Abstract
Because invertebrates lack an adaptive immune system, they had to evolve effective intrinsic defense strategies against a variety of microbial pathogens. This ancient form of host defense, the innate immunity, is present in all multicellular organisms including humans. The innate immune system of the Japanese horseshoe crab Tachypleus tridentatus, serving as a model organism, includes a hemolymph coagulation system, which participates both in defense against microbes and in hemostasis. Early work on the evolution of vertebrate fibrinogen suggested a common origin of the arthropod hemolymph coagulation and the vertebrate blood coagulation systems. However, this conjecture could not be verified by comparing the structures of coagulogen, the clotting protein of the horseshoe crab, and of mammalian fibrinogen. Here we report the crystal structure of tachylectin 5A (TL5A), a nonself-recognizing lectin from the hemolymph plasma of T. tridentatus. TL5A shares not only a common fold but also related functional sites with the γ fragment of mammalian fibrinogen. Our observations provide the first structural evidence of a common ancestor for the innate immunity and the blood coagulation systems.
Although the innate immune system of the horseshoe crab Tachypleus tridentatus includes a coagulation cascade (1), the homology to mammalian blood coagulation appears to be only formal. The crystal structure of coagulogen, the horseshoe crab clotable protein that corresponds functionally to fibrinogen, unexpectedly revealed a nerve growth factor-like fold (2). This observation suggested that there is no direct evolutionary relationship between both systems (3). On the other hand, a thorough analysis of the evolution of vertebrate fibrinogen suggested a lectin as its most likely evolutionary precursor, “the best candidates for spawning such a prototypic fibrinogen are lectins” (4), because most of the proteins bearing fibrinogen-related domains are lectins (4, 5).
In contrast to the already characterized lectins from the horseshoe crab (6), tachylectin 5A (TL5A) and its homolog TL5B occur in the hemolymph plasma and not in the hemocytes. Both lectins exhibit Ca2+-dependent hemagglutinating activity against human A, B, and O types of erythrocytes, an activity inhibited by acetyl-group-containing compounds. Hemagglutinating activity in the hemolymph plasma is strongly dependent on the presence of TL5A and TL5B. Furthermore, TL5A exhibits strong bacterial agglutinating activity against both Gram-positive and Gram-negative bacteria. Therefore, it is likely that TL5A functions as a first-line host defense protein (7).
To gain insight into the lectin activity of TL5A and to address the evolutionary issues from a structural view point, we crystallized TL5A and solved its crystal structure through multiple isomorphous replacement technique. Furthermore, we solved the structure of TL5A in complex with N-acetyl-d-glucosamine (GlcNAc) at 2.0-Å resolution (Table 1).
Table 1.
Crystallographic data statistics
| Native + GlcNAc | Native | Derivatives
|
||
|---|---|---|---|---|
| UO2(AcO)2 | Pb(AcO)2/Me3PbCl | |||
| Temperature | RT | RT | RT | RT |
| Resolution, Å | 2.0 | 2.2 | 3.1 | 2.4 |
| Completeness, % | 98.1 | 98.3 | 80.5 | 94.3 |
| Rsym∗ | 10.7 | 10.6 | 15.1 | 8.8 |
| I/σ(I) last shell | 2.0 | 2.2 | 2.1 | 2.4 |
| MIR analysis | ||||
| Number of sites | 1 | 1 | ||
| Rcullis† | 0.73 | 0.62 | ||
| Phasing power, ac/c‡ | 1.01 (1.59) | 2.67 (1.51) | ||
| Refinement | ||||
| Model | Residues 45–264, 1 GlcNAc, and 179 water molecules | |||
| Data | Native + GlcNAc, 15-2.0 Å, all reflections | |||
| R factor§ | 0.183 (Rfree = 0.198) | |||
| rms deviations | Bonds, 0.005 Å; angles, 1.40°; temperature factors, 0.86 Å2 | |||
| Average B factor, Å2 | 27.95 | |||
Data collection and phase determination by MIR. Crystal space group: I4, a = b = 108.93 Å, c = 64.06 Å, α = β = γ = 90°. RT, room temperature. ac/c, acentric and centric data.
Rsym = ∑|I − 〈I〉|/∑|I|.
Rcullis = ∑∥F(PH) ± F(P)| − |FH(calc)∥/∑|F(PH) ± F(P)|.
Phasing power, mean value of the heavy atom structure amplitudes divided by the residual lack-of-closure.
Five percent of the reflections were selected for Rfree determination.
Materials and Methods
Purification and Crystallization.
TL5A was purified from Japanese horseshoe crab (T. tridentatus) hemolymph plasma, as described (7). Tetragonal crystals were grown at room temperature by vapor diffusion using the sitting drop method. Droplets mixed from 2.5 μl of TL5A (5–7 mg⋅ml−1) and 1.5 μl of buffer (0.1 M Hepes/10% polyethylene glycol 8000/8% ethylenglycol, pH 7.0) were equilibrated against 500 μl of reservoir buffer composed of the same components. Derivatives for multiple isomorphous replacement were obtained by soaking crystals in mother liquor containing 3 mM UO2(AcO)2 and 5 mM Pb(AcO)2/Me3PbCl (1:1). Soaking time was 24 h for each derivative. The complex of TL5A with GlcNAc was prepared by soaking a native crystal in mother liquor containing 10 mM GlcNAc for 3 days.
Identification of Free SH Groups and Disulfide Bridges in TL5A.
TL5A was S-alkylated with 4-vinylpyridine in the presence of 8 M urea under nonreducing conditions. The S-alkylated protein in 50 mM sodium phosphate buffer (pH 6.8), containing 2 M urea was digested with Staphylococcal V8 protease, lysyl endopeptidase, or endoproteinase Asp-N, at 37° for 20 h. The peptides generated were separated by RP-HPLC and used for amino acid sequence analysis.
Data Collection, Phasing, and Refinement.
Data collection was conducted at room temperature on a 345 MAR-Research (Hamburg, Germany) image plate detector mounted on a Rigaku (Tokyo) RU200 rotating anode x-ray generator (λ = 1.5418 Å). All image plate data were processed with MOSFLM (8) and the CCP4 program suite (9). Heavy atom-binding sites were determined by using isomorphous difference Patterson maps calculated with CCP4 programs (9). The refinement of heavy atom parameters and calculation of multiple isomorphous replacement phases was performed with SHARP (10). Protein model building was done with O (11), and the structure was refined with CNS (12). The structure of the GlcNAc complex was solved by molecular replacement with MOLREP (9) by using the native structure model of TL5A. Statistics are given in Table 1.
Coordinates.
The atomic coordinates have been deposited in the Protein Data Bank.
Results and Discussion
Overall Fold.
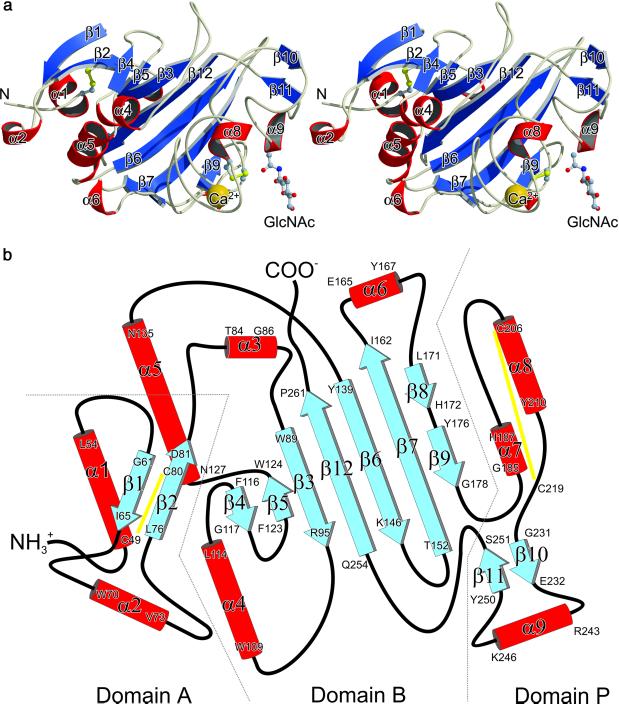
TL5A is an ellipsoidal molecule (overall dimensions ≈34 × 36 × 53 Å3), subdivided into three distinct but interacting domains (Fig. 1 a and b). The N-terminal domain A (residues Asp-45–Trp-89) comprises two short helices and a small two-stranded antiparallel β-sheet. The N-terminal helix (α1) is diagonally twisted and anchored through a disulfide bond (Cys-49–Cys-80) to the second strand of the β-sheet (β2). The second, short helix β2 connects strand β1 with strand β2. Structural elements N-terminal of Asp-45 could not be resolved because of poorly defined electron density, pointing to an increased mobility of this region. Cysteine residues Cys-6 and Cys-170 were expected to be involved in lectin homodimerization (7). However, electron density unambiguously revealed Cys-170 to be present in the free SH form. In addition, chemical-labeling experiments with 4-vinylpyridine under nonreducing conditions also showed that Cys-6 and Cys-170 are present in the free SH forms in native TL5A. Furthermore, the two disulfide bridges, Cys-49–Cys-80 and Cys-206–Cys-219, were confirmed by amino acid sequence analysis of disulfide-containing peptides derived from TL5A (data not shown).
Figure 1.
Crystal structure of TL5A from the Japanese horseshoe crab T. tridentatus. (a) Stereo view of the TL5A ribbon plot. GlcNAc is represented by a ball-and-stick model and the Ca2+ ion is represented by a golden sphere. Disulfide bridges are colored yellow. Figure was prepared with molscript (28). (b) Topology diagram showing the arrangement of secondary-structure elements in TL5A. Disulfide bridges are indicated by yellow lines. Domains named in analogy to γ-fibrinogen fragment (21).
The central and larger domain B (residues Thr-90–Ala-180 and Pro-253–Phe-264) is clamped through domains A and P and predominantly build up by a twisted seven-stranded antiparallel β-sheet (strands β3–β7, β9 and β12). Helices α4 and α5 within this domain can be interpreted as a single helix divided by a loop, which participates with a small antiparallel two-stranded β-sheet (β4, β5) to the main seven-stranded β-sheet. The central strand β12 (residues Gln-254–Pro-261) extends the C terminus of domain P back to domain B, bringing both polypeptide termini in close proximity. Finally, the C-terminal domain P (residues Gly-181–Leu-252) possesses only a few short elements of regular secondary structure, and comprises the two major functional sites within TL5A: the Ca2+-binding site and the nearby acetyl group-binding pocket.
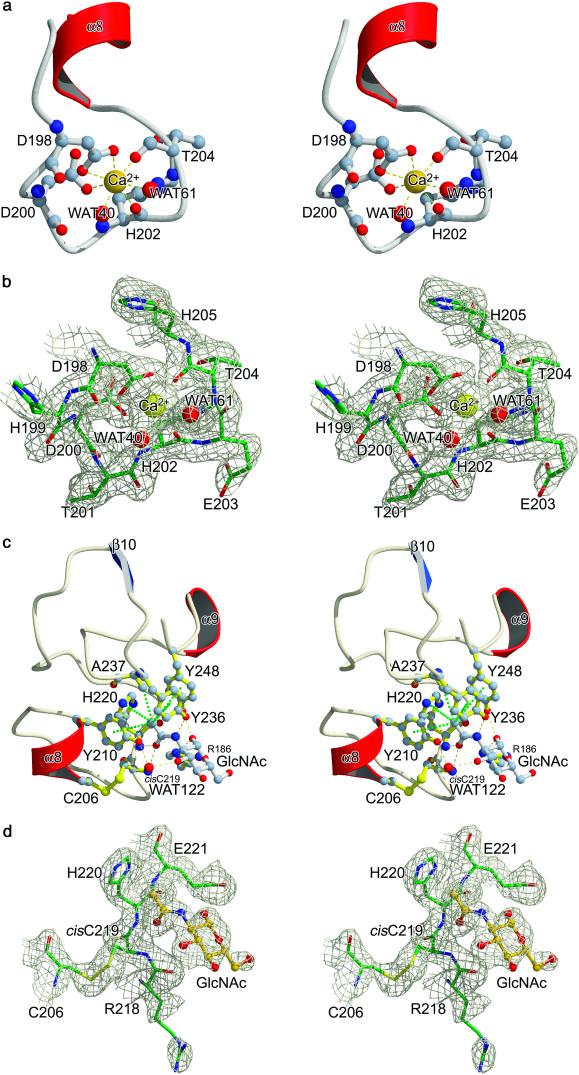
The Ca2+ ion is bound in a loop region located ≈11 Å away from the adjacent ligand binding site, coordinated by seven oxygen ligands in a pentagonal bipyramidal manner (Fig. 2 a and b). Both carboxylate oxygen atoms of Asp-198, the main chain carbonyl oxygens of His-202 and Thr-204, and one water molecule (WAT 40) form the pentagonal base, whereas WAT 61 and the side chain oxygen atom OD1 (Asp-200) occupy the vertices of the bipyramide. The bond distances are in average 2.4 Å, the characteristic value for known Ca2+-binding sites in proteins (13). Although the Ca2+-binding site is C-terminally flanked by a short helix (α8), it differs from the most common Ca2+-binding motif (EF hand- or the helix-loop-helix motif) (14). It is also worth mentioning that this short helix connects the metal-binding site to the acetyl group recognition site via disulfide bridge Cys-206–Cys-219.
Figure 2.
Major functional sites within domain P of TL5A. (a) Stereo plot of the calcium-binding site. Oxygen atoms are represented in red and nitrogen atoms in blue. Water molecules are represented by red spheres. (b) Stereo view of the 2Fo--Fc electron density map around the calcium ion. The map is contoured at 1σ. Figure was prepared with bobscript (29). (c) Structural basis of GlcNAc binding. Stereo view of the binding site, with relevant side chains labeled. Hydrogen bonds are indicated by yellow dashed lines and hydrophobic interactions by dotted green lines. Water molecules are represented by red spheres. Notice that the shortest hydrogen bond is formed between NH-atom of Cys-219 and the O atom of the GlcNAc acetamido group. (d) Stereo view of the 2Fo–Fc electron density map of the GlcNAc-binding site around the Arg-218–Cys-219 cis-peptide bond. The map is contoured at 1σ.
Ligand Binding.
Analysis of the ligand-binding pocket explains the acetyl-group specificity (7) (Fig. 2c): four aromatic side chains (Tyr-210, Tyr-236, Tyr-248, and His-220) form a funnel, with the methyl side chain of Ala-237 at its base. The hydroxyl groups of the three tyrosine residues point to the entrance of the funnel, creating a strictly hydrophobic interior and an outer polar region, which allows electrostatic interactions. The sugar methyl group mainly benefits from the hydrophobic environment and lies in the middle of this funnel, surrounded by the aromatic side chains and in close van der Waals contact with the side chain methyl group of Ala-237. An unusual cis-peptide bond between residues Arg-218 and Cys-219 generates a sharp turn and allows the backbone NH group of the latter to form a tight hydrogen bond (2.7 Å) with the carbonyl oxygen of the ligand acetamido group (Fig. 2 c and d). This cis-peptide bond hence substantially contributes to the acetyl-specificity of the binding pocket. On the other hand, the energetically unfavorable cis conformation could be compensated through this favorable interaction (15). The carbonyl oxygen of the acetamido group makes additional contacts to the main chain NH group of residue His-220, whereas the nitrogen of the acetamido group interacts with the Tyr-236 hydroxyl group. Carbohydrate recognition is furthermore supported by the 1-OH-group of GlcNAc, which forms hydrogen bonds with the guanidinium N of Arg-186 and with the hydroxyl group of Tyr-248. Finally the 3-OH group interacts via a bound water molecule (WAT 122) with the Tyr-210 side chain hydroxyl group.
Homology to the Fibrinogen γ Fragment.
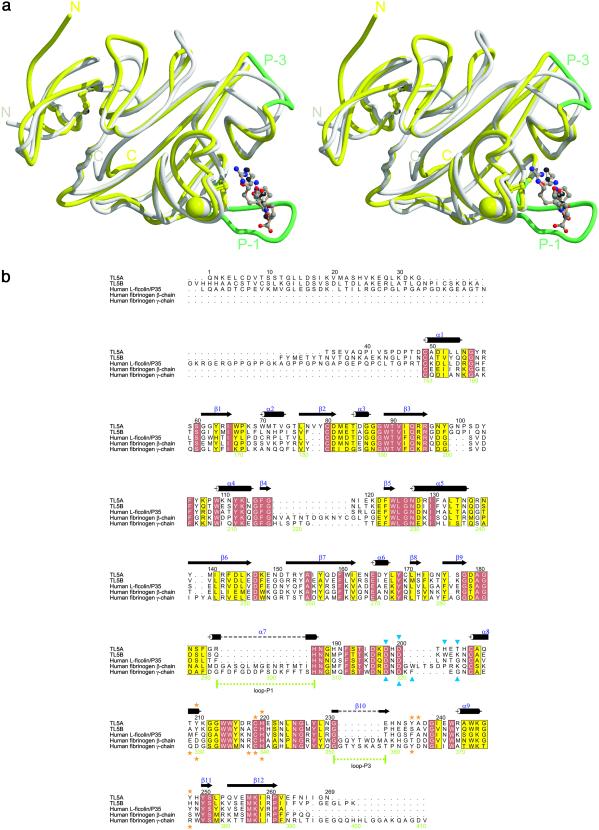
A systematic computer search performed with SCOP (16) revealed that TL5A is structurally related to the fibrinogen γ fragment (α-carbon superposition rms deviation of 1.09 Å overall) (Fig. 3a) and thus verifying our recent prediction made on the basis of amino acid sequence similarities within fibrinogen-related domains-containing proteins (7). Fig. 3b shows the corresponding sequence alignment. Even though this overall similarity is compelling, combined structural and functional similarities are necessary to establish a convincing evolutionary link between two proteins (17).
Figure 3.
Homology of TL5A and the γ chain fibrinogen fragment. (a) Superposition of the crystal structures of TL5A (gray) complexed with GlcNAc (white) and the γ chain fragment (yellow) complexed with GPRG-peptide (the A-knob, brown) (20). Ca2+ ions are represented by spheres of the same color as the structure they belong. (b) Sequence alignment of TL5A-related sequences. Black numbers on top refer to the TL5A sequence, and green numbers below refer to the fibrinogen γ chain sequence. Closed blue triangles indicate residues involved in Ca2+ binding. Orange stars indicate residues involved in GlcNAc (TL5A, above alignment), respectively A-knob binding (γ chain fragment, below). Figure was prepared with alscript (30).
The fibrinogen molecule consists of three nonidentical but homologous chains, denoted Aα, Bβ, and γ, covalently linked in an (AαBβγ)2 stoichiometry. During the final state of blood coagulation, limited proteolysis of fibrinogen occurs, releasing the short amino terminal A- and B-fibrinopeptides. The newly created N termini (designated “A-” and “B-knob”) frame the major “A”-(α chain) and “B”-(β chain) polymerization sites. Fibrin monomers (αβγ)2 rapidly polymerize through noncovalent association of A-/B-knobs into the complementary “a-/b-holes” (polymerization pockets) of another fibrin monomer. The C terminus of the γ chain (residues Val-143–Leu-392) contains, besides a Ca2+-binding site, the polymerization pocket “a,” the primary interaction site for noncovalently polymerizing fibrin. The location of this pocket around residue Tyr-363 was primarily anticipated through photoaffinity-labeling experiments (18). Three-dimensional structures of fibrinogen fragments, in complex with a model peptide that mimics the α-chain knob, resolved the primary polymerization pocket of fibrinogen at atomic level (19, 20).
As the superposition of TL5A and the fibrinogen γ fragment structure demonstrates (Fig. 3a), not only the overall three-dimensional structure, but also the Ca2+-binding site are essentially conserved. Perhaps more relevant from an evolutionary point of view, the acetyl group-recognition site in TL5A corresponds structurally to the polymerization pocket a in the fibrinogen γ fragment. Five of the seven amino acids that form the polymerization pocket a (19, 20) are structurally equivalent to amino acids in the sugar-binding site of TL5A (Fig. 3b). Furthermore the unusual Arg-218–Cys-219 cis-peptide bond in TL5A corresponds to the cis-peptide bond made in fibrinogen γ between Lys-338 and Cys-339, which is required for binding of the a-knob. The long loops P-1 and P-3 in the fibrinogen γ fragment are shortened by 14 and, respectively, 7 aa in TL5A and represent the major structural differences found in the functionally important domain P (19). Variability in this domain has been previously detected for fibrinogen-related proteins through structure-based sequence alignment (21) and points to a potential evolutionary transition from a carbohydrate to a protein-binding module (Fig. 3 a and b).
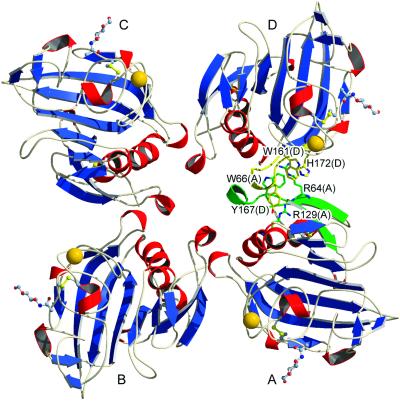
Interestingly, we have previously observed the formation of hexameric and octameric bouquet-like arrangements of purified TL5A by electron microscopy (7). Although TL5A crystallized with a monomer in the asymmetric unit, it is noteworthy that the crystallographic symmetry imposes a tetrameric arrangement (Fig. 4). The buried surface areas at the monomer–monomer interfaces (≈580 Å2) suggest that this organization could be maintained in solution and might be relevant for the physiological function of TL5A in vivo.
Figure 4.
Oligomeric arrangement of TL5A subunits around the fourfold crystallographic axis. Secondary structure elements involved in intermonomer packing are colored green (monomer A) and yellow (monomer D). In addition, some specific side chains are given with all nonhydrogen atoms. Notice in particular some planar arginine-aromatic side chain stacking similar to the ones frequently encountered in protein structures (31).
The present structural analysis indicates that each TL5A monomer possesses an independent ligand-binding site and therefore suggests that tetramers would specifically recognize surface structures on pathogens with high ligand density through multiple binding sites. The importance of multivalency for achieving high specificity and affinity has been pointed out in the case of T. tridentatus tachylectin-2 (22), mammalian mannose-binding lectin (23), and in other animal- and plant-derived lectins (24).
Finally, we mention that the innate immune system in vertebrates includes a group of proteins termed ficolins, which are involved in host defense through nonself-recognition. Similar to the role played by TL5A/B in the horseshoe crab, ficolins recognize carbohydrates on microorganisms via a C-terminal fibrinogen-like domain (25). For l-ficolin/P35, an amido group containing sugar specificity has been demonstrated (26). In addition, human l-ficolin/P35 recognizes acetyl groups as TL5A does (7). It is remarkable that human ficolins are more closely related to tachylectins 5A/5B than to the fibrinogen γ fragment (Fig. 3b) (7), emphasizing the similarity of mammalian and invertebrate innate immunity (27) and suggesting the ficolins as intermediates in the evolution from invertebrate innate immunity to the blood coagulation system.
Abbreviation
- TL5A
tachylectin 5A
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1JC9).
References
- 1.Muta, T. & Iwanaga, S. (1996) Curr. Opin. Immunol.8, 41–47. [DOI] [PubMed]
- 2.Bergner A, Oganessyan V, Muta T, Iwanaga S, Typke D, Huber R, Bode W. EMBO J. 1996;15:6789–6797. [PMC free article] [PubMed] [Google Scholar]
- 3.Bergner A, Muta T, Iwanaga S, Beisel H-G, Delotto R, Bode W. Biol Chem Hoppe-Seyler. 1997;378:283–287. doi: 10.1515/bchm.1997.378.3-4.283. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle R F, Spraggon G, Everse S J. Ciba Found Symp. 1997;212:4–17. doi: 10.1002/9780470515457.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle R F. Ann NY Acad Sci. 1983;408:13–27. doi: 10.1111/j.1749-6632.1983.tb23231.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawabata S, Iwanaga S. Dev Comp Immunol. 1999;23:391–400. doi: 10.1016/s0145-305x(99)00019-1. [DOI] [PubMed] [Google Scholar]
- 7.Gokudan S, Muta T, Tsuda R, Koori K, Kawahara T, Seki N, Mizunoe Y, Wai S N, Iwanaga S, Kawabata S. Proc Natl Acad Sci USA. 1999;96:10086–10091. doi: 10.1073/pnas.96.18.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leslie A G W. In: Crystallographic Computing 5. Moras D, Podjarny A D, Thierry J C, editors. Oxford: Oxford Univ. Press; 1991. pp. 50–61. [Google Scholar]
- 9.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 10.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 11.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 12.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 13.Glusker J P. Adv Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 14.Lewit-Bentley A, Réty S. Curr Opin Struct Biol. 2000;10:637–643. doi: 10.1016/s0959-440x(00)00142-1. [DOI] [PubMed] [Google Scholar]
- 15.Jabs A, Weiss M S, Hilgenfeld R. J Mol Biol. 1999;286:291–304. doi: 10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 16.Murzin A G, Brenner S E, Hubbard T, Chothia C J. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 17.Russell R B, Saqi M A S, Sayle R A, Bates P A, Sternberg M J E. J Mol Biol. 1997;269:423–439. doi: 10.1006/jmbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 18.Yamazumi K, Doolittle R F. Proc Natl Acad Sci USA. 1992;89:2893–2896. doi: 10.1073/pnas.89.7.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doolittle R F, Spraggon G, Everse S J. Curr Opin Struct Biol. 1998;8:792–798. doi: 10.1016/s0959-440x(98)80100-0. [DOI] [PubMed] [Google Scholar]
- 20.Pratt K P, Côté H C F, Chung D W, Stenkamp R E, Davie E W. Proc Natl Acad Sci USA. 1997;94:7176–7181. doi: 10.1073/pnas.94.14.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yee V C, Pratt K P, Côté H C F, Le Trong I, Chung D W, Davie E W, Stenkamp R E, Teller D C. Structure (London) 1997;5:125–138. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- 22.Beisel H-G, Kawabata S, Iwanaga S, Huber R, Bode W. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weis W I, Drickamer K. Structure (London) 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- 24.Rini J M. Annu Rev Biophys Biomol Struct. 1995;24:551–577. doi: 10.1146/annurev.bb.24.060195.003003. [DOI] [PubMed] [Google Scholar]
- 25.Matsushita M, Endo Y, Taira S, Sato Y, Fujita T, Ichikawa N, Nakata M, Mizuochi T. J Biol Chem. 1996;271:2448–2454. doi: 10.1074/jbc.271.5.2448. [DOI] [PubMed] [Google Scholar]
- 26.Le Y, Lee S H, Kon O L, Lu J. FEBS Lett. 1998;425:367–370. doi: 10.1016/s0014-5793(98)00267-1. [DOI] [PubMed] [Google Scholar]
- 27.Salzet M. Trends Immunol. 2001;22:285–288. doi: 10.1016/s1471-4906(01)01895-6. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 29.Esnouf R M. J Mol Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 30.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 31.Flocco M M, Mowbray S L. J Mol Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]