Abstract
A substantial fraction of the commensal microbiota is coated with IgA antibodies during homeostatic conditions. Here, we discuss the humoral strategies and mechanisms deployed in the mucosa to confront this immense diversity of bacterial antigens. Homeostatic IgA responses employ a highly polyreactive repertoire to bind a broad but taxonomically distinct subset of microbiota. These antibodies arise in the presence of limited T cell help and exhibit low rates of somatic mutation with little affinity maturation. Such processes contrast with classical paradigms of high-affinity T cell-dependent antibody responses elicited by mucosal pathogens and vaccines, yet provide a simple immunological solution to the challenge of microbiota antigenic complexity. This model raises several fundamental future questions including how polyreactive specificities are generated and selected into the IgA repertoire, how these antibodies exert their effector functions, and how they coexist or overlap with other immune responses during homeostasis and disease.
Introduction
The gastrointestinal environment presents a tremendous challenge for the immune system. Here, classical mechanisms of tolerance are challenged by the presence of a complex and dynamic mixture of largely innocuous foreign antigens from the diet and commensal microbiota as well as occasional harmful pathogens. As such, a number of unique immunological mechanisms have emerged to serve functions distinct to mucosal tissues (Honda and Littman, 2016). A homeostatic barrier consisting of mucus, antimicrobial peptides, and immunoglobulin A (IgA) antibodies maintains separation between luminal antigens and the underlying epithelium and serves as a first line of defense against both microbiota and pathogens. IgA antibodies are exceptionally abundant at mucosal surfaces: more than 80% of mammalian antibody-secreting plasma cells (PCs) reside in the gut and express the IgA isotype (Fagarasan et al., 2010). Notably, these antibodies arise prominently during homeostasis, in the absence of inflammation or immunization. However, despite its abundance, the specificity and functions of IgA in vivo have remained enigmatic. While IgA has long been known to coat the cell surface of a subset of commensal bacteria (Kroese et al., 1996; Tsuruta et al., 2009; van der Waaij et al., 1996), recent advances utilizing bacterial flow cytometry combined with high-throughput sequencing have facilitated the identification of IgA-coated bacteria and clarified the immunological mechanisms that lead to their targeting by the immune system (Bunker et al., 2017; Bunker et al., 2015; Kau et al., 2015; Kawamoto et al., 2014; Kubinak et al., 2015; Palm et al., 2014; Planer et al., 2016). Strikingly, these studies have revealed substantial differences between homeostatic responses to commensal bacteria and classical paradigms of humoral immunity to pathogens or vaccines. Here, we review the literature and propose a model in which two distinct types of humoral immunity coexist in the gastrointestinal mucosa. The first – a homeostatic response to commensals – involves natural polyreactive specificities that differentiate largely in the absence of T cell help with little somatic hypermutation or affinity maturation. The second – a protective response to pathogens – involves the production of high-affinity and specific antibodies generated in germinal centers by mechanisms that resemble systemic responses. Determining how these pathways overlap or coexist during homeostasis and disease represents a significant direction for future research.
Evolution of mucosal antibodies
The IgM isotype is a defining feature of all B cell lineages and is ancient and highly conserved in all jawed vertebrates (Flajnik and Kasahara, 2010). By contrast, IgA arose relatively recently and is present only in reptiles, birds, and mammals. Whereas mice express a single IgA subtype, humans express two subtypes termed IgA1 and IgA2. Although IgA is absent in lower jawed vertebrates, many of these organisms express specialized mucosal antibody isotypes that have arisen by convergent evolution. Bony fish express IgT in intestinal tissues, and these antibodies coat their gut microbiota (Zhang et al., 2010). Amphibians express IgX intestinal antibodies (Mussmann et al., 1996). Interestingly, mucosal antibodies show a common multimeric structure: whereas IgA is typically dimeric, IgX is pentameric and IgT is tetrameric (Mussmann et al., 1996; Zhang et al., 2010). Notably, both IgT+ and IgX+ PCs appear to differentiate in the absence of T cell help, similar to a significant fraction of the IgA repertoire in mice and the IgA2 response in humans, as we discuss in detail later (Bunker et al., 2017; Bunker et al., 2015; He et al., 2007; Macpherson et al., 2000). Together, these observations indicate that strong evolutionary pressure has driven the emergence of specialized mucosal antibodies.
Anatomy and organization of mucosal B cell responses
The largest population of IgA+ PCs is found in the small intestinal (SI) lamina propria (LP), whereas the colonic LP harbors only a minor population (Bunker et al., 2015; McWilliams et al., 1977; Walker and Isselbacher, 1977). High levels of microbiota IgA coating and free IgA are also present in the SI, with lower levels in the colon (Bunker et al., 2015; Kroese et al., 1996; Tsuruta et al., 2009). Additional minor populations of IgA+ plasma cells are detectable in extraintestinal tissues including the salivary gland, lung, lactating mammary gland (LMG), liver, and bone marrow (BM) (Bunker et al., 2017; Moro-Sibilot et al., 2016; Roux et al., 1977; Wilmore et al., 2018). Human IgA subtypes show distinct anatomical expression patterns, with IgA1 dominating in serum and IgA2 in the distal intestine (He et al., 2007).
Gut-associated lymphoid tissues (GALT) are the primary sites of IgA induction. These include Peyer’s patches (PPs), mesenteric lymph nodes (mLNs), isolated lymphoid follicles (ILFs), and the cecal patch (Craig and Cebra, 1971; Hamada et al., 2002; Macpherson and Uhr, 2004; Masahata et al., 2014; McWilliams et al., 1977; Tsuji et al., 2008). IgA may also be induced in situ in the LP (Fagarasan et al., 2001), and tertiary lymphoid structures have been observed in the LP in response to colonization with particular commensal bacteria (Lecuyer et al., 2014). Active germinal centers (GCs) are constitutively present in PPs and mLNs; these tissues therefore support both T cell-dependent (TD) and –independent (TI) pathways of IgA PC differentiation (Bunker et al., 2015; Macpherson et al., 2000). By contrast, ILFs are largely devoid of T cells and primarily support TI differentiation (Hamada et al., 2002; Tsuji et al., 2008). In vivo, there is likely considerable redundancy between these structures. Indeed, ablation of PPs or surgical removal of mLNs individually have little effect on IgA+ PC abundance (Macpherson and Uhr, 2004; Yamamoto et al., 2000). Mice lacking lymphotoxin signaling or the transcription factor retinoic acid-related orphan receptor γt (RORγt) lack all GALT tissues – these mice show a significant but incomplete reduction in IgA+ PCs (Kang et al., 2002; Tsuji et al., 2008). These GALT-independent PCs arise in lymphotoxin β-deficient mice via a TD mechanism that requires soluble lymphotoxin α3 expression by type 3 innate lymphoid cells (ILC3) (Kruglov et al., 2013), though the relevance of this pathway in wild-type (WT) mice remains unknown. Together these observations indicate that, while GALT are not strictly required for IgA PC differentiation, the vast majority of IgA PCs in vivo likely derive from these tissues.
IgA class-switch recombination, homing, maintenance, and secretion
The mechanisms that regulate class-switch recombination (CSR) to the IgA isotype, cellular migration and maintenance, and secretion of IgA antibodies have been extensively studied and reviewed – here, we discuss them only briefly (Cerutti, 2008; Kaetzel, 2005; Phalipon and Corthesy, 2003; Tuma and Hubbard, 2003). Naïve B cell precursors expressing IgM and IgD are induced to switch to the IgA isotype by cellular activation in the presence of certain factors that are constitutively present in the gut microenvironment, and the precise signals that direct class-switching differ during TD and TI pathways of activation. In all cases, BCR stimulation is necessary to induce expression of activation-induced cytidine deaminase (AID), which is essential for IgA class switching (Fagarasan et al., 2002). Additional signals through TNF-superfamily receptors promote CSR: CD40-CD40L interactions with T cells play a key role in TD responses; similar signaling in TI responses is achieved through BAFF/APRIL interactions with three potential receptors including transmembrane activator and calcium-modulating cyclophilin-ligand interactor (TACI), BAFF receptor (BAFFR), and B cell maturation antigen (BCMA) (Cerutti, 2008; Litinskiy et al., 2002). Class-switching to the IgA isotype requires induction of transcription in the Cα switch region (Sα), and this generates a substrate for AID activity and subsequent DNA recombination. Transcription at Sα can be initiated by a number of factors present in GALT tissues including transforming growth factor β1 (TGFβ1), IL-4, IL-6, IL-10, and retinoic acid (Cazac and Roes, 2000; Cerutti, 2008; Reboldi et al., 2016; Watanabe et al., 2010). Of these, TGFβ1 is likely the most important in vivo, although there may be considerable redundancy (Cazac and Roes, 2000; Reboldi et al., 2016). In humans, IgA1 or IgA2 CSR can occur in IgM+ cells; alternatively, sequential IgA1-to-IgA2 CSR can be initiated in IgA1+ cells (He et al., 2007). In vivo, many of the factors that regulate IgA CSR including BAFF and TGFβ1 activators are expressed by GALT follicular dendritic cells (fDCs), plasmacytoid DCs, and conventional DCs (Reboldi et al., 2016; Suzuki et al., 2010; Tezuka et al., 2011). Notably, the same signals that specify IgA CSR also imprint cells for homing to the intestinal LP by inducing expression of the integrin α4β7 and the chemokine receptors CCR9 and CCR10 (Kunkel et al., 2003; Mora et al., 2006). After cellular activation, IgA CSR, and gut imprinting, lymphoblasts leave the GALT via lymphatics and reenter the bloodstream. Upon circulation through the intestinal vasculature, interactions between α4β7, CCR9, and CCR10 and their ligands mucosal vascular addressin cell adhesion molecule 1 (MAdCAM-1), CCL25, and CCL27 and CCL28, respectively, direct migration into the intestinal LP.
Murine SI IgA+ PCs have an average half-life of 5 days and a maximum lifespan of 7–8 weeks (Mattioli and Tomasi, 1973). The IgA+ PC population is heterogenous and contains both short-lived major histocompatibility complex class II+ (MHCII+) cells and longer-lived MHCII− cells (Kawamoto et al., 2012); some exceptionally long-lived PCs have been identified in humans (Landsverk et al., 2017). However, the factors that regulate intestinal PC maintenance and turnover are only partially understood. Interleukin-6 (IL-6) produced by intestinal epithelial cells and eosinophils contributes to PC maintenance (Chu et al., 2014; Ng et al., 2003; Ramsay et al., 1994). B cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL) produced by intestinal epithelial cells, eosinophils, dendritic cells (DCs), and plasmacytoid DCs similarly promote PC survival (Chu et al., 2014; He et al., 2007; Huard et al., 2008; Tezuka et al., 2011; Wang et al., 2017); additional unidentified factors are also likely to contribute. Notably, most intestinal PCs express B cell receptors (BCRs) on their cell surface (Di Niro et al., 2010); while some evidence suggests that PCs can receive signals through their BCRs (Blanc et al., 2016; Pinto et al., 2013), it remains unknown whether intestinal PCs signal or internalize antigen through their BCRs in vivo, and the role of surface BCR expression in PC maintenance remains unclear.
A key pathway for IgA secretion at mucosal surfaces involves the polymeric Ig receptor (pIgR) (Kaetzel, 2005; Phalipon and Corthesy, 2003). IgA in mucosal secretions predominantly exists as a dimer connected by a small polypetide called J chain, although monomers are also detectable (Iversen et al., 2017; Koshland, 1985). pIgR is expressed on the basolateral surface of intestinal epithelial cells and binds selectively to polymeric IgA and IgM. Upon binding, antibodies are internalized and transported by transcytosis to the apical surface of the epithelial cell, where proteolytic cleavage releases the antibody bound to a highly glycosylated 80 kDa fragment of pIgR known as secretory component (SC). The complex of dimeric IgA, J chain, and SC is referred to as secretory IgA (SIgA). In numerous studies, pIgR-deficient mice have been used as models of SIgA-deficiency. However, analyses of these mice indicate only a 2–3-fold defect in SI IgA titers, a 5–10-fold decrease in fecal IgA, and no defect in breast milk IgA (Johansen et al., 1999; Rogier et al., 2014). These data suggest that alternative pathways such as paracellular transport can compensate for the loss of pIgR and may also contribute to steady-state IgA secretion (Van Itallie and Anderson, 2006).
Mechanisms of homeostatic IgA responses that target microbiota
IgA responses to microbiota occur via both TI and TD pathways (Bunker et al., 2015; Macpherson et al., 2000), and target a taxonomically distinct subset of microbiota that we consider in detail below. Precursors to IgA+ PCs include circulating naive follicular B2 cells and innate-like peritoneal B1b cells; peritoneal B1a cells that contribute to natural serum IgM responses are not observed within the IgA repertoire (Bunker et al., 2015; Macpherson et al., 2000; Reynolds et al., 2015; Roy et al., 2013). In classical models of systemic immunity, TI responses occur in response to polyvalent antigens such as bacterial polysaccharides and involve rapid cellular differentiation with little somatic hypermutation (SHM). By contrast, TD responses typically target protein antigens and involve iterative rounds of SHM and affinity selection in GCs based on cognate interactions with CD4+ T follicular helper cells (Tfh) (Victora and Nussenzweig, 2012). However, the extent to which homeostatic mucosal IgA responses resemble these processes remains unclear, and several lines of evidence suggest distinct mechanisms and regulation.
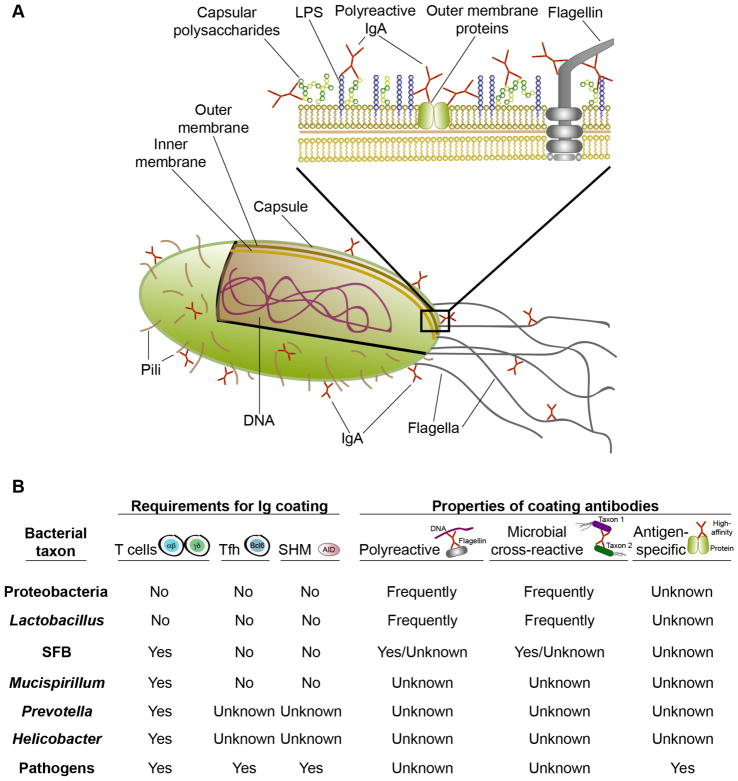
First, specific and high-affinity recognition of individual microbial antigens by homeostatic IgA antibodies has not been demonstrated. Instead, studies of monoclonal antibodies (mAbs) indicate that IgA-derived antibodies are commonly polyreactive and show low-affinity binding to numerous microbial antigens including lipopolysaccharides, DNA, flagellin, and capsular polysaccharides (Figure 1A) (Benckert et al., 2011; Bunker et al., 2017; Fransen et al., 2015; Peterson et al., 2007; Peterson et al., 2015; Quan et al., 1997; Shimoda et al., 1999). Moreover, a substantial number of natural SI IgA PCs differentiate in germ-free (GF) mice or GF mice fed an antigen-free diet (GF/AF) that are devoid of exogenous antigens; mAbs cloned from these IgA+ PCs can bind to the same bacteria normally coated with IgA in specific pathogen free (SPF) mice (Bunker et al., 2017; Fransen et al., 2015; Wijburg et al., 2006). Further, random polyreactive mAbs cloned from naïve B cells or influenza-specific responses bind the same subset of microbiota that is coated with IgA in vivo (Bunker et al., 2017). Glycan-reactive but non-polyreactive antibodies elicited by pathogens also commonly cross-react against commensal bacteria (Rollenske et al., 2018). Together, these data suggest that antibody polyreactivity and associated self-reactivity may be the predominant drivers of IgA selection, and support a model whereby IgA polyreactivity enables low-affinity binding to multiple bacterial surface molecules (Figure 1A).
Figure 1. IgA-coated bacterial taxa and mechanisms of targeting.
A) The IgA repertoire is enriched in natural polyreactive IgA antibodies that can bind multiple self and bacterial antigens with low affinity. Individual antibodies can react against lipopolysaccharides (LPS), capsular polysaccharides, flagellin, DNA, and other antigens, and may target multiple surface antigens in vivo. B) Summary of bacterial taxa that are targeted by IgA in vivo and the humoral mechanisms that lead to their coating, as determined by IgA-seq. The requirements for IgA targeting are based on Ig-seq studies of Tcrb−/−d−/−, CD4-cre Bcl-6fl/fl, or Aicda−/− mice lacking T cells, Tfh, or SHM and CSR, respectively. Properties of coating antibodies are based on polyreactivity ELISAs and Ig-seq using individual mAbs.
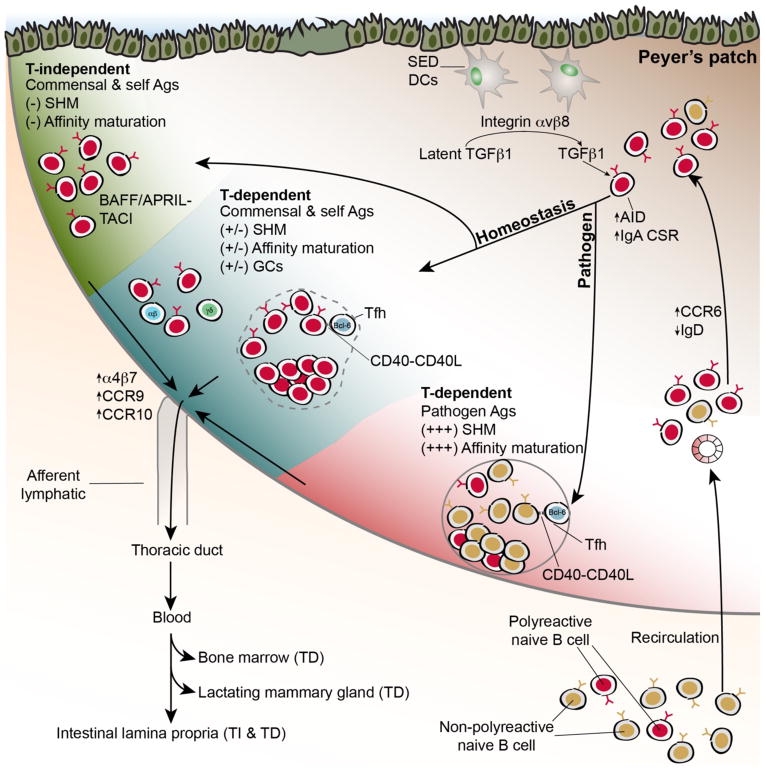
A minor fraction of murine naive B cell precursors express polyreactive specificities and can recognize microbiota in their germline configuration (Bunker et al., 2017). These polyreactive cells recirculate and are preferentially induced to divide in GALT such as the PPs, where they upregulate expression of the chemokine receptor CCR6 and downregulate IgD (Figure 2) (Bunker et al., 2017; Reboldi et al., 2016). CCR6 signaling directs their migration to the PP subepithelial dome (SED), where they receive TGFβ1 signals via interactions with CD11c+CD11b+ and CD11c+CD11b−CD8− DCs (Reboldi et al., 2016). This cellular pathway initiates IgA CSR in CCR6+ cells via a mechanism that requires DC expression of integrin αvβ8, which binds to latency-associated peptide (LAP) and liberates active TGFβ1 (Figure 2) (Reboldi et al., 2016). After receiving signals in the SED, B cells downregulate CCR6 and migrate back to the PP follicle where they continue differentiation. After splenic cell transfer, virtually all dividing B cells in the PPs express CCR6 and thus this population likely contains cells that differentiate via both TI and TD pathways, although CCR6-deficient mice show more profound TD defects (Bunker et al., 2017; Reboldi et al., 2016). Of note, the relevance of this pathway in vivo has only been demonstrated in PPs; by contrast, mLN IgA responses are largely unaffected by CCR6-deficiency (Reboldi et al., 2016) suggesting additional potential contributions of CCR6-independent pathways.
Figure 2. Mechanisms of IgA selection in Peyer’s patches.
Natural polyreactive antibodies arise at low frequencies in the naïve B cell repertoire and recirculate through secondary lymphoid organs including GALT such as PPs. Upon reaching PPs, polyreactive cells are preferentially induced to divide in a manner that may involve reactivity with self-antigens. Upregulation of CCR6 drives migration to the subepithelial dome (SED), where cells receive TGFβ1 signals that induce IgA CSR via a mechanism that requires SED DC activation of latent TGFβ1 through the integrin αvβ8. After initiating CSR, B cells migrate back to the follicle where they can differentiate through either TD or TI pathways. During homeostasis, polyreactive and microbiota-reactive specificities differentiate via either a TI pathway lacking SHM or affinity maturation, or a TD pathway that accumulates SHM but shows little affinity maturation and may include both GC and extrafollicular contributions. This contrasts with TD responses to pathogens that are typically non-polyreactive and show extensive SHM and affinity maturation in GCs. TNF-superfamily receptor-ligand interactions contribute to both TD (CD40-CD40L) and TI (BAFF/APRIL-TACI) pathways. Cells are imprinted for gut homing by induced upregulation of integrin α4β7 and the chemokine receptors CCR9 and CCR10. Lymphoblasts leave the PPs via lymphatics and transit through the thoracic duct to reenter blood circulation, which facilitates their migration to mucosal effector sites such as the intestinal LP, BM, or LMG.
SHM and affinity maturation of the homeostatic IgA response in PP and mLN GCs may also differ significantly from systemic responses. Analysis of SHM distribution and of T cell-deficient mice indicates that the SI IgA+ PC repertoire is a mixture of TI and TD specificities: in young mice and humans, ~75% of PCs are mutated and likely of TD origin, and the frequency of TD specificities increases with age (Bunker et al., 2017; Lindner et al., 2015; Lindner et al., 2012). However, while GALT GCs are clearly dependent on T cells, affinity maturation to antigens from the microbiota has not been demonstrated (Bunker et al., 2015; Casola et al., 2004; Guy-Grand et al., 1975; Macpherson et al., 2000). B cells lacking a BCR but expressing the constitutively active BCR surrogate LMP2A form GCs in GALT but not extraintestinal lymphoid tissues (Casola et al., 2004), suggesting that GC B cell differentiation can occur in the absence of cognate antigen. Moreover, careful analysis of SHM patterns in PP GC cells suggest random mutation driven by DNA sequence-intrinsic AID hotspots rather than affinity-driven selection (Yeap et al., 2015). Further, the amino acid replacement to silent mutation ratio in IgA PCs is approximately 2:1, also suggestive of random SHM in the absence of selection (Bunker et al., 2017). Many mutated IgA mAbs are polyreactive: although some studies have indicated that polyreactivity can be acquired in GCs (Mouquet et al., 2010; Tiller et al., 2007), polyreactivity of IgA mAbs was typically retained upon reversion of mutations to germline (Bunker et al., 2017). Additionally, analyses of mice lacking T cells, CD40, or GCs show largely normal IgA coating of microbiota, with the exception of a handful of rare and atypical commensals (Bergqvist et al., 2006; Bergqvist et al., 2010; Bunker et al., 2015). Perhaps the exceptionally diverse antigen burden in GALT tissues and the scarcity of T cell help for non-protein antigens precludes affinity maturation and instead selects for polyreactivity. Additionally, TI and TD pathways may be intertwined in some circumstances, as memory cells generated via TD pathways can potentially be reactivated in response to TI stimuli (Magri et al., 2017). Thus, although both TI and TD responses contribute to the homeostatic IgA repertoire, clear differences in specificity between these responses have not been documented. By contrast, IgA affinity maturation in PPs in response to vaccination or mucosal pathogens is well documented and may be mechanistically distinct from homeostatic responses (Figure 2) (Bergqvist et al., 2013).
Although homeostatic TD and TI responses do not show major differences in specificity, T cells do influence IgA responses through other mechanisms. Notably, T cell-deficient mice show a significant decrease in intestinal IgA+ PC abundance and a near complete lack of IgA+ PCs in extraintestinal tissues (Bunker et al., 2017; Bunker et al., 2015; Macpherson et al., 2000). As noted previously, a significant population of SI IgA+ PCs can differentiate in the absence of microbiota or dietary antigens; however, these IgAs are largely unmutated (Bunker et al., 2017; Lindner et al., 2012). While CCR6+ dividing cells are detectable in GF PPs, GCs are largely absent (Kubinak et al., 2015). Indeed, T cell-intrinsic sensing of microbial signals through TLRs via the universal adaptor protein myeloid differentiation primary response 88 (MyD88) may be necessary to initiate TD IgA responses (Kubinak et al., 2015). This pathway likely involves MyD88 signaling in both Tfh and forkhead box P3+ (FoxP3+) T follicular regulatory (Tfr) cells (Kubinak et al., 2015; Wang et al., 2015). The origins and specificity of mucosal Tfh/Tfr remain poorly understood, and different studies have suggested that these cells may differentiate from either naïve CD4+ T cells, Th17 cells, or T regulatory cells (Cong et al., 2009; Hirota et al., 2013; Kawamoto et al., 2014; Tsuji et al., 2009). Further, it remains unclear to what extent the interactions between GALT Tfh/Tfr and GC B cells depend on cognate antigen recognition. While high-affinity cognate recognition by Tfh is necessary to support affinity maturation in response to pathogens, the observation that IgA PCs show few signs of affinity maturation suggests that other mechanisms may operate in GALT during homeostasis. For example, these could involve low-affinity interactions with self-reactive T cells, sequential recognition by different T cell clones with distinct specificities due to internalization and presentation of diverse protein and non-protein antigens by polyreactive BCRs, and/or cytokine-driven interactions. Although the extent to which Tfh specificity influences the IgA repertoire remains unclear, other T cell factors such as PD-1 can negatively regulate TD IgA responses (Kawamoto et al., 2012). In summary, these observations support a model in which T cell-intrinsic sensing of microbiota promotes GC formation, which increases the magnitude of IgA responses, facilitates migration of extraintestinal IgAs, and randomly diversifies the IgA repertoire.
IgA-seq and the identification of IgA-coated microbiota
The commensal bacteria bound by IgA in vivo can be studied using bacterial flow cytometry of microbiota taken directly ex vivo and stained with anti-IgA detection reagents. Early studies of human and murine microbiota revealed that only a fraction of commensal bacteria are coated with IgA in vivo (Kroese et al., 1996; Tsuruta et al., 2009; van der Waaij et al., 1996). Recently, new insights have emerged from the combination of bacterial flow cytometry with high-throughput 16S gene amplicon (16S) sequencing, termed IgA-seq. This technique allows relatively unbiased profiling of the complete repertoire of IgA-bound or -unbound bacteria in vivo. Several laboratories have independently developed variants of IgA-seq and these studies have universally revealed that, despite the frequent polyreactivity of IgA antibodies, a taxonomically distinct subset of microbiota is coated with IgA antibodies in mice and humans in vivo (Bunker et al., 2017; Bunker et al., 2015; Dzidic et al., 2017; Kau et al., 2015; Kawamoto et al., 2014; Koch et al., 2016; Kubinak et al., 2015; Palm et al., 2014; Planer et al., 2016; Wilmore et al., 2018). By contrast, most microbes can become IgA-coated in monocolonized gnotobiotic mice (Geva-Zatorsky et al., 2017), likely representing an artifact of monocolonization. Additionally, while some studies have suggested that IgA antibodies may interact with bacteria in a fragment antigen-binding (Fab)-independent manner via glycans on SC (Mathias and Corthesy, 2011), such interactions have not been demonstrated in vivo and the patterns of polyclonal IgA binding to microbiota have been largely confirmed by analysis of IgA-derived mAbs expressed with a human IgG1 constant region (Bunker et al., 2017). Together, these observations suggest that IgA coats a particular subset of microbiota in vivo via interactions that are predominantly Fab-dependent. The human gut microbiota exhibits substantial inter-individual variation (Arumugam et al., 2011). Murine microbiota is relatively stable in animals that are co-housed, but can differ significantly in mice with distinct environmental histories or between animal facilities (Stappenbeck and Virgin, 2016). Moreover, substrains within a species can show considerable variation that is not typically resolved in 16S sequencing analyses (Greenblum et al., 2015). Thus, some caution is warranted in comparing and generalizing IgA-targeted bacteria identified in different studies in mice and/or humans. Below, we consider the bacterial taxa bound by IgA in vivo and the humoral mechanisms that regulate their targeting (Figure 1B).
Several studies have described substantial enrichment of multiple taxa of the phylum Proteobacteria within the IgA+ fraction (Bunker et al., 2017; Bunker et al., 2015; Planer et al., 2016; Wilmore et al., 2018). Although these organisms are relatively rare in the colon, they are often abundant in the SI, and this may explain the high frequency of IgA+ bacteria typically observed in the SI lumen (Bunker et al., 2015; Kroese et al., 1996; Tsuruta et al., 2009). Indeed, fecal IgA+ bacteria preferentially colonize the SI upon transfer to GF recipients (Bunker et al., 2015). Additionally, intestinal Proteobacteria may influence the abundance of BM IgA+ PCs and serum IgA (Wilmore et al., 2018). Proteobacteria are targeted by both TI and TD IgA antibodies in vivo, but their recognition usually does not require SHM or T cells (Bunker et al., 2017; Bunker et al., 2015). Moreover, individual antibodies typically bind multiple distinct Proteobacterial taxa. Polyreactive antibodies of various origins are frequently reactive to Proteobacteria and often bind these taxa in their germline configuration; as such, Proteobacteria-reactive antibodies can arise naturally in the SI of GF or GF/AF mice. Whether IgA binding to Proteobacteria represents active expression of microbial factors that attract these antibodies or widespread binding of polyreactive specificities remains unclear; however, individual bacterial strains typically lose their reactivity to these antibodies upon culture in vitro and regain their binding after reintroduction in vivo, suggesting that antibody binding can be modulated under different growth or environmental conditions (Bunker et al., 2017) and (JJ Bunker, unpublished observation).
IgA responses to several atypical commensals seem to uniquely require TD responses. Of these, segmented filamentous bacteria (SFB) is prototypical and is known to inhabit an unusual niche in close proximity to the ileal epithelium, where it potently stimulates IgA production as well as CD4+ Th17 cell differentiation (Ivanov et al., 2009; Klaasen et al., 1993). SFB induces PP GC hyperplasia, tertiary lymphoid structure formation in the LP, and substantial quantities of both SFB-reactive and SFB-non-reactive IgA (Klaasen et al., 1993; Lecuyer et al., 2014; Talham et al., 1999). SFB is highly coated with IgA in both SPF and monocolonized mice (Bunker et al., 2015; Jiang et al., 2001; Palm et al., 2014). Studies of AID-deficient mice that lack SHM have noted PP and ILF hyperplasia in response to SFB outgrowth, though these phenotypes have not been observed in all studies (Bunker et al., 2015; Fagarasan et al., 2002; Suzuki et al., 2004; Wei et al., 2011). While IgA coating of SFB is lost in T cell-deficient mice, the extent to which this response involves specific, high-affinity antibodies remains unclear, as SFB Ig-coating is unaltered in mice lacking AID (Aicda−/−) or GCs (CD4-cre Bcl-6fl/fl) (Bunker et al., 2015), and polyreactive IgA mAbs that cross-react with both SFB and Proteobacterial taxa in vivo have been observed (Bunker et al., 2017). Thus, SFB IgA coating apparently occurs via a non-canonical mechanism that is dependent upon T cells but neither GCs nor SHM.
In addition to SFB, several additional taxa seem to selectively elicit TD responses. These include Mucispirillum spp., Prevotella spp., and Helicobacter flexispira (Bunker et al., 2015; Palm et al., 2014). Similar to SFB, these bacteria seem to inhabit unconventional niches in close proximity to the intestinal epithelium (Palm et al., 2014; Robertson et al., 2005). While the precise humoral mechanisms that regulate the targeting of these taxa are not known, IgA coating of Mucispirillum, as with SFB, apparently requires T cells but neither SHM nor GCs (Bunker et al., 2015).
Many taxa in the microbiota are not bound by IgA antibodies in vivo (Bunker et al., 2015; Palm et al., 2014). These include most members of the phyla Bacteroidetes and Firmicutes, which are typically abundant in the colon. It remains unclear why these bacteria are not targeted by IgA antibodies. However, it seems that polyreactive antibodies, which commonly cross react with IgA-targeted taxa, do not bind most Bacteroidetes and Firmicutes in vivo (Bunker et al., 2017). This may be due to active expression of factors that prevent polyreactive antibody binding or may relate to intrinsic properties of bacterial cell surface molecules that preclude antibody binding, among other possibilities.
A number of other bacterial taxa are coated with IgA in vivo. While most Firmicutes are not bound by IgA antibodies, coating of Lactobacilli and some but not all Clostridial species have been observed (Bunker et al., 2015; Planer et al., 2016). Akkermansia mucinophila, a member of the phylum Verrucomicrobia, is highly enriched in the IgA+ fraction in humans (Bunker et al., 2015; Planer et al., 2016). Additional taxa are likely to be identified in further studies of diverse microbiota. Moreover, as we discuss later, some pathogens or opportunistic commensals may elicit strong IgA responses in the context of inflammation or dysbiosis (Kau et al., 2015; Palm et al., 2014).
Bone marrow and lactating mammary gland IgA
While the SI is the predominant site of IgA synthesis, IgA+ PCs are also found in a number of extraintestinal tissues including the bone marrow (BM) and lactating mammary gland (LMG). BM IgA+ PCs are presumably the source of most serum IgA antibodies, and the specificity of these antibodies has been analyzed by staining fecal bacteria with serum followed by IgA-seq (Koch et al., 2016; Wilmore et al., 2018). These experiments have revealed that serum IgA antibodies typically react against a similar subset of microbiota to that targeted by intestinal IgA. Notably, serum IgAs bind prominently to Proteobacterial taxa, and the relative abundance of these microbes in the gut may influence the magnitude of the BM IgA+ PC response (Bunker et al., 2017; Wilmore et al., 2018). Analysis of mAbs cloned from BM IgAs indicates that these include many polyreactive specificities that bind Proteobacterial taxa (Bunker et al., 2017). However, in contrast to the intestinal IgA repertoire, virtually all BM IgAs arise via TD responses (Bunker et al., 2017; Wilmore et al., 2018), perhaps because T cell-derived signals are required for induction of molecules such as integrin α4β1 and chemokine receptors such as CXCR4 that facilitate migration and homing to the BM (Mora and von Andrian, 2008).
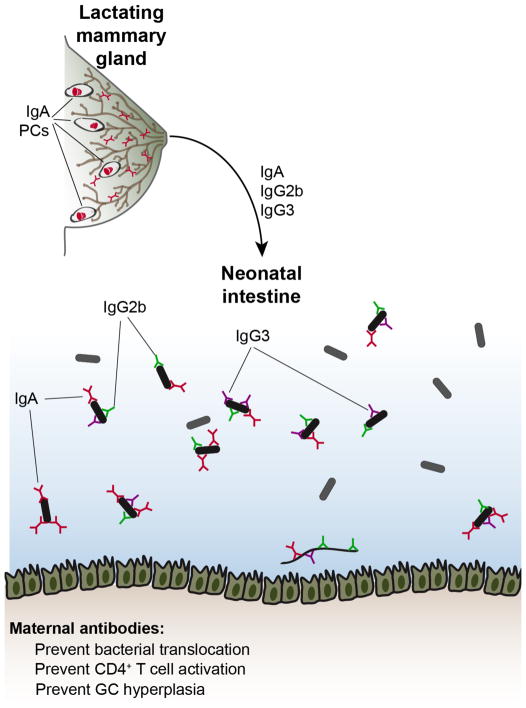
Although few IgA+ PCs are found in the mammary gland of nonpregnant females, there is a dramatic accumulation of these cells during pregnancy and postpartum lactation that wanes after lactation ceases (Figure 3) (Weisz-Carrington et al., 1977). These cells presumably secrete the IgAs that are found at high titers in breast milk (Rogier et al., 2014). As intestinal IgA+ PCs do not appear in young mice until 3–4 weeks of age (Harris et al., 2006), breast milk IgAs ostensibly serve to coat the intestinal microbiota of neonatal mice. Indeed, the LMG and SI IgA repertoires are highly similar within individual mice, implying a common origin (Lindner et al., 2015). Analysis of mAbs cloned from LMG IgAs further suggests that these antibodies resemble intestinal IgAs, with frequent polyreactivity and binding to various microbial taxa including many Proteobacteria (Bunker et al., 2017). Similar to BM IgAs, differentiation of LMG IgAs is predominantly TD and microbiota-dependent (Bunker et al., 2017). Migration of IgA+ PCs to the LMG is dependent upon CCR10 and glandular expression of its ligand CCL28 (Wilson and Butcher, 2004).
Figure 3. Maternal antibodies impact neonatal microbiota and immunity.
During postpartum lactation, IgA, IgG2b, and IgG3 antibodies are transferred to neonates via breast milk. These antibodies coat the neonatal microbiota and restrain the differentiation of CD44+ effector T cells, GCs, and IgA+ PCs in the mucosa, perhaps by preventing bacterial translocation.
IgM and IgG responses to microbiota
In addition to IgA antibodies, a variety of studies have demonstrated that IgM and IgG antibodies can also react against microbiota. There are virtually no detectable PCs expressing IgM or IgG in the murine intestine, however IgM+ and IgG+ PCs are readily detectable in the human gut (Benckert et al., 2011; Bunker et al., 2015; Magri et al., 2017). IgM and IgG coating of microbiota is observable in humans but not mice, and these antibodies coat a similar subset of microbiota to that bound by IgA (Magri et al., 2017; van der Waaij et al., 2004). IgM also coats a subset of microbiota in bony fish that predominantly express mucosal IgT (Zhang et al., 2010). While intestinal IgG+ and IgM+ PCs remain poorly characterized, their similar specificity to IgAs suggests these cells may all derive from similar precursors. It remains unclear why these cells do not undergo IgA CSR; however, the fact that they are present in humans but not mice may suggest that they differentiate during transient periods of inflammation during which IgA CSR is impaired by the presence of inflammatory cytokines. Alternatively, the observation that these cells are detectable relatively early in life may suggest that they arise spontaneously as part of normal ontogeny (Magri et al., 2017).
While IgM+ and IgG+ PCs are not detectable in the murine intestine, staining of microbiota with murine serum indicates that homeostatic IgM and IgG antibodies can bind microbiota (Koch et al., 2016; Zeng et al., 2016). Systemic IgGs that react to microbiota are further enriched under conditions that select for self-reactive and polyreactive antibodies, such as certain genetic deficiencies that alter central tolerance or anti-HIV responses (Schickel et al., 2017; Slack et al., 2009; Williams et al., 2015). In mice, homeostatic serum antibodies of the IgG2b and IgG3 isotypes are commonly reactive to microbiota and bind a similar subset to that coated with IgA; IgG1 antibodies also bind a smaller subset (Koch et al., 2016). IgG2b and IgG3 antibodies arise via a TI mechanism that is dependent upon signaling through TLR2 and TLR4. IgG2b and IgG3-expressing precursors are detectable at low frequencies in GALT and may derive from B1 cells. These antibodies commonly bind intestinal Proteobacteria and can limit their translocation to extraintestinal tissues under both steady-state and inflammatory conditions (Zeng et al., 2016). Interestingly, IgG2b and IgG3 antibodies are transmitted to neonates in breast milk and coat the neonatal microbiota (Figure 3) (Koch et al., 2016). These antibodies may also influence the metabolites found in breast milk by an unknown mechanism, and consequently modulate neonatal intestinal ILC3 and macrophage differentiation indirectly (Gomez de Aguero et al., 2016). Mice born to mothers lacking IgG2b and IgG3 show increased effector CD4+ T cell differentiation in GALT and GC hyperplasia, suggesting that these antibodies limit neonatal T cell responses (Koch et al., 2016). The precise mechanisms by which these antibodies affect microbiota and/or neonatal immunity remain unknown.
IgA memory
Pharmacological depletion of intestinal IgA+ PCs leads to rapid recall of similar specificities, suggesting that IgA+ memory B cells differentiate under homeostatic conditions (Lindner et al., 2012), although expansion of residual PCs may also explain this observation. However, IgA responses to microbiota appear to generate an atypical memory response that differs from the classical prime-boost effect observed upon systemic immunization. Studies utilizing a “reversible” GF system involving transient colonization with an auxotrophic strain of Escherichia coli suggest that repeated commensal exposure results in an additive, rather than exponential, increase in IgA titers (Hapfelmeier et al., 2010). These titers can persist for long periods in GF mice after E. coli exposure but are rapidly lost upon colonization with a complex microbiota. Repeated rounds of memory B cell reactivation, each resulting in low levels of random SHM, may explain the observation that the IgA repertoire accumulates SHM with little affinity maturation (Lindner et al., 2015). Together, these studies suggest that IgA memory cells arise under normal conditions but turn over rapidly and exhibit reduced expansion upon reactivation relative to systemic memory cells.
The cellular phenotype of IgA memory cells in GALT remains poorly defined. Memory cells are likely contained within the IgD−CCR6+ population in PPs, though this population may be heterogenous (Reboldi et al., 2016). In response to mucosal vaccination, an α4β7+CD73+PD-L2+CD80+ memory population was identified (Bemark et al., 2016). In humans, a CD19+CD27+IgA+ memory population is identifiable in blood, though the relation of these cells to intestinal responses remains unclear (Prigent et al., 2016). Further studies detailing the phenotypic and functional properties of IgA+ memory cells are warranted.
Functions of antibodies that bind microbiota
Functional consequences of IgA binding to microbiota are commonly invoked but remain poorly understood, and few if any definite functions have been demonstrated in vivo under homeostatic conditions with an intact microbiota. Assigning precise functions to IgA antibodies has been complicated by a lack of genetic models that are truly deficient in secretory antibody production. As mentioned previously, mice lacking pIgR display only partial loss of IgA secretion (Johansen et al., 1999). Mice bearing a disrupted Igμ constant region (μMT) are widely used as models of B cell deficiency, yet these mice produce largely normal titers of intestinal IgA (Macpherson et al., 2001b). Mice with a deletion in the Ig heavy chain J locus (JH−/−) lack all B cells but, due to the critical role for these cells in lymphoid organogenesis, lack most GALT and thus display numerous B cell-extrinsic deficiencies (Golovkina et al., 1999). Mice that lack IgA via disruption of the Igα constant region or deletion of AID produce a compensatory IgM response that targets the same commensal bacteria normally coated with IgA (Bunker et al., 2015). Of note, IgA deficiency is one of the most common human immunodeficiencies, ranging from 1:400 to 1:3000 in various populations (Cunningham-Rundles, 2001). Similar to IgA-deficient mice, a compensatory mucosal IgM responses arises in these patients (Barros et al., 1985; Fadlallah et al., 2018; Klemola, 1988; Magri et al., 2017). Consistent with this observation, these patients generally have few clinical symptoms. However, IgA-deficient patients do show moderately enhanced susceptibility to a variety of pathologies including recurrent respiratory infections, celiac disease, and autoimmunity (Cunningham-Rundles, 2001). In summary, genetic analyses in mice and humans have generally failed to highlight a clear functional role for homeostatic IgA antibodies.
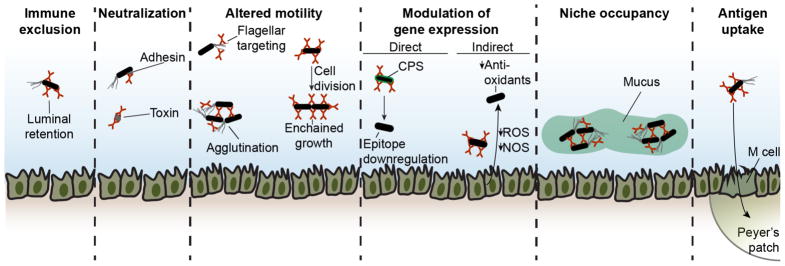
Although its functions remain elusive, the magnitude of the IgA response and the strong evolutionary pressure to secrete mucosal antibodies both suggest functional relevance. It is possible that IgA antibodies may have either beneficial or deleterious effects on IgA-targeted microbes, yet the constitutive presence of IgA-coated commensals suggests that any detrimental effects are not generally sufficient to drive extinction. Indeed, IgA binding to capsular polysaccharides may be exploited by some microbiota species in order to form clusters anchored to the mucus layer, thereby securing a niche from invasion by competing species (Figure 4) (Donaldson et al., 2018). A number of studies have suggested possible functions for these antibodies that may or may not play a role in vivo in the context of homeostatic interactions with the microbiota (Figure 4).
Figure 4. Potential functions of IgA antibodies.
Numerous potential functions for IgA antibodies have been suggested that may or may not play a role in vivo in the context of homeostatic interactions with the commensal microbiota. These include immune exclusion, neutralization, altered motility, modulation of gene expression, niche occupancy, and enhanced antigen uptake.
The protective functions of IgG and IgM antibodies typically involve opsonization and complement recruitment or binding to fragment crystallizable (Fc) receptors (Ravetch and Kinet, 1991). However, IgA is a poor stimulator of the classical complement pathway compared to other isotypes (Murphy and Weaver, 2016). An IgA Fc receptor, FcαRI, is present in humans, primates, and several other mammals but absent in mice (Bakema and van Egmond, 2011). Ligation of this receptor can lead to either activating or inhibitory effects, depending on the monomeric or dimeric nature of the IgA stimulus (Pasquier et al., 2005). FcαRI is expressed by neutrophils, eosinophils, monocytes, and macrophages that are largely absent during homeostasis but can infiltrate the gut during inflammation (Bakema and van Egmond, 2011). These observations suggest that Fc receptors and complement are unlikely to play a functional role during normal homeostasis with microbiota but may be relevant in inflammatory contexts.
One function of IgA may be immune exclusion of its targets, in which antibody binding retains antigen in the intestinal lumen until it is digested or eliminated and thereby precludes priming of other immune responses. This function has been demonstrated in the context of model antigens and specific monoclonal IgA antibodies (Stokes et al., 1975), but its relevance in vivo in the context of IgA-targeted microbiota remains untested. A second possible function is neutralization of bacterial surface antigens, such as adhesins or pili, that promote invasion and pathogenesis. While this has been demonstrated in the context of enteric pathogens (Williams and Gibbons, 1972), it remains unknown whether this mechanism is relevant to homeostasis with commensal microbes. Notably, B cells are required to prevent translocation of commensal bacteria to the mLN upon microbiota colonization of GF mice, although the mechanisms of protection remain undefined (Macpherson and Uhr, 2004).
IgA binding may restrict bacterial motility through several possible mechanisms, whose relevance in vivo in the context of microbiota and natural microbiota-reactive IgA remains unclear. Studies of monoclonal IgA antibodies specific for the pathogen Shigella flexneri have suggested that IgA can entrap bacteria within the mucus layer overlaying the intestinal epithelium (Boullier et al., 2009). Additionally, IgA may agglutinate pathogens and facilitate their clearance (Hendrickx et al., 2015). IgA antibodies generated after oral vaccination with inactivated Salmonella Typhimurium were shown to enchain daughter cells of dividing bacteria and accelerate their clearance (Moor et al., 2017). IgA may also limit motility by binding bacterial flagellins: while specific IgA recognition of commensal flagellins has not been demonstrated, many polyreactive IgAs bind flagellin with low affinity (Bunker et al., 2017; Cullender et al., 2013). In vitro studies of high-affinity mAbs to flagellin have demonstrated that these can limit bacterial motility (Cullender et al., 2013). Further, increases in flagellar gene expression are observable in the microbiota of mice lacking the flagellin innate immune sensor TLR5, and a role for IgA in this process has been suggested (Cullender et al., 2013).
Another possible function may involve direct or indirect modulation of microbial gene expression. This is supported by studies of IgA hybridomas isolated from mice monocolonized with Bacteroides thetaiotaomicron and subsequently administered to B. thetaiotaomicron-monocolonized Rag1−/− mice, which lack IgA (Peterson et al., 2007; Peterson et al., 2015). In direct modulation, IgA binding results in changes in bacterial gene expression. For example, one IgA mAb against a capsular polysaccharide antigen downregulated epitope expression and reduced bacterial fitness relative to an epitope-deficient strain (Peterson et al., 2007). A second mAb against an LPS O-antigen polysaccharide did not modulate antigen expression or microbial fitness, suggesting that this property may be variable (Peterson et al., 2015). In indirect modulation, IgA binding may alter gene expression by epithelial or other cells, which in turn secrete factors that alter bacterial gene expression. Monocolonization of Rag1−/− mice with B. thetaiotaomicron leads to upregulation of reactive oxygen and nitrogen species synthesis in the intestinal epithelium and concomitant upregulation of bacterial antioxidant enzymes; these phenotypes were abrogated in the presence of IgA monoclonal antibody (Peterson et al., 2007). This system is reductionist but informative; however, B. thetaiotaomicron is not a major target of IgA antibodies in vivo and thus it remains unclear whether these principles apply to naturally-arising IgAs in the context of an intact microbiota.
Homeostatic IgA antibodies may also play distinct functional roles in specific physiological contexts. The Proteobacteria-reactive serum IgA response that arises under normal homeostatic conditions appears to be protective against polymicrobial sepsis after intestinal injury (Wilmore et al., 2018); the mechanisms by which IgA is protective in this model remain unknown. Further, as mentioned previously, maternal antibodies transmitted in breast milk seem to attenuate intestinal immune activation in neonates (Figure 3). Mice born to B cell-deficient mothers show exaggerated intestinal IgA responses early in life, perhaps as a result of increased commensal translocation to the mLN (Harris et al., 2006; Rogier et al., 2014). Maternal IgA may also influence the composition of the neonatal microbiota via unknown mechanisms (Rogier et al., 2014). Additionally, maternal IgA may play a partially redundant role together with IgG2b and IgG3 antibodies to limit neonatal TD responses (Koch et al., 2016). Lastly, IgA binding to intestinal antigens may facilitate their uptake through epithelial microfold (M) cells and enhance priming of further responses (Figure 4) (Fransen et al., 2015).
IgA antibodies can confer protective immunity in response to mucosal pathogens in a variety of additional contexts that have been reviewed extensively elsewhere (Lycke, 2012; Macpherson et al., 2001a). Cholera toxin has been well-studied and induces a TD response that protects via neutralization (Hornquist et al., 1995). In vitro studies suggest that IgA can bind toxins in the intestinal LP and facilitate their excretion (Fernandez et al., 2003). Additionally, IgA antibodies against the HIV Envelope were strongly correlated with protection in the RV144 HIV vaccine trial (Haynes et al., 2012). While preexisting polyreactive IgA antibodies were shown to provide early protection against Salmonella Typhimurium (Wijburg et al., 2006), in the vast majority of infections the relationship between protective responses and pre-existing homeostatic IgA remains unknown. In many cases, protective responses to pathogens are likely to proceed via cellular pathways that more closely resemble systemic immunity than the homeostatic IgA response; careful studies of the pre- and post-immune repertoires and cellular processes involved should shed light on these mechanisms.
Antibodies to microbiota in inflammatory bowel diseases
Mucosal antibody responses are exaggerated in inflammatory bowel diseases (IBD) including Crohn’s disease and ulcerative colitis, which involve a complex interplay between host genetics, environmental factors, and microbiota composition (Dalal and Chang, 2014). Increased coating of fecal microbiota with IgA, IgG, and IgM has been observed in both human IBD and mouse models (Palm et al., 2014; van der Waaij et al., 2004; Viladomiu et al., 2017). IgM+ and IgG+ PCs accumulate in the inflamed gut and may exacerbate inflammation, though their specificity and contributions to pathology remains poorly understood (Kanai et al., 2006; Uo et al., 2013). IBD patients also show elevated serum IgA that reacts against flagellin and various autoantigens; whether this represents induction of specific antibodies or expansion of polyreactive specificities remains unknown (Landers et al., 2002; Lodes et al., 2004; Sitaraman et al., 2005). Mouse models of colitis induced by T cell-transfer into lymphopenic recipients suggest that B cells can protect against pathology (Gerth et al., 2004), though the mechanisms of protection and their contributions in human IBD remain unclear. Recently, several studies have suggested that IgA coating can identify disease-associated members of the microbiota in IBD or IBD-associated spondyloarthritis (Palm et al., 2014; Viladomiu et al., 2017). These studies have generally focused on single microbes or somewhat arbitrary consortia without performing systematic analysis of both IgA+ and IgA− bacteria. Given the extensive IgA coating of microbiota found under normal healthy conditions, it is unlikely that all IgA-targeted microbes in IBD patients are colitogenic. A more likely scenario is that bona fide pathogens elicit IgA responses and are present alongside a wide variety of non-pathogenic commensals in the IgA+ fraction. Therefore, IgA-seq in combination with other assays may be required for more reliable discrimination between pathogens and commensals.
Concluding Remarks
Current evidence supports a model in which two distinct types of humoral immunity coexist in the intestinal mucosa (Figure 2). The predominant pathway is intrinsically polyreactive and low affinity, largely independent of T cells and somatic mutations, and bears features of germline-encoded innate immunity; this response seems mostly involved in homeostatic interactions with commensal bacteria. The other pathway exhibits classical features of T cell dependent, hypermutated and affinity-matured adaptive responses and is predominantly triggered by pathogens. Determining how these radically different responses are integrated to protect the host while preserving a symbiotic relationship with microbiota is a major direction of future research.
From an immunological viewpoint, the enrichment of polyreactive specificities within the IgA repertoire raises questions about how cells expressing these antibodies avoid deletion during central tolerance (Wardemann et al., 2003). Future work should determine how and when these antibodies might escape from tolerance mechanisms and define the cellular pathways involved in their selection in the mucosa. Further, the structural basis of antibody polyreactivity remains poorly understood, and determining how this reactivity is achieved will require detailed structural and biochemical studies. In addition, it remains unclear to what extent polyreactivity might enable cross-reactivity to pathogens, and whether pre-existing homeostatic IgA antibodies might also contribute to protective immunity during infection. The answers to these questions may have implications for mucosal vaccination and elicitation of polyreactive broadly neutralizing antibodies against rapidly mutating viruses (Andrews et al., 2015; Mouquet et al., 2010).
From a microbiological perspective, major questions remain regarding the molecular targets of IgA. In particular, studies should focus on identifying specific antigens recognized by IgA antibodies using biochemical and genetic approaches – a matter that is complicated by a general lack of genetic tools or genomic information for many commensal bacteria. Relatedly, it will be important to define and characterize bacterial mechanisms that lead to either evasion or attraction of IgA antibodies. In addition, further work should investigate the consequences of antibody binding on microbes, and define precise mechanisms by which IgA binding alters physiology and/or fitness. The answers to these questions may shed light on the enigmatic functions of IgA and may have major implications for our understanding of its biological meaning.
While recent years have seen numerous important advances, many aspects of IgA responses remain poorly understood. Future studies exploring these questions may provide wide-ranging insights, from evolutionary aspects of immunity and commensalism to new strategies to manipulate the host or microbiota in order to prevent, treat, or cure enteric pathologies.
References
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakema JE, van Egmond M. The human immunoglobulin A Fc receptor FcalphaRI: a multifaceted regulator of mucosal immunity. Mucosal Immunol. 2011;4:612–624. doi: 10.1038/mi.2011.36. [DOI] [PubMed] [Google Scholar]
- Barros MD, Porto MH, Leser PG, Grumach AS, Carneiro-Sampaio MM. Study of colostrum of a patient with selective IgA deficiency. Allergol Immunopathol (Madr) 1985;13:331–334. [PubMed] [Google Scholar]
- Bemark M, Hazanov H, Stromberg A, Komban R, Holmqvist J, Koster S, Mattsson J, Sikora P, Mehr R, Lycke NY. Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nat Commun. 2016;7:12698. doi: 10.1038/ncomms12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benckert J, Schmolka N, Kreschel C, Zoller MJ, Sturm A, Wiedenmann B, Wardemann H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergqvist P, Gardby E, Stensson A, Bemark M, Lycke NY. Gut IgA class switch recombination in the absence of CD40 does not occur in the lamina propria and is independent of germinal centers. J Immunol. 2006;177:7772–7783. doi: 10.4049/jimmunol.177.11.7772. [DOI] [PubMed] [Google Scholar]
- Bergqvist P, Stensson A, Hazanov L, Holmberg A, Mattsson J, Mehr R, Bemark M, Lycke NY. Re-utilization of germinal centers in multiple Peyer’s patches results in highly synchronized, oligoclonal, and affinity-matured gut IgA responses. Mucosal Immunol. 2013;6:122–135. doi: 10.1038/mi.2012.56. [DOI] [PubMed] [Google Scholar]
- Bergqvist P, Stensson A, Lycke NY, Bemark M. T cell-independent IgA class switch recombination is restricted to the GALT and occurs prior to manifest germinal center formation. J Immunol. 2010;184:3545–3553. doi: 10.4049/jimmunol.0901895. [DOI] [PubMed] [Google Scholar]
- Blanc P, Moro-Sibilot L, Barthly L, Jagot F, This S, de Bernard S, Buffat L, Dussurgey S, Colisson R, Hobeika E, et al. Mature IgM-expressing plasma cells sense antigen and develop competence for cytokine production upon antigenic challenge. Nat Commun. 2016;7:13600. doi: 10.1038/ncomms13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boullier S, Tanguy M, Kadaoui KA, Caubet C, Sansonetti P, Corthesy B, Phalipon A. Secretory IgA-mediated neutralization of Shigella flexneri prevents intestinal tissue destruction by down-regulating inflammatory circuits. J Immunol. 2009;183:5879–5885. doi: 10.4049/jimmunol.0901838. [DOI] [PubMed] [Google Scholar]
- Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, Bendelac A. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358 doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, Carroll MC, Rajewsky K. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–327. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- Cazac BB, Roes J. TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–451. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Beller A, Rausch S, Strandmark J, Zanker M, Arbach O, Kruglov A, Berek C. Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity. 2014;40:582–593. doi: 10.1016/j.immuni.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SW, Cebra JJ. Peyer’s patches: an enriched source of precursors for IgA-producing immunocytes in the rabbit. J Exp Med. 1971;134:188–200. doi: 10.1084/jem.134.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham-Rundles C. Physiology of IgA and IgA deficiency. J Clin Immunol. 2001;21:303–309. doi: 10.1023/a:1012241117984. [DOI] [PubMed] [Google Scholar]
- Dalal SR, Chang EB. The microbial basis of inflammatory bowel diseases. J Clin Invest. 2014;124:4190–4196. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Niro R, Mesin L, Raki M, Zheng NY, Lund-Johansen F, Lundin KE, Charpilienne A, Poncet D, Wilson PC, Sollid LM. Rapid generation of rotavirus-specific human monoclonal antibodies from small-intestinal mucosa. J Immunol. 2010;185:5377–5383. doi: 10.4049/jimmunol.1001587. [DOI] [PubMed] [Google Scholar]
- Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, Mazmanian SK. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018 doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzidic M, Abrahamsson TR, Artacho A, Bjorksten B, Collado MC, Mira A, Jenmalm MC. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J Allergy Clin Immunol. 2017;139:1017–1025. e1014. doi: 10.1016/j.jaci.2016.06.047. [DOI] [PubMed] [Google Scholar]
- Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aan1217. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature. 2001;413:639–643. doi: 10.1038/35098100. [DOI] [PubMed] [Google Scholar]
- Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- Fernandez MI, Pedron T, Tournebize R, Olivo-Marin JC, Sansonetti PJ, Phalipon A. Anti-inflammatory role for intracellular dimeric immunoglobulin a by neutralization of lipopolysaccharide in epithelial cells. Immunity. 2003;18:739–749. doi: 10.1016/s1074-7613(03)00122-5. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S, Chiavelli A, D’Erchia AM, Sethi MK, Pabst O, et al. BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunity. 2015;43:527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Gerth AJ, Lin L, Neurath MF, Glimcher LH, Peng SL. An innate cell-mediated, murine ulcerative colitis-like syndrome in the absence of nuclear factor of activated T cells. Gastroenterology. 2004;126:1115–1121. doi: 10.1053/j.gastro.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 2017;168:928–943. e911. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkina TV, Shlomchik M, Hannum L, Chervonsky A. Organogenic role of B lymphocytes in mucosal immunity. Science. 1999;286:1965–1968. doi: 10.1126/science.286.5446.1965. [DOI] [PubMed] [Google Scholar]
- Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy-Grand D, Griscelli C, Vassalli P. Peyer’s patches, gut IgA plasma cells and thymic function: study in nude mice bearing thymic grafts. J Immunol. 1975;115:361–364. [PubMed] [Google Scholar]
- Hamada H, Hiroi T, Nishiyama Y, Takahashi H, Masunaga Y, Hachimura S, Kaminogawa S, Takahashi-Iwanaga H, Iwanaga T, Kiyono H, et al. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J Immunol. 2002;168:57–64. doi: 10.4049/jimmunol.168.1.57. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris NL, Spoerri I, Schopfer JF, Nembrini C, Merky P, Massacand J, Urban JF, Jr, Lamarre A, Burki K, Odermatt B, et al. Mechanisms of neonatal mucosal antibody protection. J Immunol. 2006;177:6256–6262. doi: 10.4049/jimmunol.177.9.6256. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Hendrickx AP, Top J, Bayjanov JR, Kemperman H, Rogers MR, Paganelli FL, Bonten MJ, Willems RJ. Antibiotic-Driven Dysbiosis Mediates Intraluminal Agglutination and Alternative Segregation of Enterococcus faecium from the Intestinal Epithelium. MBio. 2015;6:e01346–01315. doi: 10.1128/mBio.01346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Hornquist CE, Ekman L, Grdic KD, Schon K, Lycke NY. Paradoxical IgA immunity in CD4-deficient mice. Lack of cholera toxin-specific protective immunity despite normal gut mucosal IgA differentiation. J Immunol. 1995;155:2877–2887. [PubMed] [Google Scholar]
- Huard B, McKee T, Bosshard C, Durual S, Matthes T, Myit S, Donze O, Frossard C, Chizzolini C, Favre C, et al. APRIL secreted by neutrophils binds to heparan sulfate proteoglycans to create plasma cell niches in human mucosa. J Clin Invest. 2008;118:2887–2895. doi: 10.1172/JCI33760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen R, Snir O, Stensland M, Kroll JE, Steinsbo O, Korponay-Szabo IR, Lundin KEA, de Souza GA, Sollid LM. Strong Clonal Relatedness between Serum and Gut IgA despite Different Plasma Cell Origins. Cell Rep. 2017;20:2357–2367. doi: 10.1016/j.celrep.2017.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HQ, Bos NA, Cebra JJ. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun. 2001;69:3611–3617. doi: 10.1128/IAI.69.6.3611-3617.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen FE, Pekna M, Norderhaug IN, Haneberg B, Hietala MA, Krajci P, Betsholtz C, Brandtzaeg P. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J Exp Med. 1999;190:915–922. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- Kanai T, Kawamura T, Dohi T, Makita S, Nemoto Y, Totsuka T, Watanabe M. TH1/TH2-mediated colitis induced by adoptive transfer of CD4+CD45RBhigh T lymphocytes into nude mice. Inflamm Bowel Dis. 2006;12:89–99. doi: 10.1097/01.MIB.0000197237.21387.mL. [DOI] [PubMed] [Google Scholar]
- Kang HS, Chin RK, Wang Y, Yu P, Wang J, Newell KA, Fu YX. Signaling via LTbetaR on the lamina propria stromal cells of the gut is required for IgA production. Nat Immunol. 2002;3:576–582. doi: 10.1038/ni795. [DOI] [PubMed] [Google Scholar]
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beynen AC. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemola T. Immunohistochemical findings in the intestine of IgA-deficient persons: number of intraepithelial T lymphocytes is increased. J Pediatr Gastroenterol Nutr. 1988;7:537–543. doi: 10.1097/00005176-198807000-00010. [DOI] [PubMed] [Google Scholar]
- Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, Barton GM. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland ME. The coming of age of the immunoglobulin J chain. Annu Rev Immunol. 1985;3:425–453. doi: 10.1146/annurev.iy.03.040185.002233. [DOI] [PubMed] [Google Scholar]
- Kroese FG, de Waard R, Bos NA. B-1 cells and their reactivity with the murine intestinal microflora. Semin Immunol. 1996;8:11–18. doi: 10.1006/smim.1996.0003. [DOI] [PubMed] [Google Scholar]
- Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, Seleznik GM, Eberl G, Littman DR, Heikenwalder M, Tumanov AV, Nedospasov SA. Nonredundant function of soluble LTalpha3 produced by innate lymphoid cells in intestinal homeostasis. Science. 2013;342:1243–1246. doi: 10.1126/science.1243364. [DOI] [PubMed] [Google Scholar]
- Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM, Round JL. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel EJ, Kim CH, Lazarus NH, Vierra MA, Soler D, Bowman EP, Butcher EC. CCR10 expression is a common feature of circulating and mucosal epithelial tissue IgA Ab-secreting cells. J Clin Invest. 2003;111:1001–1010. doi: 10.1172/JCI17244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers CJ, Cohavy O, Misra R, Yang H, Lin YC, Braun J, Targan SR. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- Landsverk OJ, Snir O, Casado RB, Richter L, Mold JE, Reu P, Horneland R, Paulsen V, Yaqub S, Aandahl EM, et al. Antibody-secreting plasma cells persist for decades in human intestine. J Exp Med. 2017;214:309–317. doi: 10.1084/jem.20161590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Lindner C, Thomsen I, Wahl B, Ugur M, Sethi MK, Friedrichsen M, Smoczek A, Ott S, Baumann U, Suerbaum S, et al. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol. 2015;16:880–888. doi: 10.1038/ni.3213. [DOI] [PubMed] [Google Scholar]
- Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, Prinz I, Pabst O. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med. 2012;209:365–377. doi: 10.1084/jem.20111980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat Rev Immunol. 2012;12:592–605. doi: 10.1038/nri3251. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Hunziker L, McCoy K, Lamarre A. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes Infect. 2001a;3:1021–1035. doi: 10.1016/s1286-4579(01)01460-5. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Lamarre A, McCoy K, Harriman GR, Odermatt B, Dougan G, Hengartner H, Zinkernagel RM. IgA production without mu or delta chain expression in developing B cells. Nat Immunol. 2001b;2:625–631. doi: 10.1038/89775. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Magri G, Comerma L, Pybus M, Sintes J, Llige D, Segura-Garzon D, Bascones S, Yeste A, Grasset EK, Gutzeit C, et al. Human Secretory IgM Emerges from Plasma Cells Clonally Related to Gut Memory B Cells and Targets Highly Diverse Commensals. Immunity. 2017;47:118–134. e118. doi: 10.1016/j.immuni.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masahata K, Umemoto E, Kayama H, Kotani M, Nakamura S, Kurakawa T, Kikuta J, Gotoh K, Motooka D, Sato S, et al. Generation of colonic IgA-secreting cells in the caecal patch. Nat Commun. 2014;5:3704. doi: 10.1038/ncomms4704. [DOI] [PubMed] [Google Scholar]
- Mathias A, Corthesy B. Recognition of gram-positive intestinal bacteria by hybridoma- and colostrum-derived secretory immunoglobulin A is mediated by carbohydrates. J Biol Chem. 2011;286:17239–17247. doi: 10.1074/jbc.M110.209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli CA, Tomasi TB., Jr The life span of IgA plasma cells from the mouse intestine. J Exp Med. 1973;138:452–460. doi: 10.1084/jem.138.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams M, Phillips-Quagliata JM, Lamm ME. Mesenteric lymph node B lymphoblasts which home to the small intestine are precommitted to IgA synthesis. J Exp Med. 1977;145:866–875. doi: 10.1084/jem.145.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. 2017;544:498–502. doi: 10.1038/nature22058. [DOI] [PubMed] [Google Scholar]
- Mora JR, Iwata M, Eksteen B, Song SY, Junt T, Senman B, Otipoby KL, Yokota A, Takeuchi H, Ricciardi-Castagnoli P, et al. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 2006;314:1157–1160. doi: 10.1126/science.1132742. [DOI] [PubMed] [Google Scholar]
- Mora JR, von Andrian UH. Differentiation and homing of IgA-secreting cells. Mucosal Immunol. 2008;1:96–109. doi: 10.1038/mi.2007.14. [DOI] [PubMed] [Google Scholar]
- Moro-Sibilot L, Blanc P, Taillardet M, Bardel E, Couillault C, Boschetti G, Traverse-Glehen A, Defrance T, Kaiserlian D, Dubois B. Mouse and Human Liver Contain Immunoglobulin A-Secreting Cells Originating From Peyer’s Patches and Directed Against Intestinal Antigens. Gastroenterology. 2016;151:311–323. doi: 10.1053/j.gastro.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Weaver C. Janeway’s immunobiology. 9. New York, NY: Garland Science/Taylor & Francis Group, LLC; 2016. [Google Scholar]
- Mussmann R, Du Pasquier L, Hsu E. Is Xenopus IgX an analog of IgA? Eur J Immunol. 1996;26:2823–2830. doi: 10.1002/eji.1830261205. [DOI] [PubMed] [Google Scholar]
- Ng EK, Panesar N, Longo WE, Shapiro MJ, Kaminski DL, Tolman KC, Mazuski JE. Human intestinal epithelial and smooth muscle cells are potent producers of IL-6. Mediators Inflamm. 2003;12:3–8. doi: 10.1080/0962935031000096917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier B, Launay P, Kanamaru Y, Moura IC, Pfirsch S, Ruffie C, Henin D, Benhamou M, Pretolani M, Blank U, Monteiro RC. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM. Immunity. 2005;22:31–42. doi: 10.1016/j.immuni.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Peterson DA, Planer JD, Guruge JL, Xue L, Downey-Virgin W, Goodman AL, Seedorf H, Gordon JI. Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem. 2015;290:12630–12649. doi: 10.1074/jbc.M114.633800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalipon A, Corthesy B. Novel functions of the polymeric Ig receptor: well beyond transport of immunoglobulins. Trends Immunol. 2003;24:55–58. doi: 10.1016/s1471-4906(02)00031-5. [DOI] [PubMed] [Google Scholar]
- Pinto D, Montani E, Bolli M, Garavaglia G, Sallusto F, Lanzavecchia A, Jarrossay D. A functional BCR in human IgA and IgM plasma cells. Blood. 2013;121:4110–4114. doi: 10.1182/blood-2012-09-459289. [DOI] [PubMed] [Google Scholar]
- Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, Warner BB, Gordon JI. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. 2016;534:263–266. doi: 10.1038/nature17940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent J, Lorin V, Kok A, Hieu T, Bourgeau S, Mouquet H. Scarcity of autoreactive human blood IgA(+) memory B cells. Eur J Immunol. 2016;46:2340–2351. doi: 10.1002/eji.201646446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan CP, Berneman A, Pires R, Avrameas S, Bouvet JP. Natural polyreactive secretory immunoglobulin A autoantibodies as a possible barrier to infection in humans. Infect Immun. 1997;65:3997–4004. doi: 10.1128/iai.65.10.3997-4004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay AJ, Husband AJ, Ramshaw IA, Bao S, Matthaei KI, Koehler G, Kopf M. The role of interleukin-6 in mucosal IgA antibody responses in vivo. Science. 1994;264:561–563. doi: 10.1126/science.8160012. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Kinet JP. Fc receptors. Annu Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 2016;352:aaf4822. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, Kuraoka M, Kelsoe G. Natural IgM is produced by CD5- plasma cells that occupy a distinct survival niche in bone marrow. J Immunol. 2015;194:231–242. doi: 10.4049/jimmunol.1401203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005;55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, Kaetzel CS. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenske T, Szijarto V, Lukasiewicz J, Guachalla LM, Stojkovic K, Hartl K, Stulik L, Kocher S, Lasitschka F, Al-Saeedi M, et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat Immunol. 2018;19:617–624. doi: 10.1038/s41590-018-0106-2. [DOI] [PubMed] [Google Scholar]
- Roux ME, McWilliams M, Phillips-Quagliata JM, Weisz-Carrington P, Lamm ME. Origin of IgA-secreting plasma cells in the mammary gland. J Exp Med. 1977;146:1311–1322. doi: 10.1084/jem.146.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Brennecke AM, Agarwal S, Krey M, Duber S, Weiss S. An intrinsic propensity of murine peritoneal B1b cells to switch to IgA in presence of TGF-beta and retinoic acid. PLoS One. 2013;8:e82121. doi: 10.1371/journal.pone.0082121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickel JN, Glauzy S, Ng YS, Chamberlain N, Massad C, Isnardi I, Katz N, Uzel G, Holland SM, Picard C, et al. Self-reactive VH4-34-expressing IgG B cells recognize commensal bacteria. J Exp Med. 2017;214:1991–2003. doi: 10.1084/jem.20160201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda M, Inoue Y, Azuma N, Kanno C. Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer’s patches. Immunology. 1999;97:9–17. doi: 10.1046/j.1365-2567.1999.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman SV, Klapproth JM, Moore DA, 3rd, Landers C, Targan S, Williams IR, Gewirtz AT. Elevated flagellin-specific immunoglobulins in Crohn’s disease. Am J Physiol Gastrointest Liver Physiol. 2005;288:G403–406. doi: 10.1152/ajpgi.00357.2004. [DOI] [PubMed] [Google Scholar]
- Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 2016;534:191–199. doi: 10.1038/nature18285. [DOI] [PubMed] [Google Scholar]
- Stokes CR, Soothill JF, Turner MW. Immune exclusion is a function of IgA. Nature. 1975;255:745–746. doi: 10.1038/255745a0. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Maruya M, Kawamoto S, Sitnik K, Kitamura H, Agace WW, Fagarasan S. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity. 2010;33:71–83. doi: 10.1016/j.immuni.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka H, Abe Y, Asano J, Sato T, Liu J, Iwata M, Ohteki T. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity. 2011;34:247–257. doi: 10.1016/j.immuni.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, Hori S, Fagarasan S. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, Itoh K, Littman DR, Fagarasan S. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Tsuruta T, Inoue R, Nojima I, Tsukahara T, Hara H, Yajima T. The amount of secreted IgA may not determine the secretory IgA coating ratio of gastrointestinal bacteria. FEMS Immunol Med Microbiol. 2009;56:185–189. doi: 10.1111/j.1574-695X.2009.00568.x. [DOI] [PubMed] [Google Scholar]
- Tuma P, Hubbard AL. Transcytosis: crossing cellular barriers. Physiol Rev. 2003;83:871–932. doi: 10.1152/physrev.00001.2003. [DOI] [PubMed] [Google Scholar]
- Uo M, Hisamatsu T, Miyoshi J, Kaito D, Yoneno K, Kitazume MT, Mori M, Sugita A, Koganei K, Matsuoka K, et al. Mucosal CXCR4+ IgG plasma cells contribute to the pathogenesis of human ulcerative colitis through FcgammaR-mediated CD14 macrophage activation. Gut. 2013;62:1734–1744. doi: 10.1136/gutjnl-2012-303063. [DOI] [PubMed] [Google Scholar]
- van der Waaij LA, Kroese FG, Visser A, Nelis GF, Westerveld BD, Jansen PL, Hunter JO. Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2004;16:669–674. doi: 10.1097/01.meg.0000108346.41221.19. [DOI] [PubMed] [Google Scholar]
- van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–354. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaf9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker WA, Isselbacher KJ. Intestinal antibodies. N Engl J Med. 1977;297:767–773. doi: 10.1056/NEJM197710062971407. [DOI] [PubMed] [Google Scholar]
- Wang S, Charbonnier LM, Noval Rivas M, Georgiev P, Li N, Gerber G, Bry L, Chatila TA. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity. 2015;43:289–303. doi: 10.1016/j.immuni.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu L, Moore DJ, Shen X, Peek RM, Acra SA, Li H, Ren X, Polk DB, Yan F. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol. 2017;10:373–384. doi: 10.1038/mi.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Sugai M, Nambu Y, Osato M, Hayashi T, Kawaguchi M, Komori T, Ito Y, Shimizu A. Requirement for Runx proteins in IgA class switching acting downstream of TGF-beta 1 and retinoic acid signaling. J Immunol. 2010;184:2785–2792. doi: 10.4049/jimmunol.0901823. [DOI] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Weisz-Carrington P, Roux ME, Lamm ME. Plasma cells and epithelial immunoglobulins in the mouse mammary gland during pregnancy and lactation. J Immunol. 1977;119:1306–1307. [PubMed] [Google Scholar]
- Wijburg OL, Uren TK, Simpfendorfer K, Johansen FE, Brandtzaeg P, Strugnell RA. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J Exp Med. 2006;203:21–26. doi: 10.1084/jem.20052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RC, Gibbons RJ. Inhibition of bacterial adherence by secretory immunoglobulin A: a mechanism of antigen disposal. Science. 1972;177:697–699. doi: 10.1126/science.177.4050.697. [DOI] [PubMed] [Google Scholar]
- Williams WB, Liao HX, Moody MA, Kepler TB, Alam SM, Gao F, Wiehe K, Trama AM, Jones K, Zhang R, et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmore JR, Gaudette BT, Gomez Atria D, Hashemi T, Jones DD, Gardner CA, Cole SD, Misic AM, Beiting DP, Allman D. Commensal Microbes Induce Serum IgA Responses that Protect against Polymicrobial Sepsis. Cell Host Microbe. 2018 doi: 10.1016/j.chom.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E, Butcher EC. CCL28 controls immunoglobulin (Ig)A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J Exp Med. 2004;200:805–809. doi: 10.1084/jem.20041069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Rennert P, McGhee JR, Kweon MN, Yamamoto S, Dohi T, Otake S, Bluethmann H, Fujihashi K, Kiyono H. Alternate mucosal immune system: organized Peyer’s patches are not required for IgA responses in the gastrointestinal tract. J Immunol. 2000;164:5184–5191. doi: 10.4049/jimmunol.164.10.5184. [DOI] [PubMed] [Google Scholar]
- Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, et al. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]