Abstract
Comprehensive genomic characterization of prostate cancer has identified recurrent alterations in androgen signaling, DNA repair, and PI3K among others. However, larger and uniform genomic analysis may reveal additional recurrently mutated genes at lower frequencies. Here we aggregate and uniformly analyze exome sequencing data from 1013 prostate cancers. We identify and validate a new class of E26 transformation-specific (ETS) fusion negative tumors defined by mutations in epigenetic regulators, as well as alterations in pathways not previously implicated in prostate cancer, such as the spliceosome pathway. We find that the incidence of significantly mutated genes (SMGs) follows a long-tail distribution, with many genes mutated in less than 3% of cases. We identify a total of 97 SMGs, including 70 not previously implicated in prostate cancer, such as the ubiquitin ligase CUL3 and the transcription factor SPEN. Finally, comparing primary and metastatic prostate cancer reveals a set of genomic markers that may inform risk stratification.
Keywords: Prostate cancer, whole exome sequencing, cancer genomics, mutational significance
The genomic landscape of primary and metastatic prostate cancer has been robustly assessed through whole exome sequencing (WES) of tumors and matched germline samples. These studies have identified multiple recurrently altered genes and pathways, including androgen signaling, DNA repair, and phosphoinositide 3-kinase (PI3K)/AKT signaling1,2. Additionally, they revealed genomically distinct classes of prostate cancer, defined by ETS transcription family fusions3 or mutations in SPOP4, FOXA14, or IDH12. Nevertheless, prostate cancer harbors significant interpatient genomic heterogeneity, and power analyses have suggested that larger WES studies may reveal additional statistically significant mutated genes occurring at lower frequencies, indicating that the spectrum of novel prostate cancer genes is incompletely defined5. As the aggregation and uniform meta-analysis of WES data has been transformative to research and clinical interpretation of germline genetics6, we hypothesized that mutational significance analysis using statistical and biological frameworks in a large and uniformly analyzed WES cohort may similarly identify novel genes and pathways to refine the genomic landscape of prostate cancer.
We assembled and uniformly analyzed whole exome sequencing data from 1013 tumor and matched germline prostate cancers (680 primary and 333 metastatic tumors) (Supplementary Table 1)1,2,4,7–9 that passed joint quality control parameters (Fig. 1a, Supplementary Fig. 1, Supplementary Table 2, Supplementary Note, Methods). Patient characteristics, including age at diagnosis, Gleason score, and metastatic site, are shown in Table 1 and Supplementary Table 3. The mean non- synonymous mutational load for primary and metastatic prostate cancers was 1.36 mutations/Mb and 2.93 mutations/Mb, respectively (Supplementary Fig. 2). As previously reported2, mutational load was significantly higher in metastatic tumors (p<0.001, estimated 1.43 mut/MB higher mutational load adjusted for differences in tumor sequencing depth and tumor purity) (Supplementary Fig. 2, Methods). Additionally, copy-number burden was significantly higher in metastatic tumor (p<0.001). In primary tumors, increased age and higher Gleason score was associated with higher mutation burden (p = 0.02, p < 0.001) and copy-number burden (p < 0.001, p < 0.001) (all adjusted for tumor purity and tumor sequencing depth) (Supplementary Fig. 2)10.
Figure 1.
Mutational significance in 1013 prostate cancers. (a) Uniform alignment, mutation calling, and significance analysis. (b) Recurrently mutated genes (n = 97). Genes are ordered by frequency, and mutations are stratified by mutation type and, for missense mutation, by recurrence. Recurrence is defined via cancerhotspots.org, OncoKB.org, and COSMIC; truncating mutations are defined as frameshift, nonsense, splice, nonstop. (c) Mutations in epigenetic regulators and chromatin remodelers are significantly enriched in ETS-negative tumors. p-values are calculated using a two-tailed Fisher’s exact test and shown for ETS fusions compared to all epigenetic mutations (including those co-occurring with SPOP and CUL3) and for ETS fusions compared to non-overlapping mutations in epigenetic modifiers only. (d) Cohort-wide view of mutations in epigenetic regulators and chromatin remodelers, which affect 20% of samples. Samples are shown from left to right (only the 202 tumors with alterations are shown, out of 1013), and gene alterations are color-coded by mutation type and, for missense mutations, by assumed driver status; mutations are assumed to be drivers if they have been previously reported and entered into COSMIC or annotated in OncoKB or variants of unknown significance (VUS).
Table 1.
Cohort characteristics. Baseline demographic and clinical data for the aggregate cohort, including age, Gleason score, metastatic site (if applicable).
| Primary Tumors (n=680) |
Gleason Score | 6 | 103 |
| 3+4 | 208 | ||
| 4+3 | 143 | ||
| 8–10 | 196 | ||
| Unknown | 30 | ||
| Age at Diagnosis | Median | 62 | |
| Unknown (n) | 80 | ||
| Metastatic Tumors (n=333) |
Metastatic Site | Bone | 80 |
| Lymph Node | 82 | ||
| Lung | 7 | ||
| Soft Tissue | 2 | ||
| Other | 26 | ||
| Unknown | 107 |
Mutational significance analysis of point mutations and short insertion/deletions using MutSig2CV11 and additional biological significance filters (Methods) identified 97 significantly recurrently mutated genes (SMGs) (Fig. 1a,b; Supplementary Fig. 3, 4a-c; Supplementary Table 4). As predicted by prior power analyses5, the majority of these new SMGs occurred in less than 5% of the overall cohort and could only be discovered in cohorts with over 900 samples (Supplementary Table 5). SMGs include well known prostate cancer genes1,2,4,7–9, such as AR, SPOP, FOXA1, TP53, and PTEN (Fig. 1b and Supplementary Table 4). We identified 70 SMGs previously implicated in cancer, but not previously reported as significantly altered in prostate cancer1,2,4,7–9 and an additional 9 SMGs not previously identified as recurrently altered in any cancer type (Fig. 1b and Supplementary Table 4)5,12.
We then integrated focal copy-number events and available ETS fusion data to stratify these findings by pathway and function and developed a categorized set of significantly mutated genes in prostate cancer. Through this approach, we identified 20% of prostate cancer samples with mutations, frequently truncating, in epigenetic modifiers or chromatin remodeling genes (Fig. 1c,d and Supplementary Table 6). Within this class of tumors, 5% had mutations in genes that encode SWI/SNF nucleosome remodeling complex members (Fig. 1d), including ARID1A (1.6%), ARID4A (1%), ARID2 (1.3%), SMARCA1 (1.1%) similar to observations made in other tumor types12,13. In primary tumors, mutations in epigenetic regulators and chromatin modifiers are significantly associated with higher Gleason score (10% Gleason 3+4, 22% Gleason 8–10, p=0.001 Fisher’s exact test). Furthermore, upon examination of the subset of our cohort for which ETS fusion status was available (n=765), we found that alterations in epigenetic regulators and chromatin remodelers were significantly more common in tumors that lack an ETS fusion (p=1e-04, Fisher’s exact test), and in tumors without previously known drivers (ETS fusion, IDH1/SPOP/CUL3, or FOXA1) (p = 0.007, Fisher’s exact test) (Fig. 1c,d and Supplementary Table 6).
Our analysis also identified recurrently mutated genes in the ubiquitin protease (USP) and ligase gene family, of which SPOP is a member, with mutations found in USP28 (1.4%), USP7 (1.2%), CUL3 (1.3%) (Fig. 2a). CUL3 encodes part of a cullin-RING-based (BTB-CUL3-RBX1) E3 ubiquitin ligase complex with SPOP14,15, and mutations may affect degradation of prostate cancer tumorigenesis regulators including AR, SRC-3, and TRIM2416,17. CUL3 mutations were primarily in a hotspot, p.Met299Arg, and were mutually exclusive with SPOP mutations (Fig. 2a), although this cohort size was not sufficiently powered to establish statistical significance. CUL3 mutant tumors also exhibited copy-number profiles similar to those of SPOP-mutant tumors, with losses at chromosomes 5q, 6q and 13 (Fig. 2b and Supplementary Fig. 5)18. To confirm this finding in an orthogonal cohort, we identified nine additional somatic CUL3 mutations in an independent cohort of advanced prostate cancers (1.3% in the MSK-IMPACT data19,20), including three p.Met299Arg mutations (Supplementary Fig. 6a).
Figure 2.
Ubiquitin and splicing pathways in prostate cancer. (a) Mutations in CUL3 are exclusively missense mutations, and five tumors show the recurrent p.Met299Arg mutation. CUL3 mutations are mutually exclusive with SPOP mutations, and including USP28, USP7, 12% of tumors harbor alterations in members of the ubiquitin pathway. (b) CUL3-mutant tumors show copy-number profile similar to those of SPOP-mutant tumors. Chromosomes are shown from left to right, samples from top to bottom. Regions of loss are indicated by shades of blue, and gains are indicated by shades of red. (c) Mutations in members of the splicing pathway are found in 4% of tumors, including oncogenic mutations in SF3B1 and U2AF1.
In addition, the splicing pathway was altered in 4% of prostate tumors (Fig. 2c), most notably through hotspot mutations in SF3B1 (1.1%) and U2AF1 (0.5%). Mutations in SF3B1 mostly clustered around the highly conserved HEAT repeats in the C-terminus (Fig. 2c), similar to other cancer types21,22. This alteration is thought to disrupt the recognition and binding of 3’ splice sites23.
We also identified SMGs in previously known prostate cancer pathways, including AR signaling, WNT/beta-catenin, PI3K, and RAS/MAPK. Within the AR/hormone signaling pathway, our analysis identified SPEN, which encodes a hormone inducible transcription repressor, mutated in 2.4% of this cohort, mostly through truncating mutations (Fig. 3a,b). The SPEN protein known to repress the estrogen receptor via NCOR2, by recruiting histone deacetylases and SRA, an RNA co-activator, interaction24,25. SPEN is activated via estrogen, and potentially other hormones25, and its overexpression is associated with response to tamoxifen in breast cancer25,26. SPEN mutations were significantly enriched in metastatic samples (q=0.008, Fisher’s exact test) and clonal (Fig 3a), suggestive of SPEN being a driver in advanced disease.
Figure 3.
SPEN mutations and WNT pathway alterations. (a) The majority of SPEN mutations are truncating and clonal in metastatic samples. (b) Oncoprint highlighting the distributions of SPEN mutations with alterations in members of the AR signaling. (c) Alterations in WNT/CTNNB1 pathway are found in 10% of tumors, primarily with loss of function mutations in APC and missense mutations in CTNNB1. (d) CTNNB1 mutations cluster primarily in hotspots in the N-terminal domain. (e) 3D structure of CTNNB1 showing novel mutations clustered around the CTNNB1-interacting domain of AXIN (highlighted in light gray).
The PI3-Kinase pathway was altered in 25% of our samples, primarily due to homozygous loss and truncating mutations in PTEN (16%). Our analysis identified a novel prostate cancer gene in the PI3-kinase pathway, PIK3R2 (1%), which, like PIK3R1, encodes a PI3K regulatory subunit27. One of the PIK3R2 mutations, p.Asp557Tyr, is paralogous to the known oncogenic p.Asp560Tyr mutation in PIK3R1 (Supplementary Fig. 6b), and was also found in our validation cohort.
Genomic alterations in the WNT/CTNNB1 pathway were found in 10% of the cohort (Fig. 3c and Supplementary Table 6). For CTNNB1, while the majority of mutations clustered in the N-terminal domain (Fig. 3d), three residues, including a novel p.Lys335Ile hotspot cluster around the CTNNB1 interacting domain of AXIN (Fig. 3e, CTNNB1 binding domain of AXIN highlighted in light gray). The RAS/RAF/MAPK pathway was altered in 5% of samples (Supplementary Table 6), including SMGs in KRAS and BRAF, mostly due to established hotspot mutations not previously enriched for significance in prostate cancer.
As previously reported, we observed a significant number of inactivating alterations in DNA repair genes (16% of samples, Supplementary Table 6). Novel prostate cancer specific SMGs in this pathway included MRE11A and PALB2. CDK12 was mutated primarily by truncating mutations (p<0.001, binomial test), as previously observed in ovarian cancer. Of note, CDK12 missense variants significantly clustered in the kinase domain (p<0.001, binomial test) (Supplementary Fig. 6c), suggesting a putative functional relevance. Furthermore, 15 of 31 CDK12 mutant tumors (as well as 27 of 56 samples in the validation cohort) harbored two mutations in the gene, suggestive of frequent biallelic inactivation of the gene. Broadly, these results expand on SMGs in known cancer pathways not previously implicated in prostate cancer, and further delineate the genomic heterogeneity of mutations in the long tail of this disease.
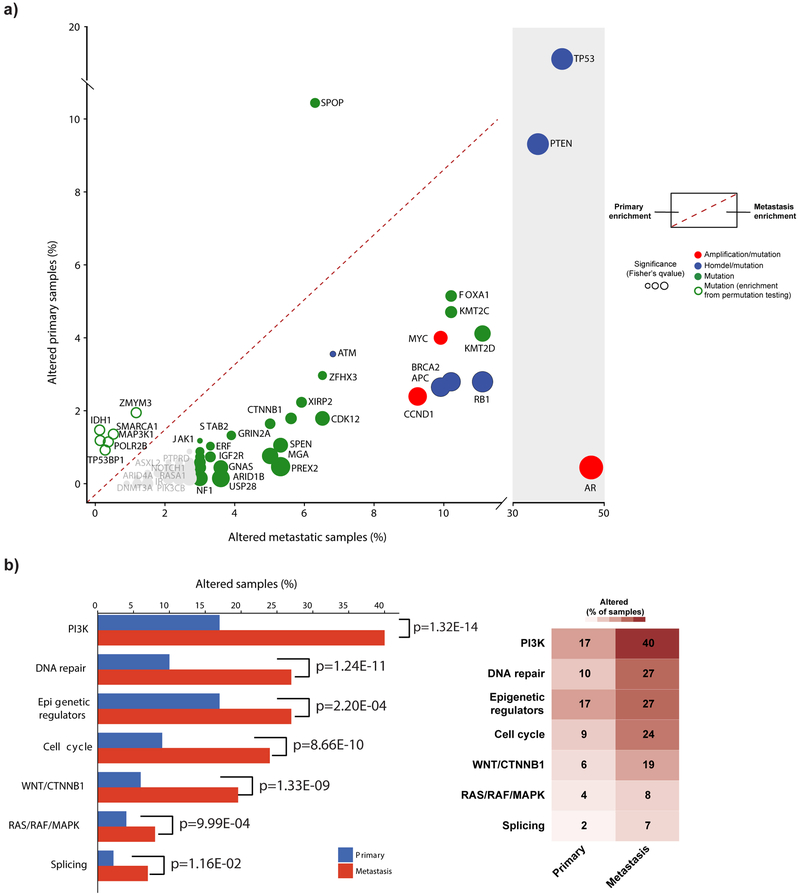
Finally, we conducted a systematic comparison of primary and metastatic tumors to identify which events are associated with advanced disease (Fig. 4a, Methods). Genes with enrichment in metastatic samples include TP53, AR, PTEN, RB1, FOXA1, APC, and BRCA2 (Fig. 4a). Alterations in epigenetic regulators, including KMT2C and KMT2D are also significantly enriched in metastatic tumors, and in aggregate define a genomic signature of high risk disease. Conversely, mutations in SPOP were significantly enriched in primary tumors (Fig. 4a). After correction for differences in mutational load, IDH1 and ZMYM3 mutations were also enriched in primary tumors (p=0.01, mutation rate-adjusted permutation test). At the pathway level, PI3K, DNA repair, Cell cycle, WNT/CTNNB1, and epigenetic regulators were significantly more frequently altered in metastatic compared to primary tumors (p<0.0001 Fisher’s exact test, Fig. 4b and Supplementary Table 7).
Figure 4.
Enrichment of genomic alterations in metastatic tumors. (a) Most genomic alterations are enriched in metastatic disease. Alteration percentages in metastatic samples (n=333) are shown on the x-axis, primary samples (n=680) on the y-axis. The significance of enrichment (two-sided Fisher’s test q-value or weighted permutation test) is shown by the size of the dots. Genes in bold have a significant enrichment of mutations using Fisher’s test and weighted permutation test correcting for mutation burden. (b) Pathway alteration frequencies in metastatic disease compared to primary disease. A sample was considered altered in a given pathway if at least a single gene in the pathway had a genomic alteration. p-values indicate the level of significance (two-sided Fisher’s exact test).
Within a given cancer type, the ability to redefine mutational significance with rapidly expanding sample sizes may identify new biologically and clinically relevant genes and pathways not previously appreciated. This study has leveraged this strategy to identify novel driver genes and pathways potentially implicated in the pathogenesis of prostate cancer. While many of the significantly altered genes and pathways are mutated at low frequencies, given the incidence of prostate cancer these alterations still impact large patient populations. In addition, whereas expanded analysis of primary indolent prostate cancer suggests near saturation for gene discovery28, this analysis, which includes more advanced cases, has revealed new biologically and clinically relevant events and creates an opportunity to prospectively assess a metastasis-associated genomic marker for clinical stratification in localized prostate cancer.
Combined statistical and biological significance analysis enabled a focused assessment of the SMGs identified herein, and efforts to functionally characterize this long tail of SMGs in prostate cancer may inform their relative phenotypic effects on oncogenicity, metastatic potential, and response characteristics to known or emerging prostate cancer therapeutics. Indeed, many of the genes identified through statistical analysis alone are of unknown function and suggest that even larger sample sizes paired with functional analysis will be necessary to discriminate which are relevant to prostate cancer oncogenesis. Subsequent studies that harmonize even larger prostate cancer molecular cohorts through uniform genomic analysis may also orthogonally validate these findings and further mitigate technical differences, such as stochastic effects of sequencing on variant detection, when analyzed in aggregate. Overall, our analysis demonstrates the utility of uniform genomic analysis in a single cancer type at a larger scale than previously reported, thereby redefining the molecular landscape of prostate cancer and providing rationale to revisit mutational significance in other cancer types as data generation scales by orders of magnitude.
ONLINE METHODS
Cohort collection and quality control
Samples were included in this study if tumor and matched germline WES raw sequencing data (BAM or Fastq files) were accessible and met downstream quality control characteristics (see Quality Control, below). These cohorts were identified through review of the literature and expert review (Supplementary Table 1). All cohorts had institutional review board approval for access from the original studies, listed in the citation. We obtained the WES BAM files from all samples. All samples underwent uniform alignment through the same version of the PICARD pipeline. Details of versions and parameters for all tasks within the PICARD pipeline are provided in the Supplementary Note. All tumor samples were required to have at least 50x mean target coverage and all paired normals were required to have at least 30x mean target coverage. Mean target coverage across the cohorts for tumors was 104.7x and for normals was 103.8x. ContEst was used to estimate the level of contamination with foreign DNA29. All samples had Contest scores lower than 5%, and the mean Contest value was 0.6%.
Clinical Data
All clinicopathological annotations were obtained from the original papers1,2,4,7–9. All primary tumors were treatment-naive; all metastatic tumors were castration resistant.
Variant Calling
To restrict the analysis to consider sites in the common pool of bases covered the bait sets used in the respective source projects, an intersected BED file was created using the bedtools intersect tool (Supplementary Table 8; Supplementary Fig. 7)
(http://bedtools.readthedocs.io/en/latest/content/tools/intersect.html). Single nucleotide variants (SNVs) were called with MuTect (version 1.1.6)30, using the intersected BED file. Unfiltered MuTect mutation call are located in Supplementary Table 9.
Artifacts introduced by DNA oxidation during sequencing or formalin fixation process were removed when appropriate31. Specifically regarding artifacts from formalin fixation, formalin fixation introduces multiple types of DNA damage including deamination, which converts cytosine to uracil and leads to downstream mispairing in PCR: C>T / G>A. Because deamination occurs prior to ligation of palindromic Illumina adapters, likely deamination artifacts will have a read orientation bias. We then use this read orientation to identify artifacts and calculate a Phred scaled Q-score for FFPE artifacts32.
To further reduce low confidence mutations with potential strand bias, we performed a Fisher’s exact test on each called mutation site in aggregate to identify variants occurring significantly more frequently in one read direction than in the other. A false discovery rate threshold, measured by Benjamini-Hochberg, of <0.0001 was used. In addition, all SNVs were required to have an allelic fraction of >= 0.01 to be called.
Insertions and deletions (indels) were called with Strelka (version 1.0.11)33. SNVs and indels were also filtered through a large panel of normals to extract additional poor calls. Any mutations in hotspot genes, defined by cancerhotspots.org34, initially called by MuTect but subsequently filtered out were rescued for the final variant list. When possible, we used ERG fusions calls defined as per the original source data2,4. For the 126 additional TCGA samples that were not part of the TCGA manuscript, we derived ERG fusion status via mRNA expression levels, inferring from samples with outlier expression of ERG likely contain an ERG fusion2.
Exome-wide copy-number ratios were inferred from coverage information using ReCapSeg (http://gatkforums.broadinstitute.org/gatk/categories/recapseg). For 303 prostate cancer samples that were analyzed by TCGA, we compared the segmented copy-number profiles generated by ReCapSeg to those from SNP6 data2 (Supplementary Fig. 8). We generated a scatter plot to compare the segment means of matched segments >200KB from the SNP6 and the ReCapSeg data, resulting in a pearson correlation of 0.92. Significant focal copy-number alterations were identified from segmented data using GISTIC 2.035. In addition, we called the allelic copy-number of well known prostate cancer genes, accounting for purity and ploidy, obtained from FACETS (version 0.5.10)36 (genes examined: TP53, APC, PTEN, RB1, BRCA2, CDKN1B, FANCA, ATM, AR). We performed manual review of copy number calls for selected oncogenes and tumor suppressors. All data is available for visualization and analysis in the cBioPortal for Cancer Genomics at http://www.cbioportal.org/37.
Mutation and Copy-Number Burden
Mutational burden was calculated as the number of mutations over the number of bases covered per sample and is reported as mutations per megabase. Copy-number burden was calculated as fraction of genome altered using copy-number segments with >|0.2|, as previously defined2. A multivariate linear regression adjusting for purity and coverage was used to evaluate the difference in mutational and copy-number burden in metastatic and primary tumors. Additional information is provided in the Supplementary Note.
Mutational Significance Analysis
All mutations that passed QC were analyzed using Mutsig2CV5 to identify significantly mutated genes (SMGs). Mutsig2CV integrates three separate significance algorithms: MutsigCV, MutsigFN, which looks at the functionality of a mutation in a gene, and MutsigCL, which looks at the clustering of mutations within the gene, specifically looking for hotspot mutations. Both MutsigFN and MutsigCL measure significance based on permutations. Significantly mutated genes (SMGs) fell within two different categories: 1) q values less than 0.1 and altered in at least 10 samples, 2) q values between 0.1 and 0.25, altered in 10 samples, and in known cancer genes5,38. Additionally, genes with low median allelic fraction (<0.1) were removed from the SMG list. Genes whose length was >1500aa (except for cancer genes5,38 or if the gene had a fraction of truncating variants larger than 50% of total mutations, indicating a putative tumor suppressor) were also removed from the SMG list. Genes with low expression in prostate cancer (median expression below bottom tertile TCGA RNAseq2) were also removed from the SMG list. Finally, genes with at least five oncogenic variants (according to OncoKB, http://oncokb.org), but were not previously included in the SMG list, were added to the SMG list39.
Comparison of genomic alterations between primary and metastatic tumors
Enrichment analysis of mutations and copy-number alterations observed in metastatic tumors compared to primary tumors was performed by tabulating the frequency of mutations or copy-number events observed in either metastatic or primary prostate cancer and performing a two-sided Fisher’s exact test on a set of biologically relevant cancer genes (n=650 genes)5,38. Multiple hypothesis test correction was performed using Benjamini-Hochberg method. To adjust for differences due to increased mutation load in metastatic tumors, we also performed a modified Fisher’s exact test; a permutation test where the probability of mutation in each sample is weighted by the mutation rate in that sample, and a simulation of 10,000 permutations performed with a two-sided p-value calculated as the proportion of those permutations with the observed or more extreme outcome. This directly corrects for differential observed mutation rates between primary and metastatic tumors, and represents the null hypothesis that mutations are equally likely to be found in primary vs. metastatic tumors, adjusting for differences in mutation rate. We were able to perform this mutational-rate based adjustment in genes where the only events were mutations. In cases where functional events included both gene mutations and copy-number changes (e.g. PTEN), we performed only a Fisher’s Exact test.
Clonality analysis
Clonality of mutations was estimated as cancer cell fraction (CCF)40, and implemented in the FACETS algorithm36. Additional information is provided in the Supplementary Note.
Statistical Analysis
Two-tailed Fisher’s exact test was used to assess enrichment of alterations in epigenetic regulators and chromatin remodelers in ETS-negative tumors. Association of mutation burden and fraction genome altered with metastasis status, age at diagnosis and Gleason score were evaluated using Mann Whitney Wilcoxon test and permutation test. All statistical analysis were performed using R version 3.3.1 (https://www.r-project.org).
Validation Datasets
To validate mutations detected in this study cohort, we queried cancer panel data from two sources: 1) Foundation Medicine, 204 patients with prostate cancer, as published41. Mutation calling for this cohort was obtained as previously described and data is available in phs001179. 2) Clinical sequencing data from 706 samples from Memorial Sloan Kettering patients (MSK-IMPACT)19,20. Mutation calling for this cohort was obtained as previously described and data is available from the paper or at cBioPortal.org.
Data Availability
BAM files are accessible as described for the original cohorts (Supplementary Table 1). In addition, all mutation calls and clinical annotation were deposited into cBioPortal for analysis and visualization: http://www.cbioportal.org/study?id=prad_p1000.
Code Availability
Bioinformatics tools used in the analysis of this data set are publicly available. Any that are not are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients for participating in this study. We also thank the Broad Cancer Genome Analysis and Data Sciences groups for analysis methodology and computational support.
FUNDING: This work was supported by the SU2C-PCF Prostate Cancer International Dream Team, Prostate Cancer Foundation Young Investigator Awards (B.S.T., C.P., N.S., E.M.V.A.), Prostate Cancer Foundation-V Foundation Challenge Award (E.M.V.A., P.S.N., J. S. d.B.), NIH K08CA188615 (E.M.V.A.), NCI P50-CA097186 and NCI P50-CA92629 SPOREs in Prostate Cancer, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, a National Cancer Institute Cancer Center Core Grant (P30-CA008748), and the Robertson Foundation (B.S.T, N.S.).
Footnotes
COMPETING FINANCIAL INTEREST
Dr. Van Allen is a consultant for Tango Therapeutics and Genome Medical.
REFERENCES
- 1.Robinson D et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomlins SA et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Barbieri CE et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet 44, 685–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran H et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med 22, 369–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baca SC et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hieronymus H et al. Copy number alteration burden predicts prostate cancer relapse. Proc. Natl. Acad. Sci. U. S. A 111, 11139–11144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theurillat J-PP et al. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346, 85–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng C et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc. Natl. Acad. Sci. U. S. A 110, 6997–7002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan W-C et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 20, 214–228 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Groner AC et al. TRIM24 Is an Oncogenic Transcriptional Activator in Prostate Cancer. Cancer Cell 29, 846–858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boysen G et al. SPOP mutation leads to genomic instability in prostate cancer. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abida W et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zehir A et al. Mutational Landscape of Metastatic Cancer Revealed from Prospective Clinical Sequencing of 10,000 Patients. Nat. Med (2017). doi: 10.1038/nm0817-1004c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolatshad H et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 29, 1092–1103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciriello G et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papaemmanuil E et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHugh CA et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev 15, 1140–1151 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Légaré S et al. The Estrogen Receptor Cofactor SPEN Functions as a Tumor Suppressor and Candidate Biomarker of Drug Responsiveness in Hormone-Dependent Breast Cancers. Cancer Res 75, 4351–4363 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Kuchay S et al. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol 15, 472–480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser M et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature (2017). doi: 10.1038/nature20788 [DOI] [PubMed] [Google Scholar]
- 29.Cibulskis K et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27, 2601–2602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costello M et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 41, e67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Allen EM et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med 20, 682–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Chang MT et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mermel CH et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen R & Seshan VE FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravarty D. OncoKB: A Precision Oncology Knowledge Base. Journal of Clinical Oncology Precision Oncology (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGranahan N et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med 7, 283ra54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmaier RJ et al. High-Throughput Genomic Profiling of Adult Solid Tumors Reveals Novel Insights into Cancer Pathogenesis. Cancer Res (2017). doi: 10.1158/0008-5472.CAN-16-2479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BAM files are accessible as described for the original cohorts (Supplementary Table 1). In addition, all mutation calls and clinical annotation were deposited into cBioPortal for analysis and visualization: http://www.cbioportal.org/study?id=prad_p1000.