB RNPs or HiFi HBB RNPs and then were immediately transduced with an HBB-specific rAAV6 (UbC-GFP flanked with HBB homology arms). Cells were harvested and then analyzed for GFPhigh expression (which is a measure of homologous recombination) via FACS (n =11, number of data points within each group, all from different cord blood donors from at least 3 independent experiments). (d) CD34+ HSPCs from 3 healthy donors were electroporated as described above with WT or HiFi HBB-RNPs. HBB (on-target), the top 7 bioinformatically determined and GUIDE-seq validated regions (see Supplementary Fig. 4b) were PCR-amplified with sequencing primers utilized in deep sequencing MiSeq runs. Bars represent mean ± s.e.m., n=3 independent experiments performed in three CD34+ cord blood donors. (e) Cord blood-derived CD34+ HSPCs were electroporated with WT or HiFi Cas9 HBB RNPs, cultured for 72-96hr post electroporation, gDNA was harvested, and HBB (on target) or a highly complementary off-target site (off-target 1) were PCR amplified. PCR products were gel extracted and INDEL frequencies were calculated using TIDE software (Bars represent mean ± s.e.m., n =6, number of data points within each group, all from different cord blood donors from at least 3 independent experiments performed at different times). (f) HSPCs were electroporated with 300 μg mL−1 WT Cas9 or 300–600 μg mL−1 HiFi Cas9 and half of the cells were transduced with an UbC-GFP ssAAV6 donor and the other half were cultured. Following 72-96 hr post-electroporation, cells were harvested and analyzed for HR by GFP FACS analysis or off-target INDELs by TIDE. Median is depicted, n=5, number of data points within each group, all from different cord blood donors from at least 3 independent experiments performed at different times). *P<0.05, **P<0.01, ***P<0.001, ****P < 0.0001, NS (not significant) = P ≥ 0.05, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.
Since INDELs are only one measure of a DSB repair (NHEJ), we also tested the capacity of HiFi Cas9 to promote HR in HSPCs. AAV6 homologous donor DNA templates are an efficient way to deliver donor templates for sensitive primary immune cells, including HSPCs and T-Cells15,35–40. Guides were complexed with either WT, eSpCas9(1.1), SpCas9-HF1 or HiFi Cas9, electroporated into HSPCs, then transduced with gene-specific AAV6 donors. The HiFi Cas9 was significantly more active at promoting HR in HSPCs (>50% of WT activity at all loci tested) than all of the other high-fidelity Cas9 variants. Of note, the eSpCas9(1.1) again was more active than the SpCas9-HF1 Cas9 (Fig. 5b and Supplementary Fig. 6b). We also examined the performance of the HypaCas9 mutant28 in targeting HBB with RNP delivery, which likewise showed a significant reduction in on-target editing efficiency (Supplementary Fig. 6c). Furthermore, we tested the on-target activity of the newly described evoCas941 (M495V/Y515N/K526E/R661Q) at the EMX1 site with the RNP delivery method and again found severely reduced INDEL activity compared to WT and HiFi Cas9 (Supplementary Fig. 6d). These results imply that the HiFi Cas9 is currently the most active and specific SpCas9 available when delivered as an RNP complex.
The HBB guide site is a good candidate to evaluate the specificity of HiFi Cas9 in HSPCs since 1) HiFi Cas9 facilitated levels of HR similar to that of WT Cas9 at the on-target locus, 2) the HBB guide has a known off-target site that persists with RNP delivery, and 3) the HBB gene is of immediate clinical relevance. Across 11 independent experiments, the HiFi Cas9 mediated a median of 28% HR in comparison to 30% HR for the WT Cas9 (Fig. 5c and Supplementary Fig. 6e). While maintaining nearly identical on-target INDEL activity, the HiFi Cas9 reduced INDEL frequency at the dominant off-target site by 20 fold (Figs. 5d, 5e and Supplementary Figs. 4b and 6c). Any reduced HiFi efficiencies could be fully rescued by increasing the concentration of HiFi Cas9 used without adversely affecting specificity or cell viability (Fig. 5f and data not shown). Our rigorous quality characterization assays ensure that the reduced RNP activity of the 3 previously published improved-fidelity Cas9 mutants was not influenced by protein purity, stability or storage effects, we performed extensive quality characterization assays on the recombinant proteins (Supplementary Fig 7). These stability studies also support that the HiFi variant has properties well suited to translational uses, as single lots of protein are stable over time.
Gene knockouts in primary T-cells are currently being pursued for therapeutic applications. For example, knockout of the TRAC locus is used when generating off-the-shelf allogeneic CAR-T cells42 and knockout of CCR5 can establish HIV resistance43. Therefore, we tested if the R691A HiFi Cas9 mutant was as active as WT Cas9 in primary human T-cells. Human T-cells were isolated from buffy coats and were electroporated with CCR5 or TRAC gene-specific WT or HiFi RNPs. The HiFi Cas9 showed similar high activity in primary T-cells as it did in HSPCs (Supplementary Fig. 8a). We also show that AAV6 can be used as a donor template to knock in a CD19-specific CAR-T into the TRAC locus (Supplementary Fig 8b), creating potentially non-immunogenic allogeneic CAR-T cells39,44. These results show that the HiFi Cas9 can be used in the context of T-cell RNP-based therapeutic genome editing, another indication where a reduced risk for off-target activity is desirable.
The R691A HiFi Cas9 delivered as RNP mediates high-level Glu6Val correction while reducing off-target activity in sickle cell disease-CD34+ HSPCs
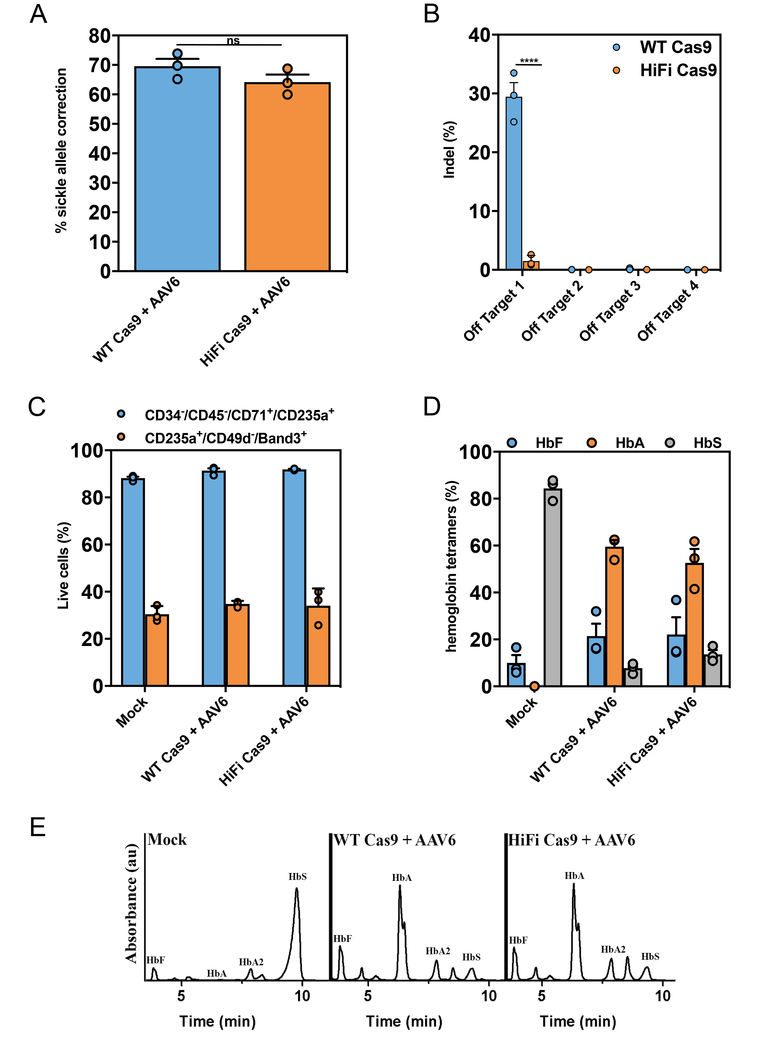
Sickle cell disease (SCD) is an autosomal recessive red blood cell disorder that is genetically caused by the nucleotide change of an adenine (A) to thymidine (T) in codon 6 of HBB leading to substitution of glutamic acid by valine (“Glu6Val”) in the beta-globin protein. It is postulated that HR-mediated HBB gene correction in autologous HSPCs would be a safe and definitive curative approach for SCD. Accordingly, several reports have shown efficient Glu6Val correction in SCD-HSPCs, although correction efficiency can sometimes be linked with off-target INDELs in the same edited population15,16,45,46.
SCD-HSPCs were targeted with a sickle correction ssAAV6 donor15 by either electroporating HiFi Cas9 or WT Cas9 RNPs. Compared to WT Cas9 editing, the HiFi Cas9 converted a nearly equivalent number of Glu6Val alleles (~70% vs ~64%), while reducing off-target cutting by 20 fold (Figs. 6a and 6b). Mock electroporated, WT Cas9 HBB targeted and HiFi Cas9 HBB targeted SCD-HSPCs were differentiated down the erythrocyte lineage47 and tested for hemoglobin tetramers via cation-exchange HPLC. Equivalent frequencies of erythrocytes (CD34−/CD45−/CD71+/CD235a+)48 and reticulocytes (CD235a+/CD49d−/Band3+)49 were generated between the HiFi and WT Cas9 targeted samples (Fig. 6c). Hemoglobin tetramer analyses confirmed the high frequency of Glu6Val correction using HiFi Cas9 by showing that >50% of hemoglobin tetramers produced in targeted differentiated erythrocytes were adult hemoglobin (HbA) (Figs. 6d and 6e).
Figure 6. R691A HiFi Cas9 delivered as RNP mediates robust Glu6Val HBB gene correction in sickle cell disease-CD34+ HSPCs while significantly reducing off-target activity.
(a) Sickle cell disease (SCD) patient-derived HSPCs were isolated from peripheral blood and subjected to Glu6Val gene correction methods using HBB RNPs (either WT (blue) or HiFi (orange) Cas9) and a gene corrective ssAAV6. Gene correction frequencies were analyzed by ddPCR™ as described in materials and methods (Bars represent mean ± s.e.m., n =3, number of data points within each group, all from different SCD patients). NS (not significant) = P ≥ 0.05, Two-tailed Student’s T-test. (b) SCD-HSPCs were targeted for Glu6Val HBB gene correction as described above and off-target editing was analyzed by NGS. Bars represent mean ± s.e.m., n=3 SCD patients. ****P < 0.0001, NS (not significant) = P ≥ 0.05, two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test. (c) Glu6Val targeted WT or HiFi SCD-HSPCs were differentiated down the erythroid lineage ex vivo. At 14 days post-differentiation, cells were harvested and evaluated for the percentage of erythrocytes (CD34−/CD45−/CD71+/CD235a+) and reticulocytes (CD235a+/CD49d−/Band3+) by FACS-based immunophenotypic analyses. Bars represent mean ± s.e.m., n=3 SCD patients. (d) Glu6Val targeted erythrocyte differentiated HSPCs were subjected to cation-exchange HPLC analysis of steady-state hemoglobin tetramers. The normalized percentages of fetal (HbF), adult (HbA) and sickle (HbS) hemoglobin are plotted as a function of total hemoglobin tetramers on the HPLC plot (as presented in Fig. 6f). Hemoglobin tetramers were quantified by measuring the area under the curve (AUC) of the absorbance peaks. Bars represent mean ± s.e.m., n=3 SCD patients. (e) Representative HPLC plot from the data presented in Fig. 6d showing Glu6Val correction in SCD-HSPCs results in HbA protein production when using both the WT and HiFi Cas9 HBB RNPs. Experiment is representative of the 3 experiments performed in Fig. 6d with similar results.
DISCUSSION
Using an unbiased, high-throughput Cas9 mutant screen to select for high on-target and low off-target activity, we identified a Cas9 variant (R691A) that displays an improved on-to-off-target ratio when delivered as a ribonucleoprotein (RNP) complex. The HiFi Cas9, when delivered as an RNP and electroporated into human primary blood stem and effector cells, stimulates robust gene targeting at therapeutically-relevant loci, including correction of the SCD-causing Glu6Val mutation in patient-derived cells.
While the previously identified improved-specificity Cas9 mutants do not function as well as the WT or R691A HiFi mutant using RNP delivery methods (Figs. 3b, 3c, 5a, 5b and Supplementary Fig. 6), these earlier studies do offer important mechanistic insights into how Cas9/sgRNA complexes recognize the target (and off-target) and lead to a DSB. For example, it was originally hypothesized that the eSpCas9(1.1) and SpCas9-HF1 mutants lowered off-target cleavage levels by reducing the relative affinity of the mutant Cas9 nuclease to the substrate DNA. The mutations present in eSpCas9(1.1) affect interactions between the protein and the non-targeted DNA strand, while the mutations present in SpCas9-HF1 affect interactions between the protein and the targeted DNA strand. It was theorized that these amino-acid substitutions would further destabilize any mismatches between the gRNA and the substrate DNA26,27. In contrast, it was recently shown that these mutants do not have a decreased affinity for substrate, leading to a new theory that these mutations instead affect transition of Cas9 structure from an inactive conformation to an active conformation and that this transition occurs less efficiently in the presence of mismatches between the RNA guide and the DNA target28. These new data led to development of a new variant, HypaCas9, which included 4 mutations (N692A/M694A/Q695A/H698A) in the REC3 domain. In the present study, the most potent and specific mutation identified was R691A and the second best mutation identified was N692A, which is identical to one of the 4 sites mutated in the HypaCas9 mutant. It therefore seems likely that the R691A HiFi Cas9 improves specificity through a similar mechanism of inhibiting the transition from the inactive to active state in the presence of sgRNA target mismatches. Interestingly, when the R691 and N692 REC3 point mutations were combined (Fig. 3a and Supplementary Fig. 2e), a reduction in on-target activity was observed when used in RNP format. We suspect that the combination of multiple mutations into a single Cas9 variant results in over-engineering of the protein that often lowers on-target catalytic activity when delivered in the RNP format. We propose that the sustained overexpression of both enzyme and sgRNA that results when using plasmid-based systems overcomes this loss of activity and accounts for the discrepancies seen in performance between RNP-based and plasmid-based delivery of these mutants. Experiments with plasmid-based expression of the Cas9 mutants previously described in the literature support this conclusion (Supplementary Fig. 2c and 2d). It will be interesting to see future work that fully characterizes the mechanisms, structure and conformational changes of the R691A HiFi Cas9 mutant when bound to the target sequence.
Ex vivo RNP-mediated Cas9 genome editing of HSPCs and T-cells will likely be some of the first Cas9 editing programs to enter clinical trials within the next few years. While safety and efficacy can only be truly tested in Phase I clinical trial in humans, using the best possible reagents is important for risk mitigation. We show that the R691A HiFi Cas9 has similar on-target efficiency but is more specific than WT Cas9 for mediating HBB gene targeting in primary human HSPCs. Similar studies done examining sites at CCR5, IL2RG, HEXB and TRAC showed on-target activity reduced by as much as ~40%, which was nevertheless still 3-10 times higher than the activity seen using eSpCas9(1.1) and SpCas9-HF1 targeting with AAV6 (Fig. 5b and Supplementary Fig. 6). Future experiments will examine whether increasing the amount of HiFi Cas9 can rescue this reduced targeting as it did for HBB (Fig. 5f) and will also test different combinations of NLS signals to determine if activity can be improved by improving nuclear localization. While the present work focused primarily on the utility of the HiFi Cas9 in RNP formats, specificity concerns exist when using all methods of Cas9 delivery for any in vivo application. Results obtained from the HEK293-HiFiCas9 cell line that show a reduction in OTEs, even with continuous HiFi Cas9 expression, suggest that the HiFi mutation may similarly have benefit if translated to use in AAV viral vectors50 where it will also be constitutively expressed. This benefit should also hold true when packaging HiFi Cas9 mRNA51,52 or RNPs53 into nanoparticles, or even when complexed with cell-penetrating peptides54 for tissue-specific delivery. We anticipate that future studies will investigate the utility of HiFi Cas9 in these therapeutically relevant contexts.
In conclusion, the new R691A HiFi Cas9 variant will have broad applicability in CRISPR/Cas9 genome editing approaches for both research and clinical purposes.
METHODS
Bacterial strains and plasmid construction
Bacterial strain DH5α (F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1) was used for routine cloning purposes, and strain BW2514155 (F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR)580, λ−, galU95, ΔuidA3::pir+, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514) was used for the Cas9 mutagenesis screen. Bacterial strain BL21(DE3) (F– ompT gal dcm lon hsdSB(rB–mB–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS)) was used for Cas9 protein expression and purification. Relevant plasmids, primers, and gBlocks® gene fragments used in this study are listed in Supplementary Tables 1, 2, and 3, respectively. The toxin plasmid pIDTV-CcdB was created by subcloning gBlocks® (Integrated DNA Technologies, Inc, Coralville, IA, USA) gene fragments encoding lacYA177C and araC-PBAD-ccdB into pUC19. The VEGFA3 or EMX1 on-target sites were introduced into pIDTV-CcdB with site-directed mutagenesis resulting in plasmids pIDTV-CcdB-V3 and pIDTV-CcdB-EMX1, respectively. The Cas9 and VEGFA3 sgRNA expression and off-target site plasmid pIDTV-V3sg-off1 was made by subcloning PCR-amplified cas9 and the V3off1 gBlocks® fragment into the NcoI/SacI/PstI sites of pACYCduet-1 (Millipore Sigma, Burlington, MA, USA). The Cas9 and EMX1 sgRNA expression and EMX1 off-target site plasmid pIDTV-EXsg-off1 was made by subcloning PCR-amplified (WT or mutant) cas9 and the EXoff1 gBlocks® fragment into the NcoI/SacI/PstI sites of pACYCduet-1 (Millipore Sigma, Burlington, MA, USA). The commercially-available Alt-R® S.p. Cas9 Expression Plasmid (Integrated DNA Technologies, Inc) was used for all plasmid-based Cas9 experiments in human cells. Mutations were introduced into the Alt-R® S.p. Cas9 Expression Plasmid by site-directed mutagenesis (Supplementary Table 2). Unless otherwise indicated, bacteria were grown in LB medium containing the following antibiotics as necessary: carbenicillin (50 μg mL−1), kanamycin (50 μg mL−1), and chloramphenicol (25 μg mL−1). For toxin induction during Cas9 mutant selection, arabinose was added to a final concentration of 0.5% (w/v).
E. coli dual selection Cas9 mutant screen
The basic bacterial selection scheme performed was adapted from a previously described positive selection system for identifying Cas9 mutants in E. coli31. E. coli strain BW25141(DE3) was transformed with the pIDTV-CcdB-V3 or pIDTV-CcdB-EMX1 toxin plasmids, and single colonies were verified to permit both carbenicillin resistance and arabinose-dependent toxicity. These colonies were used to make electrocompetent cells for screening using ice cold 10% glycerol. A mutant Cas9 library was generated by first amplifying the cas9 gene with low-fidelity PCR using the GeneMorph II random mutagenesis kit (Agilent, Santa Clara, CA, USA), introducing 5–7 mutations per kilobase. The resulting fragments were then subcloned into the pIDTV-V3sg-off1 plasmid to generate a mutant library with less than 1% WT Cas9 as determined by Sanger sequencing. Approximately 250,000 transformants were screened using strain BW25141(DE3) pIDTV-CcdB-V3 yielding 875 surviving colonies for an effective survival rate of 0.35%, significantly above the 0.01% background survival rate of the VEGFA3 on/off-target site pair. Plasmid DNA from survivors was pooled and cas9 mutants were subcloned into pIDTV-EXsg-off1 to create a low complexity library, which was then used to perform a secondary screen with on/off-target site pairs from EMX1 (Fig. 2b). Background survival with the EMX1 plasmids indicating that this on/off-target site pair was more stringent than the VEGFA3 primary screen (data not shown). We screened 20,000 transformants to be > 99% confident that all 875 possible plasmids within the pool were represented at least one time, and 163 survivors were isolated in total (Supplementary Fig. 1).
Isolation and culture of HEK293 cells
The human embryonic kidney cell line HEK293 was used for the majority of HiFi Cas9 development studies. HEK293 cells were cultured in Dulbecco’s Modified Essential Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA). For routine genome editing experiments cells were cultured to an average 80–90% confluency and dissociated with trypsin.
Genome editing by plasmid lipofection in HEK293 cells
Plasmid-based genome editing experiments in HEK293 cells were performed with 0.5 μL per well TransiT-X2® (Mirus Bio, Madison, WI, USA) lipid transfection reagent, 0.1 μg per well Cas9 human expression plasmid, and 30 nM 1:1 (crRNA:tracrRNA) Alt-R® (Integrated DNA Technologies, Inc) gRNA complex prepared in a volume of 50 μL total using Opti-MEM™ reduced serum media (Thermo Fisher Scientific) according to the manufacturer’s specifications. Transfections were performed in 96-well plates with 50,000 HEK293 cells in a total volume of 150 μL using DMEM with 10% FBS, and DNA was extracted 48 hrs post-transfection using QuickExtract™ DNA extraction solution (Epicentre Biotechnologies, Madison, WI, USA) according to the manufacturer’s specifications. Mutation detection was performed (unless otherwise indicated) with T7 Endonuclease I using the Alt-R® Genome Editing Detection Kit (Integrated DNA Technologies, Inc) according to the manufacturer’s instructions. For the T7EI assays, both “cells only” and “no gRNA” controls were performed for every experiment to assess background levels of INDELs. Our T7EI experiments were performed only with amplicons that gave no detectable background under these conditions. The targeting sequence for each Cas9 crRNA is listed in Supplementary Table 4.
Genome editing by RNP lipofection in HEK293 cells
RNP lipofection genome editing experiments in HEK293 cells were performed with 1.2 μL per well Lipofectamine® RNAiMAX lipid transfection reagent and 10 nM (1:1:1, Cas9:crRNA:tracrRNA) Alt-R® (Integrated DNA Technologies, Inc) Cas9 RNP complex as described previously56. Guide RNAs were prepared according to the manufacturer’s instructions and RNPs were formed by the addition of purified Cas9 protein to gRNAs at an equimolar ratio in Opti-MEM™ with 50 μL total volume. Complexes were allowed to form for 10 min at room temperature prior to lipid complex formation. Transfections were performed with 35,000 HEK293 cells in 96-well plates in a volume of 150 μL total using DMEM with 10% FBS, and DNA was extracted 48 hrs post-transfection using QuickExtract DNA extraction solution (Epicentre Biotechnologies) according to the manufacturer’s specifications. Mutation detection was performed (unless otherwise indicated) with T7 Endonuclease I using the Alt-R® Genome Editing Detection Kit (Integrated DNA Technologies, Inc) according to the manufacturer’s instructions. PCR cycling parameters for each amplicon consisted of an initial 5 minute incubation at 95° C, followed by 30 cycles at 98° C for 20 seconds, 64–68° C for 30 seconds, and 72° C for one minute. The annealing temperature for each amplicon varied for each primer set and is indicated in Supplementary Table 2. The targeting sequence for each Cas9 crRNA is listed in Supplementary Table 4.
Genome editing by RNP electroporation in HEK293 cells
RNP electroporation genome editing experiments in HEK293 cells were performed using the Lonza 96-well Shuttle™ System (Lonza, Basel, Switzerland), 4 μM (1:1.2:1.2, Cas9:crRNA:tracrRNA) Alt-R® (Integrated DNA Technologies, Inc) Cas9 RNP complex, and 4 μM Alt-R® Cas9 Electroporation Enhancer (Integrated DNA Technologies, Inc) as described previously56. Chemically synthesized guide RNAs and crRNAs were prepared according to the manufacturer’s instructions and RNPs were formed by the addition of purified Cas9 protein to gRNAs in 1X phosphate buffered saline (PBS). Complexes were allowed to form for 10 min at room temperature prior to electroporation. RNP complexes (5 μL) and 3.5 × 105 HEK293 cells (20 μL) were mixed and electroporated according to the manufacturer’s specifications using protocol 96-DS-150. DNA was extracted 48 hrs post-transfection as described above. The targeting sequence for each Cas9 crRNA is listed in Supplementary Table 4.
Isolation of HEK293-PCMV-Cas9 stable cell lines
Human codon-optimized SpCas9, as well as the flanking 5’- and 3’ nuclear localizing sequences and 5’-V5 tag were digested from the GeneArt® CRISPR Nuclease Vector (Thermo Fisher Scientific) using NotI/EcoRI, and this fragment was subcloned into the pcDNA™3.1(−) mammalian expression vector (Thermo Fisher Scientific) resulting in pIDTG-Hs-Int-SpCas9. This plasmid was linearized with the restriction enzyme PvuI (New England Biolabs, Ipswich, MA, USA) at a single site in the ampicillin resistance gene (bla) and transfected into HEK293 cells by lipofection using Lipofectamine® 2000 (Thermo Fisher Scientific) for random chromosomal integration. Neomycin-resistant cells were selected using DMEM containing 500 μg mL−1 G418 (Gibco), and a monoclonal population of cells was isolated by limiting dilution. The resulting stable cell line was determined to have a single copy of the cas9 gene by Droplet Digital™ PCR (Bio-Rad, Hercules, CA, USA), the correct cas9 sequence by Sanger sequencing, and the inserted fragment was mapped to the X chromosome by inverse PCR57. The resulting HEK293-PCMV-Cas9 stable cell line was used as a starting point to make the HiFi R691A substitution to ensure that WT and HiFi cell lines would be otherwise as close to genetically identical as possible with similar expression levels. The R691A substitution was introduced into the cas9 gene by HR following Cas9 cleavage by transfecting gRNA and HR donor oligonucleotide by lipofection. A crRNA was used that targets a TGG PAM site located 3 nt 5’ adjacent to the R691-CGA arginine codon, and a ssDNA donor oligo containing symmetrical, 40 nt homology arms corresponding to the non-targeting strand (Supplementary Table 2). Monoclonal cell populations were isolated by limiting dilution, and a clone expressing the R691A variant of Cas9 was successfully isolated. The WT and HiFi stable cell lines were confirmed to produce equivalent steady-state Cas9 expression levels (Supplementary Fig. 4c). Western blots were performed to compare Cas9 expression levels between WT and HiFi HEK293 stable cell lines using a method that has been described previously58. The Anti-beta-actin (Cat# ab8226) and Anti-SpCas9 (Cat# ab191468) were both obtained from Abcam. Reactive bands in a Western Blot were confirmed to be of the correct molecular weight and were dependent on the presence of the corresponding gene.
Expression and Purification of Cas9 Proteins
A gBlocks® gene fragment (Integrated DNA Technologies, Inc) encoding E. coli codon-optimized S. p. cas9 with a single 5’ SV40 NLS and dual 3’ SV40 NLS sequences was subcloned into the NcoI/NotI restriction sites of pET28b (Novagen) and the resulting plasmid was called pIDTV-NLS-C9–2NLS-H. Mutations were introduced into this plasmid with site-directed mutagenesis protocols that have been described previously59. E. coli Bl21(DE3) carrying pIDTV-NLS-C9–2NLS-H was grown to mid-log phase at 37 °C (OD600-0.6), at which time the culture was chilled to 18 °C and 1 mM IPTG was added. Following shaking incubation (250 rpm) at 18 °C for 24 hrs, cells were collected by centrifugation and lysed at 4 °C with the Emulsiflex®-C3 high pressure homogenizer (Avestin, Ottawa ON, Canada). Cas9 was purified from clarified lysates using immobilized metal affinity chromatography (IMAC) and heparin chromatography protocols that have been described for Cas9 previously60. Purified proteins in this study were rigorously studied through commercial-grade analytical and quality control procedures. Briefly, proteins were analyzed for purity via denaturing SDS-PAGE and RP-HPLC and were determined be > 95% pure. Every protein was quality controlled to contain <10 EU/mg of endotoxin, be free of residual host cell DNA beyond the limits of detection with qPCR, and to be free of contaminating DNase/RNases. Proteins were stored at 10 mg mL−1 (61 μM) at −20 °C in a standard enzyme storage buffer (20 mM Tris-HCl pH 7.4, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 50% glycerol) to avoid multiple freeze-thaw cycles. WT Cas9 and the mutants analyzed in these studies are stable for at least 4 months at −20 °C in storage buffer, whereas repeated freeze-thaw cycles tended to cause precipitation and loss of activity over time. Proteins were determined to be of equal activity from the time of initial purification to the completion of work presented herein (Supplementary Fig. 7).
NGS-based sequencing of global editing in HEK293 cells
The unique target-specific domain of each sequencing primer for both on- and off-target studies is listed in Supplementary Table 5. NGS-based sequencing of on-target loci was performed as described previously56. The GUIDE-seq method was used to identify global editing events in an unbiased fashion. The procedure was performed as described previously except for the source of Cas9 and gRNA complex. In this study, we used the HEK293-PCMV-Cas9-WT and HEK293-PCMV-Cas9-HiFi stable cell lines as the source of Cas9. The Alt-R® gRNA complexes (10 μM – 1:1 – crRNA:tracrRNA) were delivered by electroporation into HEK293 cells along with 0.5 μM GUIDE-seq dsDNA donor fragment. DNA was extracted 72 hrs after electroporation using the GeneJET Genomic DNA purification kit (Thermo Fisher Scientific). NGS library preparation, sequencing, and operation of the GUIDE-seq software was performed as described previously10. To better quantify editing percentages at off-target sites found using GUIDE-seq, multiplex PCR coupled to amplicon NGS was performed using rhPCR with blocked-cleavable primers. This amplification technology requires that the primer properly hybridize to a target site before amplification. Mismatches between target and primer prevent unblocking, thereby increasing specificity and eliminating primers dimers33. This approach enables efficient production of highly multiplex PCR amplicons in a single tube. Data were analyzed using a custom-built pipeline. PCR amplicons were sequenced on an Illumina MiSeq instrument (v2 chemistry, 150bp paired end reads) (Illumina, San Diego, CA, USA). Data were demultiplexed (Picard tools v2.9; https://github.com/broadinstitute/picard); forward and reverse reads were merged into extended amplicons (flash v1.2.11)61; reads were aligned against the GRCh38 genomic reference (bwa mem v0.7.15), and were assigned to targets in the multiplex primer pool (bedtools tags v2.25)62. Reads with any base quality score <10 were filtered out. At each target, editing was calculated as the percentage of total reads containing an INDEL or SNP within a 10bp window of the cut site.
In vitro culture of human CD34+ HSPCs
CD34+ HSPCs were harvested and cultured as previously described15,63,64. Briefly, HSPCs were used from either fresh cord blood (CB) (kindly provided by Binns Family program for Cord Blood Research), isolated from the peripheral blood of SCD patients, or frozen plerixafor mobilized CD34+ (AllCells). HSPCs were cultured at 250,000–500,000 cells/mL in StemSpan SFEM II (Stemcell Technologies) supplemented with SCF (100 ng mL−1), TPO (100 ng mL−1), Flt3-Ligand (100 ng mL−1), IL-6 (100 ng mL−1), UM171 (35 nM), 20 mg mL−1 streptomycin, 20 unit/mL penicillin and CB and SCD-derived with StemRegenin1 (SR1) (0.75 μM). Cells were cultured at 37 °C, 5% CO2 and 5% O2.
Isolation, culture and genome editing of primary human T cells
Buffy coats were obtained from the Blood Center of the Stanford University School of Medicine and T cells isolated through Ficoll density gradient centrifugation followed by magnetic enrichment using the Pan T cell isolation kit (Miltenyi, Bergisch Gladbach, Germany). Cells were cultured at 37°C, 5% CO2 in X-Vivo 15 (Lonza) supplemented with 5% human serum (Sigma-Aldrich, St. Louis, MO, USA) and 100 IU/mL human recombinant IL-2 (Peprotech, Rocky Hill, NJ, USA) and 10 ng mL−1 human recombinant IL-7 (BD Biosciences, San Jose, CA, USA). T cells were activated using immobilized Anti-CD3 (clone OKT3, Tonbo Biosciences) and soluble anti-CD28 (clone CD28.2, Tonbo Biosciences) for 3 days before electroporation. The cells were resuspended in electroporation solution, mixed with the pre-complexed RNP and electroporated using a 4D-Nucleofector (Lonza) using program EO-115. To assess homologous recombination, the cells were transduced with rAAV after electroporation. Genomic DNA was harvested 4 days after electroporation using QuickExtract™ DNA Extraction Solution (Epicentre). To assess the phenotype, cells were stained with Anti-CD3-PE (clone UCHT1, BioLegend, San Diego, CA, USA) and Anti-CD271 (NGFR)-APC (clone ME20.4, BioLegend) and analyzed on an Accuri C6 (BD Biosciences) or a CytoFLEX (Beckman Coulter, Indianapolis, IN, USA) flow cytometer. Data was analyzed using FlowJo® (FlowJo LLC, Ashland, OR, USA).
AAV6 vector cloning, production and purification
AAV vector plasmids were cloned in the pAAV-MCS plasmid (Agilent Technologies, Santa Clara, CA) containing ITRs from AAV serotype 2 (AAV2). All of the constructs had at least one homology arm directly next to the break if not both of them. For the HBB targeting construct, it contained the left homology arm (LHA, 540bp), UbC-GFP (approx. 2200bp) and then the right homology arm (RHA, 400bp). For the IL2RG targeting vector, it contained the LHA (400), SFFV-GFP (approx. 1300bp) and a RHA (414bp). For the CCR5 targeting construct, it contained the LHA (400bp), EF1α-GFP (approx. 2000bp) and RHA (400bp). For the HEXB targeting construct, it contained a LHA (500bp), Iduronidase cDNA-flag3x, SFFV-GFP (approx. 1300bp), and a RHA (500bp). For the TRAC targeting construct, it contained the LHA (400bp), EF1α-CD19 CAR (approx. 2700bp) and the RHA (400bp). TRAC donors used in T cell experiments contained LHA (400bp), T2A-CD19-T2A-tNGFR (approx. 2500bp), and RHA (400bp). rAAV6 vectors were produced as described with a few alterations65. Briefly, 293T cells (Life Technologies, Carlsbad, CA, USA) were seeded at 13–15 million cells per dish in ten 15 cm dishes one day before they were transfected. Each 15 cm dish was transfected using standard PEI transfection with 6 μg ITR-containing plasmid and 22 μg pDGM6 (gift from David Russell, University of Washington, Seattle, WA, USA), which contains the AAV6 cap genes, AAV2 rep genes, and Ad5 helper genes. Cells were incubated for 48–72 hr until rAAV6 was harvested from cells by three freeze-thaw cycles followed by 45 min incubation with Benzonase (Thermo Fisher Scientific) at 250 U/mL. AAV6 vectors were purified on an iodixanol density gradient by ultracentrifugation at 48,000 rpm for 2.25 hr at 18°C. Full AAV6 capsids were extracted at the 40–58% iodixanol interface and then were dialyzed three times in 1× PBS (for 2h each) with 5% sorbitol in the last dialysis (for 2h) using a 10K MWCO Slide-A-Lyzer G2 Dialysis Cassette (Thermo Fisher Scientific). Vectors were added with F-68 pluronic acid to a final concentration of 0.001%, aliquoted, and then stored at 80°C until further use. rAAV6 vectors were tittered using quantitative PCR or ddPCR to measure number of vector genomes as previously described66
Electroporation and AAV6 transduction
The single guide (sgRNAs) used in this study for editing primary hematopoietic stem and progenitor cells as well as effector T-cells were purchased from TriLink BioTechnologies (San Diego, CA, USA) and HPLC-purified. The modified sgRNAs contain 2’-O-methyl-3’-phosphorothioate modifications at the three terminal nucleotides of the 5’ and 3’ ends as described previously12. The genomic target sequences for the guides were as follows: HBB: 5’-CTTGCCCCACAGGGCAGTAA-3’, CCR5: 5’-GCAGCATAGTGAGCCCAGAA-3’, IL2RG: 5’-GGTAATGATGGCTTCAACA-3’, HEXB; 5’-CCCCAGCCCGCACAGCTCCA-3’, and TRAC: 5’-GAGAATCAAAATCGGTGAAT-3’. All Cas9 proteins (WT and mutants) were obtained from Integrated DNA Technologies. RNP complexes were made by mixing Cas9 with sgRNA at a molar ratio of 1:2.5 (Cas9:sgRNA) at 25ºC for 10 minutes prior to electroporation. CD34+ cells were electroporated with RNP at a concentration of 300 μg mL−1 (unless stated otherwise). CD34+ cells were resuspended in either P3 buffer (Lonza) or buffer 1M67 and electroporated using the Lonza 4D Nucleofector (with program DZ100). Cells were plated at 250,000–500,000 cells/mL in cytokine-supplemented media and then rAAV6 was delivered onto cells immediately (within 15 minutes) upon plating after electroporation ranging vector genomes/cell from 50,000–100,000 as tittered by Q-PCR or 5,000–10,000 as tittered by ddPCR.
Gene targeting analysis by fluorescence activated cell sorting (FACS)
After cells were targeted with WT or mutant Cas9 RNP and gene-specific AAV6 vectors, the cells were harvested at 2–4 days post electroporation and the percentage of cells was measured by transgene expression. HBB (GFP reporter), CCR5 (GFP reporter), and TRAC (CD19 CAR or tNGFR reporter) targeting efficiencies were carried out by FACS. Cells were analyzed for viability and GFP expression using either the Accuri C6 flow cytometer (BD Biosciences) or a FACS Aria II SORP (BD Biosciences). For CD19 CAR analysis, targeted cells (and negative controls) were washed and stained for L-Biotin (GenScript, Piscataway, NJ, USA) at 3 ug mL−1 for 20 min at 4 °C, washing, and secondary staining was performed using Streptavidin-APC (BD Biosciences).
Gene targeting analysis by digital droplet PCR (ddPCR)
The percentage of HEXB and IL2RG targeted alleles were measured by ddPCR™. All gDNA samples used in ddPCR were first extracted using QuickExtract™ DNA Extraction Solution (Epicentre). IL2RG targeted gDNA was then digested in HINDIII-HF and HEXB was digested in XbaI following the manufacturer’s instructions (New England Biolabs). 1–3 μL of gDNA sample was then used in 25 μL ddPCR™ Supermix for Probes (No dUTP) (BioRad) following the manufacturer’s instructions with integration specific and reference primer/probes that were purchased at a ratio of 3.6:1. Amplification was performed using in-out PCR (one primer binding to transgene insert in the genomic locus and other primer binding to the specific locus outside of the homology arm). Droplet samples were prepared using 20 μL of the PCR mix and 70 μL droplet generation oil, and 40 μL of the droplet sample. PCR cycling conditions are as follows: 98°C (10 minutes), [94°C (30 seconds), 57 or 60°C (30 seconds), 72°C (2 minutes)] (X40–50 cycles), 98°C (10 minutes). Finally, droplets were analyzed according to the manufacturer’s instructions using the QX200™ Droplet Digital™ PCR System (Bio-Rad). The primers used to amplify integrated transgenes and references are as follows: HEXB forward: 5’-GGGAAGACAATAGCAGGCAT-3’, HEXB reverse: 5’-GTCTAAGCCCTTCGGGTAAC-3’, HEXB probe: 5’ FAM-CTAGTGAGCTGTGCGGGCTG-BHQ-3’, IL2RG forward: 5’-AAGGGGGAGGATTGGGAAG-3’, HEXB reverse: 5’-TCAGAAGGAGGAGGCCAAG-3’, HEXB probe: 5’-HEX-GCATGCTGGGGATGCGGTGGGC-BHQ-3’. The primers used to amplify the reference genes CCRL2 and IL2RG (upstream of cut site) were as follows: CCRL2 forward: 5’-GCTGTATGAATCCAGGTCC-3’, CCRL2 reverse: 5’-CCTCCTGGCTGAGAAAAAG-3’, CCRL2 probe 5’ HEX-TGTTTCCTCCAGGATAAGGCAGCTGT-BHQ-3’, IL2RG (ref) forward: 5’-GGGAAGGTAAAACTGGCAAC-3’, IL2RG (ref) reverse: 5’-GGGCACATATACAGCTGTCT-3’, IL2RG (ref) probe 5’ FAM-CCTCGCCAGTCTCAACAGGGACCCAGC-BHQ-3’.
Glu6Val allele correction analysis by nested-ddPCR
HBB edited SCD-HSPCs were harvested 2 days post electroporation (4 days in culture) and then analyzed for correction frequencies of the Glu6Val mutation (as well as the synonymous nucleotide changes) by using a nested-ddPCR approach. An ‘In-Out’ PCR approach (as described above) was carried out to only amplify the chromosomal HBB locus, excluding the episomal AAV6 from analysis. The following primers were used to generate an HBB-specific 1.4kb band: forward (out): 5’-AGGAAGCAGAACTCTGCACTTCA-3’ and reverse (in): 5’-AGTCAGTGCCTATCAGAAACCCAAGAG-3’. The PCR product was purified and then diluted to 10 fg/μL in nuclease-free water and then ddPCR was carried out as described above with the PCR product generated as the template (20 fg total amplicon). This scheme used a 2-probe set on the sample amplicon, where one probe (HEX) binds the HR sequence and the other probe (FAM) is a reference sequence downstream of the Cas9 break site. To calculate the percentage of alleles that had HR, we divided the Poisson-corrected copies/μL HEX (HR)/FAM (Ref). The following primer/probes were used in the ddPCR reaction: HR probe (HEX)-5’-TGACTCCTGAGGAAAAATCCGCAGTCA-3’, reference probe (FAM)-5’-ACGTGGATGAAGTTGGTGGTGAGG-3’, nested-forward 5’-TCACTAGCAACCTCAAACAGAC-3’ and nested-rev 5’-CCTGTCTTGTAACCTTGATACC-3’.
Measuring INDEL frequencies by TIDE (tracking of INDELs by decomposition)
INDEL frequencies were quantified using the TIDE software68 and sequenced PCR products obtained by PCR of genomic DNA extracted at 2–4 days after electroporation as previously described12. Mock electroporated cells were always used as a negative control for calculating INDEL frequencies with TIDE. The primers used to amplify the genes of interest were as follows: HBB; forward: 5’-CCAACTCCTAAGCCAGTGCCAGAAGAG-3’, reverse: 5’-AGTCAGTGCCTATCAGAAACCCAAGAG-3’, IL2RG; forward: 5’-TCACACAGCACATATTTGCCACACCCTCTC-3’, reverse: 5’-TGCCCACATGATTGTAATGGCCAGTG-3’, CCR5; forward: 5’-GCACAGGGTGGAACAAGATGG-3’, reverse: 5’-CACCACCCCAAAGGTGACCGT-3’, HEXB; forward: 5’-TAGCCATCCCGTGTTTGAGGTT-3’, reverse: 5’-AGAGCGTGGGTTCAGTCCATGT-3’, TRAC; forward: 5’-ATCACGAGCAGCTGGTTTCT-3’, reverse: 5’-CCCGTGTCATTCTCTGGACT-3’.
HBB sgRNA Off-Target Identification
Bioinformatic prediction of potential off-target sites for HBB (R-02) was carried out using the webtool COSMID69 with up to 3 mismatches allowed in the 19 PAM proximal bases and the PAM sequence NRG. GUIDE-Seq was used for experimental identification of potential off-target sites. Briefly, a plasmid expressing R-02 and Cas9 was nucleofected into U2OS cells as previously described10. Genomic DNA was extracted 3 days after nucleofection, sheared using a Covaris M220 focused ultrasonicator to an average size of 500 bp. The DNA was prepared and sequenced on an Illumina MiSeq and the resulting sequencing data analyzed using the standard pipeline10 with a reduced gap penalty.
Quantification of putative HBB off-target activity in CD34+ HSPCs by deep sequencing
DNA flanking the potential off-target sites was amplified by two rounds of PCR to introduce Illumina sequencing adaptors and sample barcodes. All PCR amplicons were normalized to 20 nM, pooled, and then quantified using the PerfeCTa® NGS Quantification kit (Quantabio, Beverly, MA, USA). Samples were prepared for the Illumina MiSeq according to manufacturer’s instructions and sequenced using 2 × 250bp paired end reads. CRISPR-Cas9 genome editing outcomes from deep sequencing data was analyzed as previously described70. Mock electroporated negative controls were used as to subtract background INDELs when performing the aforementioned analyses. The limit of detection in these experiments is based on the inherent error rate of the Illumina platform (up to 0.4%), which has previously been described for genome editing experiment analyzed by NGS71–73.
In vitro differentiation of SCD-CD34+ HSPCs into erythrocytes and reticulocytes
SCD patient-derived HSPCs were cultured over 14–16 days following targeting at 37 °C and 5% CO2 in SFEM II media according to previously established protocols47,49. Media was supplemented with 100 U mL−1 penicillin/streptomycin, 10 ng mL−1 SCF, 1 ng mL−1 IL-3 (PeproTech), 3U mL−1 erythropoietin (eBiosciences) 200 μg mL−1 transferrin (Sigma-Aldrich), 3% AB serum (heat inactivated from Atlanta Biologicals, Flowery Branch, GA, USA), 2% human plasma (umbilical cord blood), 10 μg mL−1 insulin (Sigma Aldrich) and 3U mL−1 heparin (Sigma-Aldrich). In the first phase of erythroid differentiation, corresponding to days 0–7 (day 0 being day 4 after electroporation), cells were cultured at 100,000 cells/mL. In the second phase, corresponding to days 7–10, cells were maintained at 100,000 cells/mL and IL-3 was discontinued from culture media. In the third and final phase, days 11–16, cells were cultured at 1,000,000 cells/mL and transferrin was increased to 1 mg mL−1. Cells were then immunophenotyped for essential erythrocyte cell surface markers as described below.
Immunophenotyping of differentiated erythrocytes
HSPC were differentiated as described above and then were harvested at day 14-16 for analyses of erythrocyte lineage specific markers. Cells were spun down and washed and then erythrocyte differentiation of edited (WT and HiFi Cas9) and non-edited HSPCs was assessed by flow cytometry using the following antibodies: hCD45 V450 (HI30, BD Biosciences), CD34 FITC (8G12, BD Biosciences), CD71 PE-Cy7 (OKT9, Affymetrix), and CD235a PE (GPA) (GA-R2, BD Biosciences). Reticulocyte differentiation was analyzed using the following antibodies: CD235a PE (GPA) (GA-R2, BD Biosciences), CD49d BV421 (9F10, BD Biosciences), and Band3 APC (a kind gift from Dr. Anumpama Narla and Dr. Mohan Narla).
HPLC analysis of steady-state hemoglobin tetramers
To lyse erythrocytes, water equal to 3 volume of the packed cells was added and mixed with the cells by pipetting up and down. The mixture was incubated at room temperature for 15 min followed by 30 s in a sonicating water bath to aid the lysis. The lysate was then separated from the erythrocyte ghosts by centrifugation at 13,000 rpm for 5 min. HPLC analysis of hemoglobins was the performed. Hemoglobins in their native form were analyzed on a weak cation-exchange PolyCAT A column (100 × 4.6-mm, 3 µm, 1000Å) (PolyLC Inc., Columbia, MD, USA) using a Shimadzu UFLC system at room temperature. Mobile phase A (MPA) consists of 20 mM Bis-tris + 2 mM KCN, pH 6.96. Mobile phase B (MPB) consists of 20 mM Bis-tris + 2 mM KCN + 200 mM NaCl, pH 6.55. Clear hemolysate was diluted 4 times in MPA and then 20 µl was injected onto the column. A flow rate of 1.5 mL/min and the following gradient were used in time (min)/%B: (0/10%; 8/40%; 17/90%; 20/10%; 30/stop).
Statistical tests used in this study
All the statistical tests used in this study were completed with Prism7 GraphPad Software. The specific statistical tests used for each comparison are specifically denoted in the figure legends. For making multiple comparisons, we used two-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test. For comparing the average of two means, we used the Student’s T-test to reject the null hypothesis (P<0.05).
Supplementary Material
ACKNOWLEDGEMENTS
M.H.P. gratefully acknowledges the support of the Amon Carter Foundation, the Laurie Kraus Lacob Faculty Scholar Award in Pediatric Translational Research and NIH grant support R01-AI097320 and R01-AI120766. We thank the Binns Program for Cord Blood Research at Stanford University for cord-blood-derived CD34+ HSPCs and also for SCD-HSPCs. Sickle cell disease patients consented on the use of CD34+ HSPCs for research with the accompanying IRB approval. We thank Sruthi Mantri for collecting and purifying the HSPCs. We thank Kim Lennox for critical review of the manuscript.
Footnotes
Life science reporting summary can be found on the online version of this manuscript.
Supplementary information is available in the online version of this manuscript.
DATA AVAILABILITY
The authors declare that the data supporting the findings of this study are available within the paper. The sequencing data is available at NCBI under SRA accession: SRP150376.
COMPETING FINANCIAL INTERESTS
M.H.P. is a consultant and has equity interest in CRISPR Tx, but CRISPR Tx had no input or opinions on the subject matter described in this manuscript. C.A.V., G.R.R., R.T., A.M.J., M.A.C., N.M.B., M.S.M., S.Y., and M.A.B. are employees of Integrated DNA Technologies (IDT), which sells reagents similar to some described in the manuscript.
REFERENCES
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA & Charpentier E Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES & Zhang F Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagdas YS, Chen JS, Sternberg SH, Doudna JA & Yildiz A A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci Adv 3, eaao0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sternberg SH, LaFrance B, Kaplan M & Doudna JA Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kass EM & Jasin M Collaboration and competition between DNA double-strand break repair pathways. FEBS letters 584, 3703–3708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porteus MH Towards a new era in medicine: therapeutic genome editing. Genome biology 16, 286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dever DP & Porteus MH The changing landscape of gene editing in hematopoietic stem cells: a step towards Cas9 clinical translation. Curr Opin Hematol 24, 481–488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron P, et al. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods 14, 600–606 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hendel A, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundry MC, et al. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep 17, 1453–1461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gundry MC, et al. Technical considerations for the use of CRISPR/Cas9 in hematology research. Exp Hematol 54, 4–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dever DP, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeWitt MA, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med 8, 360ra134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hultquist JF, et al. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep 17, 1438–1452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumann K, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A 112, 10437–10442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, Kim D, Cho SW, Kim J & Kim JS Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatti RA, Meuwissen HJ, Allen HD, Hong R & Good RA Immunological reconstitution of sex-linked lymphopenic immunological deficiency. Lancet 2, 1366–1369 (1968). [DOI] [PubMed] [Google Scholar]
- 21.Johnson FL, et al. Bone-marrow transplantation in a patient with sickle-cell anemia. N Engl J Med 311, 780–783 (1984). [DOI] [PubMed] [Google Scholar]
- 22.Lucarelli G, et al. Allogeneic marrow transplantation for thalassemia. Exp Hematol 12, 676–681 (1984). [PubMed] [Google Scholar]
- 23.Naldini L Gene therapy returns to centre stage. Nature 526, 351–360 (2015). [DOI] [PubMed] [Google Scholar]
- 24.De Ravin SS, et al. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci Transl Med 9(2017). [DOI] [PubMed] [Google Scholar]
- 25.Traxler EA, et al. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med 22, 987–990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang X, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208, 44–53 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Rees HA, et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 8, 15790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z & Zhao H A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic acids research 33, e154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobosy JR, et al. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol 11, 80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haeussler M, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome biology 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol 33, 1256–1263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic acids research 44, e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Ravin SS, et al. Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat Biotechnol 34, 424–429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sather BD, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med 7, 307ra156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyquem J, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiroli G, et al. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci Transl Med 9(2017). [DOI] [PubMed] [Google Scholar]
- 41.Casini A, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol 36, 265–271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborn MJ, et al. Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases. Mol Ther 24, 570–581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370, 901–910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacLeod DT, et al. Integration of a CD19 CAR into the TCR Alpha Chain Locus Streamlines Production of Allogeneic Gene-Edited CAR T Cells. Mol Ther 25, 949–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoban MD, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoban MD, et al. CRISPR/Cas9-Mediated Correction of the Sickle Mutation in Human CD34+ cells. Mol Ther 24, 1561–1569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dulmovits BM, et al. Pomalidomide reverses gamma-globin silencing through the transcriptional reprogramming of adult hematopoietic progenitors. Blood 127, 1481–1492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romero Z, et al. beta-globin gene transfer to human bone marrow for sickle cell disease. J Clin Invest (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu J, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JB, et al. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin H, et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci U S A 113, 2868–2873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24, 1020–1027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haldimann A, Daniels LL & Wanner BL Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol 180, 1277–1286 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobi AM, et al. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods 121–122, 16–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams M, Rainville IR & Nicklas JA Use of inverse PCR to amplify and sequence breakpoints of HPRT deletion and translocation mutations. Environ Mol Mutagen 39, 22–32 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Lennox KA, Vakulskas CA & Behlke MA Non-nucleotide Modification of Anti-miRNA Oligonucleotides. Methods Mol Biol 1517, 51–69 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Munteanu B, Braun M & Boonrod K Improvement of PCR reaction conditions for site-directed mutagenesis of big plasmids. J Zhejiang Univ Sci B 13, 244–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karvelis T, et al. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol 10, 841–851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magoc T & Salzberg SL FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bak RO, et al. Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AAV6. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bak RO & Porteus MH CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Rep 20, 750–756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan IF, Hirata RK & Russell DW AAV-mediated gene targeting methods for human cells. Nat Protoc 6, 482–501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aurnhammer C, et al. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum Gene Ther Methods 23, 18–28 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Chicaybam L, Sodre AL, Curzio BA & Bonamino MH An efficient low cost method for gene transfer to T lymphocytes. PLoS One 8, e60298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brinkman EK, Chen T, Amendola M & van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic acids research 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cradick TJ, Qiu P, Lee CM, Fine EJ & Bao G COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids 3, e214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee CM, Cradick TJ & Bao G The Neisseria meningitidis CRISPR-Cas9 System Enables Specific Genome Editing in Mammalian Cells. Mol Ther 24, 645–654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol 14, R51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quail MA, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guell M, Yang L & Church GM Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics 30, 2968–2970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.