Abstract
Mycobacterium bovis infection in cattle persists in Mexico, posing a threat to human health. Control of bovine tuberculosis, through the National Program Against Bovine Tuberculosis, has led to the decrease of disease prevalence in most of the country, except for high dairy production regions. Genotyping of M. bovis has been performed mainly by spoligotyping and variable number tandem repeats (VNTR), but higher resolution power can be useful for a finer definition of the spread of the disease. Whole genome sequencing and spoligotyping was performed for a set of 322 M. bovis isolates from different sources in Mexico: Baja California, Coahuila, Estado de Mexico, Guanajuato, Hidalgo, Jalisco, Queretaro and Veracruz, from dairy and beef cattle, as well as humans. Twelve main genetic clades were obtained through WGS and genetic diversity analysis. A clear differentiation of the Baja California isolates was seen as they clustered together exclusively. However, isolates from the central states showed no specific clustering whatsoever. Although WGS proves to have higher resolving power than spoligotyping, and since there was concordance between WGS and spoligotyping results, we consider that the latter is still an efficient and practical method for monitoring bovine tuberculosis in developing countries, where resources for higher technology are scarce.
Introduction
Mycobaterium bovis is known to infect a wide variety of mammals, including the badger [1], white-tailed deer [2], brush-tailed possum [3], dogs, cats, goats, buffaloes, and pigs [4], which can act as spillover hosts to more economically important species such as dairy and beef cattle, causing bovine tuberculosis (bTB). The importance of this pathogen escalates due to its ability to infect and cause serious disease in humans [5]. In developed countries, such as Australia, Canada, the USA and New Zealand have either eradicated the disease or very nearly eradicated it, with wild fauna reservoirs preventing total eradication [6]. In Mexico, however, bTB is endemic, and although 85.8% of the national territory shows a prevalence under 0.5% [7], high milk production regions have prevalences as high as 16% [8].
To this date, studies performed on bovine tuberculosis in Mexico consist mainly on the characterization of strains through spoligotyping and VNTR [8–12], and host-pathogen interactions for immunological studies [13,14]. Furthermore, the disease in cattle is monitored through the National Program for the Control and Eradication of Bovine Tuberculosis [15], which is based on test-and-slaughter, slaughterhouse surveillance and movement restrictions for infected herds. In addition, efforts towards the control and eradication of bTB in Mexico are encouraged to improve international trade of livestock and minimize the risk to public health.
Great breakthroughs have been made with the introduction of whole genome sequencing strategies for the study of outbreaks of M. tuberculosis in humans [16–18], through which it has been possible to detect transmission chains. If this could be achieved in cattle, principal sources of infection could be identified and control efforts can be proposed in more specific directions. A few studies have already used whole genome sequencing to gain insights into the local spread and persistence of the disease in some countries [19–21]. However, in developing countries, where resources are scarce, whole genome sequencing is difficult to implement. Spoligotyping is easy to perform, cheap and requires a low amount of DNA, which is why to this date it has shown to be a useful tool for large-scale epidemiological studies, with such success that some authors consider it to be the “gold standard” [22,23]. Therefore, the aim of this study is to perform spoligotype and WGS analysis on a sub-population of M. bovis isolates from different sources in Mexico in order to compare the usefulness of each technique in this particular setting.
Materials and methods
The study was performed mainly in Queretaro, Mexico (20.588056, -100.388056), at the Autonomous University of Queretaro. Bacterial isolates were collected in Aguascalientes (22.021667, -102.356389), Baja California (29.95, -115.116667), Coahuila (27.302222, -102.044722), Estado de Mexico (19.354167, -99.630833), Guanajuato (21.033333, -101.241667), Hidalgo (20.478333, -98.863611), Jalisco (20.566667, -103.676389), Queretaro and Veracruz (19.190278, -96.153333). Whole genome sequencing was performed at the National Veterinary Services Laboratories, in Ames, Iowa, USA (42.034534, -93.620369).
Bacterial isolates
A total of 322 M. bovis isolates collected from cattle (n = 320) and humans (n = 2) between 1997 and 2015 in different parts of Mexico: Aguascalientes (AGS = 28), Baja California (BCA = 25), Coahuila (COA = 10), Estado de Mexico (EDOMX = 29), Guanajuato (GTO = 8), Hidalgo (HGO = 12), Jalisco (JAL = 33), Queretaro (QRO = 176), and Veracruz (VER = 1) were included in the study. This collection of isolates had previously been obtained from: bTB suspicious lesions collected from bovine carcasses at slaughterhouses and sputum samples from TB suspicious patients. After isolation from culture in Stonebrink medium [24], colonies were stored at -80° C. The protocol of this project was approved by the Bioethics Committee of the School of Natural Sciences of the Autonomous University of Queretaro, with registry number 83FCN2016.
DNA extraction
DNA extraction was performed directly from M. bovis colonies. Briefly, colonies were thawed at room temperature for 1 hour. Then, about 100 μg of cell culture were homogenized for DNA extraction by the phenol-chloroform method (CTAB-chloroform method) [25]. Next, 50 μl of lysozyme (10 mg/ml) (Sigma Aldrich, St. Louis, Missouri, USA) was added and the sample homogenized and incubated at 37° C for 1 hour. Then 100 μl of 10% SDS was added followed by a 10 μl of a proteinase K (10 mg/ml) (Invitrogen, Carlsbad, California, USA) solution. The mixture was incubated in a warm bath at 65° C for 30 minutes. After incubation, 100 μl of 5 M NaCl was added and the mixture homogenized. This was followed by the addition of 40 μl of CTAB solution (Sigma Aldrich, St. Louis, Missouri, USA) at 10% and vortexed. The new mixture was incubated again at 65° C for 30 minutes. Following this, 400 μl of a phenol-chloroform-isoamyl alcohol reagent (volume ratio 24:24:1) was added and the mixture vortexed for 10 seconds and centrifuged at 13000 rpm for 10 minutes. From this mixture, 500 μl of aqueous phase was transferred to a 1.5 ml Eppendorf tube, where 600 μl of absolute isopropilic alcohol were added. The DNA pellet was then precipitated at -20° C for 2 hours. The product was centrifuged at 13000 rpm for 15 minutes. The supernatant was removed, and the pellet washed with 500 μl of ethanol (70%) and vortexed for 10 seconds, followed by two centrifugations at 13000 rpm. The pellet was collected and dried at room temperature for 30 minutes. The DNA pellet was then stored and frozen in 50 μl of ultra pure nuclease-free water. Final DNA concentration was 20 ng/μl, quantified with a Nanodrop™ 2000 (Thermofisher Scientific, Waltham, Massachussetts, USA).
Whole genome sequencing
To obtain the whole genome sequences of the 322 M. bovis isolates, 20 ng of total DNA were used to perform sequencing on a MiSeq instrument (Illumina, San Diego, CA) using 2x250 paired-end chemistry and the Nextera XT library preparation kit (Illumina, San Diego, CA), according to manufacturer’s instructions. The bioinformatics pipeline used for this study was created by the National Veterinary Services Laboratories belonging to the United States Department of Agriculture (see https://github.com/USDA-VS; Iowa, USA), and has been used in numerous studies [18,20,26]. Quality of reads was analyzed by the software FASTQC [27]. Reads were aligned to the reference genome AF2122/97, NCBI accession number NC_0002945, using BWA and Samtools [28,29]. A depth of coverage of 100X was targeted. BAM files were processed based on Genome Analysis Toolkit (GATK)’s best practices workflow. SNPs were called using GATK’s HaplotypeCaller outputting them to variant call files (VCF) [30,31] Results were filtered using a minimum QUAL score of 150 and AC = 2. VCF files of closely related samples were grouped and SNPs were outputted to three formats: an aligned FASTA file; a formatted Excel worksheet; and a maximum likelihood phylogenetic tree created with RaxML [32] optimized for visualization using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). The tree was built using a GTR-CAT model with input taken as an alignment file containing only informative and validated SNPs. SNPs were also visually validated using Integrative Genomics Viewer [33,34]. Visualization of reads aligned to the reference genome AF2122/97 was performed, checking SNP’s location and coverage (S1 Fig).
Spoligotyping and phylogenetic analysis
Spoligotypes were obtained in silico through the whole-genome sequence analysis performed through the NVSL in-house pipeline, as it determines the absence or presence of the spacer units in the mycobacterial genome. In order to determine evolutionary relationships among spoligotypes, a spoligoforest was built using SpolTools [35–37]. The spoligoforest provides a visualization of the probable relationships among spoligotypes in a given sample. The method makes use of a model that considers mutation by irreversible deletions of spacers and assigns probabilities to the lengths of these deletions. Solid edges are relationships between spoligotypes that are found to be unique in the data set, and the principle is that a spoligotype is inferred to arise from only one specific parent spoligotype; edges that are broken lines are chosen among multiple edges that have a probability measure greater than or equal to 0.5, and dotted lines have a probability measure less than 0.5. The size of each node is an increasing function of the number of isolates (i.e., the cluster size); edges between nodes reflect evolutionary relationships between spoligotypes with arrowheads pointing to descendants. Cladograms to establish phylogenetic relationships were made using MIRU-VNTR Plus [38,39].
Results
Whole genome sequences of 322 M. bovis isolates from nine different states of Mexico were obtained. From these, 320 (99%) were from cattle: 217 (68%) dairy and 13 (4%) beef. No information about breed could be confirmed for 90 (28%) isolates. Two isolates (1%) were obtained from humans. The SRA sequences were deposited under NCBI Bioproject PRJNA449507. The supplemental file S1 Table lists the relevant metadata for the isolates included in this study.
Spoligotyping
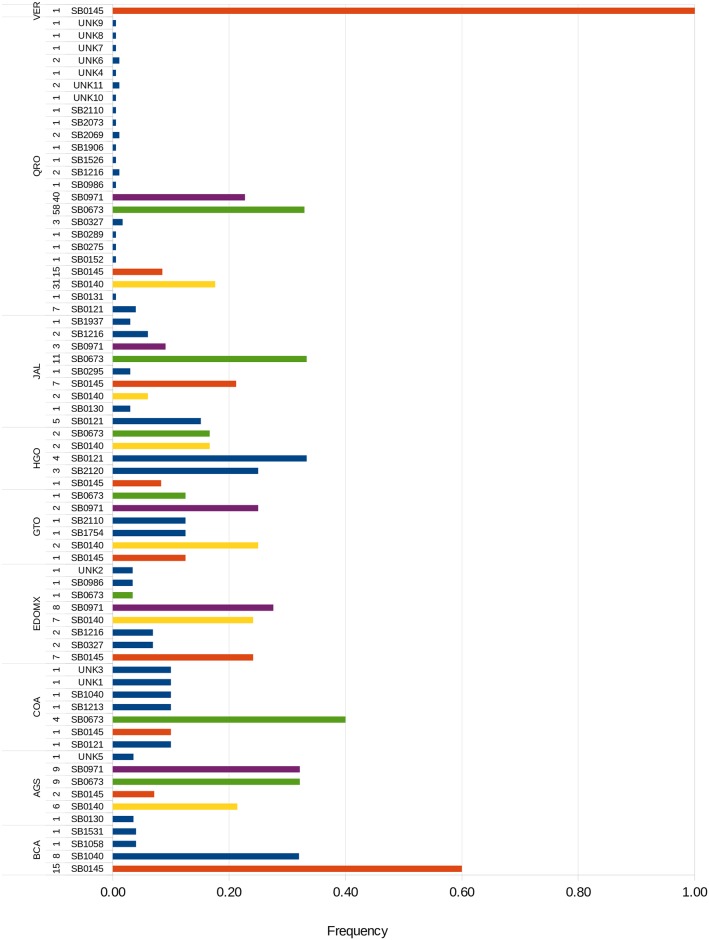
From 322 isolates, 28 recognized spoligotype patterns were identified, 11 were not found in the Mycobacterium bovis Spoligotype Database [40], therefore defined as “unknown” (UNK). The most frequent spoligotypes were SB0673 (85), SB0971 (60), SB0140 (50), and SB0145 (50), which included about 75% of all isolates. The distribution and frequency of each spoligotype by state is shown in Fig 1. Most spoligotypes are present in almost every state included in the study. SB0145 had the widest distribution of all, found in all the states. Unkown spoligotypes were found in COA, AGS, EDOMX, and QRO. There were two spoligotypes found exclusively in Baja California, SB1058 and SB1531. QRO had the largest genetic diversity, accounting for 24 of the 39 (61%) spoligotypes found. BCA had the lowest genetic diversity with only two spoligotypes. Spoligotypes were shared between dairy and beef cattle, as well as humans. Human isolates matched spoligotypes SB0673 and SB0971.
Fig 1. M. bovis spoligotypes.
Spoligotype frequency and distribution found among the 322 M. bovis isolates. Number of isolates are presented to the left of each spoligotype. Overall most frequent spoligotypes are highlighted, green: SB0673, purple: SB0971, yellow: SB0140, and orange: SB0145. More information regarding each isolate can be found in S1 Table.
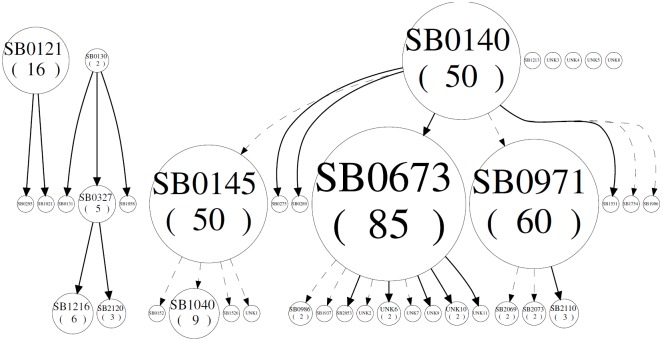
To establish relationships between the strains, a spoligoforest was generated using the hierarchal method with a ranked layout where spoligotypes that are inferred to be derived from another spoligotype are placed below the inferred parent. In Fig 2, spoligotype SB0140 is the ancestral genotype from which most of the other spoligotypes originated, including the most frequent spoligotypes SB0673, SB0971 and SB0145. At the same time, each of these gave rise to the other less frequent extant spoligotypes. However, spoligotypes SB0130 and SB0121 form two other separate genetic groups which show no relationship with the rest. Five spoligotypes, SB1213, UNK3, UNK4, UNK5 and UNK8, had no relationship to the rest. Moreover, spoligotype SB0673, the most frequent one, is parent of nine strains, one more than the ancestral SB0140, which only gave rise to eight daughter strains.
Fig 2. Spoligoforest of M. bovis isolates from Mexico.
Evolutionary analysis by spoligoforest of the M. bovis spoligotypes obtained in this study (SpolTools).
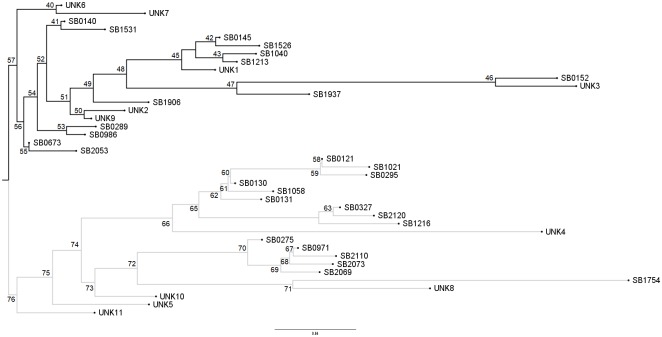
To further resolve the genetic relationships of these strains, a neighbor-joining phylogenetic tree was generated through the MIRU-VNTRplus web application for spoligotype data (Fig 3). There are two major clades, one formed by SB0140 and its daughter strains SB0673 and SB0145, and another formed by SB0121, SB0130 and SB0971, whose relationship to SB0140 was weak through the spoligoforest analysis, represented by the dashed line. Relationships for the unknown spoligotypes were resolved, UNK3 was placed within the SB0140 clade and SB1213, UNK4, UNK5 and UNK8 were placed within the SB0121 clade.
Fig 3. Cladogram of M. bovis spoligotypes from Mexico.
Phylogenetic relationships of M. bovis spoligotypes with Neighbor-joining method using MIRU-VNTRplus.
SNP based genotyping and phylogenetic analysis
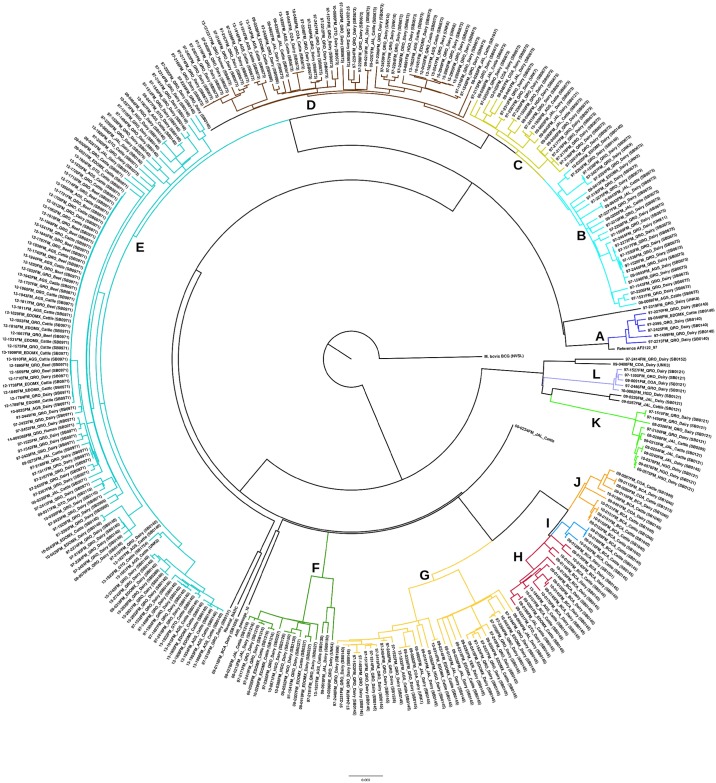
Based on SNP data, M. bovis isolates were divided into 12 main genetic groups (Table 1 and Fig 4). The groups with the largest number of isolates were Groups E (107) and D (51). Groups D and E included isolates from dairy and beef cattle, as well as the human isolates. Group G was the most widely distributed, found in 8 of the 9 states included in the study. Group I was the least distributed, found only in BCA. Again, QRO showed the largest genetic diversity, with 9 of the 12 major SNP groups (75%). VER, on the other hand, showed the lowest genetic diversity with only major SNP Group G. Frequency and distribution of the major SNP Groups are in Table 2.
Table 1. SNP loci, number of isolates and spoligotypes found in each SNP Group (A-L) obtained through whole-genome SNP based analysis.
(SNP sites refers to the highest number of SNPs found in each group, i.e. Group A isolates had up to 175 SNPs with respect to the reference genome AF2122/97).
| Major SNP Group | SNP sites | No. of isolates | Spoligotype |
|---|---|---|---|
| A | 175 | 6 | SB0140 |
| B | 572 | 28 | SB0140, SB0673, UNK9, UNK11, UNK2 |
| C | 378 | 21 | SB0121, SB0673, UNK7 |
| D | 750 | 51 | SB0673, SB0986, SB1937, UNK6, UNK10 |
| E | 1418 | 107 | SB0140, SB0275, SB0289, SB0971, SB1754, SB2069, SB2073, SB2110 |
| F | 451 | 16 | SB0130, SB0327, SB1216, SB2120, UNK4 |
| G | 986 | 32 | SB0145, UNK1 |
| H | 123 | 11 | SB0145 |
| I | 54 | 4 | SB0145, SB1531 |
| J | 190 | 11 | SB0145, SB1040, SB1213 |
| K | 95 | 11 | SB0121, SB0145, SB0295 |
| L | 63 | 5 | SB0121 |
Clustering of isolates did not result from host species, breed or year of isolation. However, there is evidence of clustering by geographic location with respect to the isolates from BCA, which form three SNP Groups, H, I and J. At leas one “unknown” strain is found within each group.
Fig 4. Phylogeny of M. bovis isolates.
Radial representation of molecular phylogenetic analysis by Maximum Likelihood (RAxML) method based on whole-genome SNP sequences of 322 isolates of M. bovis from Mexico. Groups are represented by colors: A (blue), B (cyan), C (light green), D (brown), E (light blue), F (green), G (yellow), H (red), I (bright blue), J (orange), K (lime), L (lila).
Table 2. Distribution of each major SNP Group by state.
Number of isolates corresponding to a specific group within a state is indicated in parenthesis (). It is possible that the total number of isolates per state does not equal the sum of the isolates per group because not all isolates fell within a group, some were outliers in the phylogeny (i.e. 09-0234FM_JAL_Cattle).
| Origin | AGS | BCA | COA | EDOMX | GTO | HGO | JAL | QRO | VER |
|---|---|---|---|---|---|---|---|---|---|
| Total number of isolates per state* | 28 | 25 | 10 | 29 | 8 | 12 | 33 | 176 | 1 |
| SNP Group | B (1) C (1) D (6) E (16) F (1) G (1) H (1) |
H (11) I (5) J (8) |
C (2) D (2) G (1) J (3) L (1) |
A (1) B (2) D (2) E (4) F (4) G (6) H (1) |
D (1) E (6) H (1) |
C (1) D (1) E (2) F (3) G (1) K (3) L (1) |
B (3) C (5) D (4) E (4) F (3) G (5) H (1) K (6) |
A (5) B (21) C (12) D (35) E (69) F (6) G (18) K (4) L (3) |
G (1) |
Discussion
Bovine tuberculosis in Mexico remains an endemic disease [9], which leads to the restriction of international trade of cattle and cattle products, and most importantly, poses a threat to public health. To be able to monitor the disease, genetic profiling of the pathogen is necessary. Spoligotyping and WGS are efficient for the molecular characterization of M. bovis, though the latter provides more information relevant to the pathogen’s biology [41], while spoligotyping is useful for diagnosis and strain characterization [42]. Unfortunately, WGS is still a high-cost and complex technology that many developing countries can’t yet fully incorporate it into routine disease surveillance activities. In this study we aimed at comparing both methods in order to evaluate the capacity of spoligotyping to stand against WGS to and continue to be a valid method for bovine tuberculosis surveillance.
In Mexico, typing of M. bovis has been done by RFLP, RAPD-PCR, spoligotyping, and MIRU-VNTR [8,10,11,43,44]. Spoligotyping is cheap, easy to perform, it requires minimal amount of DNA and there is an international database where spoligotypes can be reported, stored and compared [Smith et al., 2012]. However, against WGS, this method has poor discriminatory power and it is prone to homoplasy, where unrelated strains evolve independently. In our results, spoligotyping grouped the strains into 28 genetic groups, of which the most frequent were SB0673, SB0971, SB0140, SB0145 and SB0121. These spoligotypes have been found previously at high frequency in Mexico [9,45–49]. In order to establish the relationships among the spoligotypes and classify groups of related strains, a spoligoforest (Fig 2) and a cladogram (Fig 3) were built. Overall, there are two main clades of M. bovis, SB0140 and SB0121, and their daughter strains. These correspond to groups A-J and K-L, respectively. A recent study in Eritrea [50] clustered M. bovis isolates into two groups that also corresponded to spoligotype pattern. In Uruguay [51], similar results were obtained, they found concordance between clustering by spoligopyte and WGS, obtaining three main M. bovis clades in the territory of Uruguay.
Through whole genome sequencing, however, it is possible to follow the evolution of a strain according to the SNPs it acquires over time. This way, strains that are a few SNP differences apart could be considered directly related. From our study have been able to see how strains of spoligotypes SB0971 and SB0673 have diverged into numerous subgroups. Spoligoforest analysis also places SB0971 and SB0673 as parent strains of a few spoligotypes (Fig 2), though resolution is poor. Due to the higher resolving power of WGS, spoligotypes overlap among two or more different groups in the SNP phylogeny (Fig 4), such as SB0673, which can be seen in Groups B, C and D. In Table 1, the same is seen for other spoligotypes: SB0121 in Groups C, K and L; SB0145 in Groups G through K; SB0121 in Groups K and L; and SB0140, because it is an ancestral genotype from which the majority derived, in Groups A, B and E. Finally, it is important to highlight that cattle and humans share spoligotypes SB0673 and SB0971, and SNP-types D and E, respectively. Other authors have found similar results with respect to human and bovine isolates [48,52], suggesting clear transmission between hosts.
It is clear that the discriminatory power is much higher with WGS than with spoligotyping; however, the cost is also considerably higher and hard to accomplish for large amounts of isolates in large-scale studies in developing countries. WGS is worth the cost for studies of evolution or for tracing back sources of infection in the presence of outbreaks. Perhaps in large-scale epidemiological studies where the objective is only to monitor disease prevalence, such as in medium to high-burden countries like Mexico, where there is great genetic diversity, spoligotyping is efficient and accurate. Although spoligotyping is not considered cutting-edge technology any more, it is still used worldwide for the study of bovine tuberculosis, mainly in developing countries where resources for WGS technology are scarce [50,53–60].
According to the number of spoligotypes and WGS typing groups, it seems that spoligotyping is more discriminatory than WGS, since there are 28 spoligotypes and only 12 major SNP groups. In WGS, every isolate that does not match perfectly should be identified as different, so there may be only 28 spoligotypes, but since each isolate has a different SNP profile, then there are 322 genotypes. In this study, we counted a group as one that contained all isolates that were within sharing a common ancestor by 10 to 20 SNPs. For example, major SNP group H includes 11 isolates, grouped into spoligotype SB0145, but it can be divided into 6 subgroups, where three subgroups have two isolates each, and the rest are single-isolate subgroups. In some cases, WGS is so sensitive that it provides very detailed information that can be complex to interpret. For this reason, spoligotyping may be adequate for large-scale epidemiological studies.
Conclusion
Spoligotyping and WGS complement each other. WGS is an excellent tool for detecting sources of infection and transmission chains during outbreak investigations, but it remains a costly and complex procedure for developing countries to incorporate into routine disease surveillance activities. Spoligotyping, though not of recent date, continues to be a valid technique for the molecular characterization of M. bovis strains worldwide. WGS requires important resources, financial, computational and technical. Spoligotyping is easy and cheap to perform, so it is still a legitimate method for large scale epidemiological studies. In Mexico, more studies should be performed that include isolates from all of the states in order to get a more precise picture of the dynamics of bovine tuberculosis in the country.
Supporting information
Reads are aligned to the reference genome AF2122/97, coverage is represented as a gray bar chart in the upper track of each strain and SNPs are shown as colored bars.
(TIF)
Metrics and metadata for the 322 M. bovis whole genome sequences used in this study.
(XLS)
Acknowledgments
We thank the United States Department of Agriculture, with special consideration to SRA, TS, and the team at the Mycobacteria and Brucella Section of the National Veterinary Services Laboratories of the United States Department of Agriculture (Ames, Iowa, USA) for their continuing collaboration.
Data Availability
All SRA sequence files are available from the NCBI SRA database under BioProject ID PRJNA449507.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Smith NH, Dale J, Inwald J, Palmer S, Gordon S V, Hewinson RG, et al. The population structure of Mycobacterium bovis in Great Britain: Clonal expansion. Proc Natl Acad Sci [Internet]. 2003;100(25):15271–5. Available from: http://www.pnas.org/content/100/25/15271.abstract 10.1073/pnas.2036554100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.le Roex N, van Helden PD, Koets AP, Hoal EG. Bovine TB in livestock and wildlife: what’s in the genes? Physiol Genomics [Internet]. 2013;45(15):631–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23757394 10.1152/physiolgenomics.00061.2013 [DOI] [PubMed] [Google Scholar]
- 3.Buddle B, de Lisle G, Griffin J, Hutchings S. Epidemiology, diagnostics, and management of tuberculosis in domestic cattle and deer in New Zealand in the face of a wildlife reservoir. N Z Vet J [Internet]. 2014;0169(July):1–23. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24992203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thoen CO, Kaplan B, Thoen TC, Gilsdorf MJ, Shere JA. Zoonotic tuberculosis. A comprehensive ONE HEALTH approach. Medicina (B Aires). 2016;76(3):159–65. [PubMed] [Google Scholar]
- 5.Olea-Popelka F, Muwonge A, Perera A, Dean AS, Mumford E, Erlacher-Vindel E, et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—a call for action. Lancet Infect Dis [Internet]. World Health Organization. Published by Elsevier Ltd/Inc/BV. All rights reserved.; 2017;17(1):e21–5. Available from: 10.1016/S1473-3099(16)30139-6 [DOI] [PubMed] [Google Scholar]
- 6.KAO RR, PRICE-CARTER M, ROBBE-AUSTERMAN S. Use of genomics to track bovine tuberculosis transmission. Rev Sci Tech l’OIE [Internet]. 2016. April 1 [cited 2018 Jan 23];35(1):241–68. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27217181 [DOI] [PubMed] [Google Scholar]
- 7.Milian-Suazo F, Salman MD, Black WC, Triantis JM, Ramirez C, Payeur JB, et al. Molecular epidemiologic analysis of Mycobacterium bovis isolates from Mexico. Am J Vet Res [Internet]. 2000;61(1):90–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10630786 [DOI] [PubMed] [Google Scholar]
- 8.Milian-Suazo F, Harris B, Díaz CA, Romero Torres C, Stuber T, Ojeda GA, et al. Molecular epidemiology of Mycobacterium bovis: Usefulness in international trade. Prev Vet Med. 2008;87(3–4):261–71. 10.1016/j.prevetmed.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 9.Milian-Suazo F, Garcia-Casanova L, Robbe-Austerman S, Canto-Alarcon GJ, Barcenas-Reyes I, Stuber T, et al. Molecular relationship between strains of M. bovis from Mexico and those from countries with free trade of cattle with Mexico. PLoS One. 2016;11(5):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nava Vargas A, Milián Suazo F, Cantó Alarcón GJ, Rubio Venegas Y, Guerrero Solorio R, Rodríguez Hernández E, et al. Genetic diversity based on MIRU-VNTR profile of isolates of Mycobacterium bovis from Mexican cattle. Prev Vet Med [Internet]. Elsevier B.V.; 2016;131:75–8. Available from: 10.1016/j.prevetmed.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 11.Zumárraga MJ, Arriaga C, Barandiaran S, Cobos-Marín L, de Waard J, Estrada-Garcia I, et al. Understanding the relationship between Mycobacterium bovis spoligotypes from cattle in Latin American Countries. Res Vet Sci. 2013;94(1):9–21. 10.1016/j.rvsc.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 12.Orduña P, Cevallos MA, de León SP, Arvizu A, Hernández-González IL, Mendoza-Hernández G, et al. Genomic and proteomic analyses of Mycobacterium bovis BCG Mexico 1931 reveal a diverse immunogenic repertoire against tuberculosis infection. BMC Genomics [Internet]. 2011;12(1):493 Available from: http://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-12-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milian-Suazo F, Escalera AMA, Torres RMG. A review of M. bovis BCG protection against TB in cattle and other animals species. Prev Vet Med. 2003;58(1–2):1–13. [DOI] [PubMed] [Google Scholar]
- 14.SENASICA. Guía Para El Seguimiento Epidemiológico De La Tuberculosis Bovina. 2016;41. [Google Scholar]
- 15.Hasnain SE, O’Toole RF, Grover S, Ehtesham NZ. Whole genome sequencing: A new paradigm in the surveillance and control of human tuberculosis. Tuberculosis [Internet]. Elsevier Ltd; 2015;95(2):91–4. Available from: 10.1016/j.tube.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 16.Kato-Maeda M, Ho C, Passarelli B, Banaei N, Grinsdale J, Flores L, et al. Use of Whole Genome Sequencing to Determine the Microevolution of Mycobacterium tuberculosis during an Outbreak. PLoS One. 2013;8(3):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee RS, Behr MA. The implications of whole-genome sequencing in the control of tuberculosis. Ther Adv Infect Dis [Internet]. 2016;3(2):47–62. Available from: http://journals.sagepub.com/doi/10.1177/2049936115624630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser L, Carstensen M, Shaw S, Robbe-Austerman S, Wunschmann A, Grear D, et al. Descriptive epidemiology and whole genome sequencing analysis for an outbreak of bovine tuberculosis in beef cattle and white-tailed deer in northwestern Minnesota. PLoS One. 2016;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trewby H, Wright D, Breadon EL, Lycett SJ, Mallon TR, McCormick C, et al. Use of bacterial whole-genome sequencing to investigate local persistence and spread in bovine tuberculosis. Epidemics [Internet]. Elsevier B.V.; 2016;14:26–35. Available from: 10.1016/j.epidem.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsao K, Robbe-Austerman S, Miller RS, Portacci K, Grear DA, Webb C. Sources of bovine tuberculosis in the United States. Infect Genet Evol [Internet]. Elsevier B.V.; 2014;28:137–43. Available from: 10.1016/j.meegid.2014.09.025 [DOI] [PubMed] [Google Scholar]
- 21.Perea Razo CA, Milian Suazo F, Reyes IB, Gallegos SS, Rodriguez Hernandez E, Villalva SF, et al. Whole Genome Sequencing for Detection of Zoonotic Tuberculosis in Queretaro, Mexico. J Anc Dis Prev Remedies [Internet]. 2017;05(02). Available from: https://www.esciencecentral.org/journals/whole-genome-sequencing-for-detection-of-zoonotic-tuberculosis-in-queretaro-mexico-2329-8731-1000158.php?aid=86618 [Google Scholar]
- 22.Coscolla M, Gagneux S. NIH Public Access. Semin Immunol. 2014;6(26):431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagielski T, Van Ingen J, Rastogi N, Dziadek J, Mazur PK, Bielecki J. Current methods in the molecular typing of mycobacterium tuberculosis and other Mycobacteria. Biomed Res Int. Hindawi Publishing Corporation; 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payeur JB, Jamagin JL, Marquardt JG, Schaper LA, Martin BM. Manual of laboratory methods in veterinary mycobacteriology for isolation and identification of mycobacteria. In: National Veterinary Services Laboratories, Veterinary Services APHIS, editor. Ames, Iowa: United States Department of Agriculture; 1993. p. 14–23. [Google Scholar]
- 25.de Almeida IN, da Silva Carvalho W, Rossetti ML, Dalla Costa ER, de Miranda SS. Evaluation of six different DNA extraction methods for detection of Mycobacterium tuberculosis by means of PCR-IS6110: preliminary study. BMC Res Notes. 2013;6(561):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thacker TC, Palmer M V., Robbe-Austerman S, Stuber TP, Waters WR. Anatomical distribution of Mycobacterium bovis genotypes in experimentally infected white-tailed deer. Vet Microbiol [Internet]. Elsevier B.V.; 2015;180(1–2):75–81. Available from: 10.1016/j.vetmic.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 27.Andrews S. FastQC: a quality control tool for high throughput sequence data [Internet]. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics [Internet]. Oxford University Press; 2009. July 15 [cited 2018 Jul 6];25(14):1754–60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19451168 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo MA, Banks E, Poplin R, Garimella K V, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet [Internet]. NIH Public Access; 2011. May [cited 2018 Jul 6];43(5):491–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21478889 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res [Internet]. Cold Spring Harbor Laboratory Press; 2010. September [cited 2018 Jul 6];20(9):1297–303. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20644199 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma [Internet]. NIH Public Access; 2013. [cited 2018 Jul 6];43(1110):11.10.1–33. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25431634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics [Internet]. Oxford University Press; 2014. May 1 [cited 2018 Jul 6];30(9):1312–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24451623 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson ER, Walker TM, Pallen MJ. Genomics and outbreak investigation: from sequence to consequence. Genome Med [Internet]. 2013;5(4):36 Available from: http://genomemedicine.biomedcentral.com/articles/10.1186/gm440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform [Internet]. Oxford University Press; 2013. March 1 [cited 2018 Jul 6];14(2):178–92. Available from: https://academic.oup.com/bib/article-lookup/doi/10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka MM, Francis AR. Detecting emerging strains of tuberculosis by using spoligotypes. 2006. [cited 2018 Jul 6];103(41):15266–71. Available from: www.pnas.orgcgidoi10.1073pnas.0603130103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang C, Reyes JF, Luciani F, Francis AR, Tanaka MM. spolTools: Online utilities for analyzing spoligotypes of the Mycobacterium tuberculosis complex. Bioinformatics. 2008;24(20):2414–5. 10.1093/bioinformatics/btn434 [DOI] [PubMed] [Google Scholar]
- 37.Reyes JF, Francis AR, Tanaka MM. Models of deletion for visualizing bacterial variation: an application to tuberculosis spoligotypes. BMC Bioinformatics [Internet]. BioMed Central; 2008. November 27 [cited 2018 Jul 6];9:496 Available from: http://www.ncbi.nlm.nih.gov/pubmed/19036166 10.1186/1471-2105-9-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol [Internet]. American Society for Microbiology (ASM); 2008. August [cited 2018 Jul 6];46(8):2692–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18550737 10.1128/JCM.00540-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res [Internet]. Oxford University Press; 2010. July [cited 2018 Jul 6];38(Web Server issue):W326–31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20457747 10.1093/nar/gkq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith NH, Upton P. Naming spoligotype patterns for the RD9-deleted lineage of the Mycobacterium tuberculosis complex; www.Mbovis.org. Infect Genet Evol [Internet]. Elsevier B.V.; 2012;12(4):873–6. Available from: 10.1016/j.meegid.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 41.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun [Internet]. 2014;5:4812 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4166679&tool=pmcentrez&rendertype=abstract 10.1038/ncomms5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milián-Suazo F, Pérez-Guerrero L, Arriaga-Díaz C, Escartín-Chávez M. Molecular epidemiology of human cases of tuberculosis by Mycobacterium bovis in Mexico. Prev Vet Med. 2010;97(1):37–44. 10.1016/j.prevetmed.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 44.Perumaalla VS, Adams LG, Payeur JB, Jarnagin JL, Baca DR, Güemes FS, et al. Molecular epidemiology of Mycobacterium bovis in Texas and Mexico. J Clin Microbiol. 1996;34(9):2066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodwell TC, Kapasi AJ, Moore M, Milian-Suazo F, Harris B, Guerrero LP, et al. Tracing the origins of Mycobacterium bovis tuberculosis in humans in the USA to cattle in Mexico using spoligotyping. Int J Infect Dis [Internet]. International Society for Infectious Diseases; 2010;14(SUPPL. 3):e129–35. Available from: 10.1016/j.ijid.2009.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobos-Marín L, Montes-Vargas J, Zumarraga M, Cataldi A, Romano MI, Estrada-Garcia I, et al. Spoligotype analysis of Mycobacterium bovis isolates from Northern México. Can J Microbiol. 2005;51(11):996–1000. 10.1139/w05-083 [DOI] [PubMed] [Google Scholar]
- 47.Santillan-Flores MA, Flores J, Arriaga-Diaz C, Romero-Torres C, Suárez-Güemes F, Espitia C. Polymorphism of the PE domain of PE/PE_PGRS sequences in clinical isolates of Mycobacterium bovis in Mexico. Vet Microbiol [Internet]. Elsevier; 2006. July 20 [cited 2018 Jul 6];115(4):364–9. Available from: https://www.sciencedirect.com/science/article/pii/S0378113506000897?via%3Dihub 10.1016/j.vetmic.2006.02.021 [DOI] [PubMed] [Google Scholar]
- 48.Sandoval-Azuara SE, Muñiz-Salazar R, Perea-Jacobo R, Robbe-Austerman S, Perera-Ortiz A, López-Valencia G, et al. Whole genome sequencing of Mycobacterium bovis to obtain molecular fingerprints in human and cattle isolates from Baja California, Mexico. Int J Infect Dis [Internet]. Elsevier; 2017. October 1 [cited 2018 Jan 30];63:48–56. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28739421 10.1016/j.ijid.2017.07.012 [DOI] [PubMed] [Google Scholar]
- 49.Gutiérrez Reyes JA, Casanova LG, Torres CR, Gallegos SLS, Alarcón GJC, Pezzat MM, et al. Population structure of Mycobacterium bovis isolates from cattle in Mexico. Prev Vet Med [Internet]. Elsevier; 2012. September 1 [cited 2018 Jul 6];106(1):1–8. Available from: https://www.sciencedirect.com/science/article/pii/S016758771200164X?via%3Dihub 10.1016/j.prevetmed.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 50.Ghebremariam MK, Hlokwe T, Rutten VPMG, Allepuz A, Cadmus S, Muwonge A, et al. Genetic profiling of Mycobacterium bovis strains from slaughtered cattle in Eritrea. PLoS Negl Trop Dis. 2018;12(4):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lasserre M, Fresia P, Greif G, Iraola G, Castro-Ramos M, Juambeltz A, et al. Whole genome sequencing of the monomorphic pathogen Mycobacterium bovis reveals local differentiation of cattle clinical isolates. BMC Genomics [Internet]. BioMed Central; 2018. December 2 [cited 2018 Feb 23];19(1):2 Available from: https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-017-4249-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez-Guerrero L, Milián-Suazo F, Arriaga-Díaz C, Romero-Torres C, Escartín-Chávez M. Epidemiología molecular de las tuberculosis bovina y humana en una zona endémica de Querétaro, México. Salud Publica Mex. 2008;50(4):286–91. [DOI] [PubMed] [Google Scholar]
- 53.Andrievskaia O, Turcotte C, Berlie-Surujballi G, Battaion H, Lloyd D. Genotypes of Mycobacterium bovis strains isolated from domestic animals and wildlife in Canada in 1985–2015. Vet Microbiol [Internet]. Elsevier; 2018;214(October 2017):44–50. Available from: 10.1016/j.vetmic.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 54.Ghavidel M, Mansury D, Nourian K, Ghazvini K. The most common spoligotype of Mycobacterium bovis isolated in the world and the recommended loci for VNTR typing; A systematic review. Microb Pathog [Internet]. Elsevier Ltd; 2018;118(September 2017):310–5. Available from: 10.1016/j.micpath.2018.03.036 [DOI] [PubMed] [Google Scholar]
- 55.Jang Y, Ryoo S, Lee H, Kim N, Lee H, Park S, et al. Isolation of Mycobacterium bovis from Free-Ranging Wildlife in South Korea. J Wildl Dis [Internet]. 2017;53(1):181–5. Available from: http://www.bioone.org/doi/10.7589/2015-11-295 [DOI] [PubMed] [Google Scholar]
- 56.Yahyaoui-Azami H, Aboukhassib H, Bouslikhane M, Berrada J, Rami S, Reinhard M, et al. Molecular characterization of bovine tuberculosis strains in two slaughterhouses in Morocco. BMC Vet Res. BMC Veterinary Research; 2017;13(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Djemal SE, Siala M, Smaoui S, Kammoun S, Marouane C, Bezos J, et al. Genetic diversity assessment of Tunisian Mycobacterium bovis population isolated from cattle. BMC Vet Res. BMC Veterinary Research; 2017;13(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Egbe NF, Muwonge A, Ndip L, Kelly RF, Sander M, Tanya V, et al. Molecular epidemiology of Mycobacterium bovis in Cameroon. Sci Rep. Springer US; 2017;7(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amato B, Di Marco Lo Presti V, Gerace E, Capucchio MT, Vitale M, Zanghì P, et al. Molecular epidemiology of Mycobacterium tuberculosis complex strains isolated from livestock and wild animals in Italy suggests the need for a different eradication strategy for bovine tuberculosis. Transbound Emerg Dis. 2018;65(2):e416–24. 10.1111/tbed.12776 [DOI] [PubMed] [Google Scholar]
- 60.Machado A, Rito T, Ghebremichael S, Muhate N, Maxhuza G, Macuamule C, et al. Genetic diversity and potential routes of transmission of Mycobacterium bovis in Mozambique. PLoS Negl Trop Dis [Internet]. 2018;12(1):e0006147 Available from: http://dx.plos.org/10.1371/journal.pntd.0006147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reads are aligned to the reference genome AF2122/97, coverage is represented as a gray bar chart in the upper track of each strain and SNPs are shown as colored bars.
(TIF)
Metrics and metadata for the 322 M. bovis whole genome sequences used in this study.
(XLS)
Data Availability Statement
All SRA sequence files are available from the NCBI SRA database under BioProject ID PRJNA449507.