Abstract
Purpose
Proton magnetic resonance spectroscopic imaging (MRSI) is a non-invasive modality capable of generating volumetric maps of in vivo tissue metabolism without the need for ionizing radiation or injected contrast agent. MRSI has been shown to be a viable imaging modality for studying several neuropathologies. However, a key hurdle in the routine clinical adoption of MRSI is the presence of spectral artifacts that can arise from a number of sources, possibly leading to false information.
Methods
A deep learning model was developed that was capable of identifying and filtering out poor quality spectra. The core of the model used a tiled convolutional neural network (CNN) that analyzed frequency-domain spectra to detect artifacts.
Results
When compared to a panel of MRS experts, our CNN achieved high sensitivity and specificity with an area under the curve of 0.95. A visualization scheme was implemented to better understand how the CNN made its judgement on single-voxel or multi-voxel MRSI, and the CNN was embedded into a pipeline capable of producing whole-brain spectroscopic MRI volumes in real-time.
Conclusion
The fully-automated method for assessment of spectral quality provides a valuable tool to support clinical MRSI or spectroscopic MRI studies for use in fields such as adaptive radiation therapy planning.
Keywords: MR spectroscopic imaging, spectroscopic MRI, machine learning, deep learning
Introduction
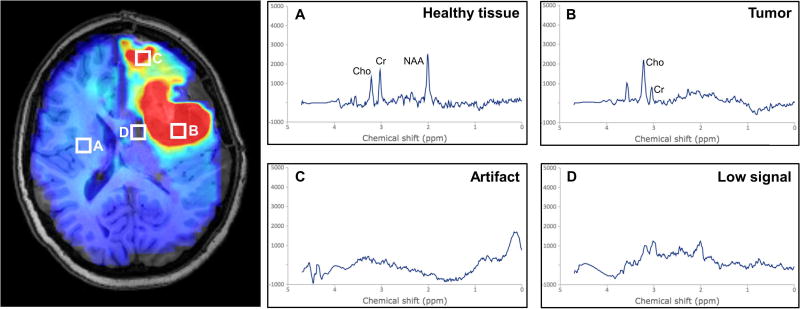
Proton magnetic resonance spectroscopy imaging (MRSI) is a quantitative imaging modality that measures endogenous metabolite concentrations in vivo (1,2). Recent advances in MRSI protocols, such as the development of the 3D echo-planar spectroscopic imaging (EPSI) sequence, and improved post-processing methods have enabled whole-brain acquisition with higher spatial resolutions, thereby improving utility for diagnostic and potentially radiotherapy treatment planning applications (3–6). Three metabolites of key importance in the evaluation of patients with glioblastoma, specifically choline (Cho), N-acetylaspartate (NAA), and the Cho/NAA ratio. The Cho/NAA ratio is widely used for depiction of tumor volumes and infiltration due to increased contrast as a result of the opposite changes of these metabolites in the tumor (3). A key challenge to the analysis and interpretation of tumor volume based on EPSI data is the presence of artifacts due to poor spectral quality (7). Artifacts arise from magnetic field inhomogeneities, subject movement, or improper water and lipid suppression, yielding reduced peak signal-to-noise ratios and distorted and broadened line shapes that lead to difficulties in quantification of the metabolite peaks (7,8). Visually, artifacts may appear as foci of hyper- or hypo-intense signal (Fig. 1), which can lead to false interpretation. For treatment planning purposes, it is especially important to obtain accurate volumetry of target pathology. Currently, confirmation of true metabolic abnormality requires manual review of spectra by experts. However, with several thousand spectra in an EPSI data set, manual review of whole-brain spectroscopy volumes is impractical. To adopt whole-brain EPSI data into the clinical workflow, it is therefore necessary to develop automated methods for assessment of spectral quality.
Figure 1.
Artifacts in MRSI arise due to several causes and can lead to false interpretation of pathology. (A) Healthy tissue shows a relatively low choline to N-acetylaspartate ratio (Cho/NAA), whereas (B) tumor shows an elevated ratio, appearing as hyperintense on a Cho/NAA map. Artifacts can arise in tissue boundaries and in areas with poor lipid or water suppression, and can result in either (C) hyperintense lesions or (D) dropout of signal.
Several approaches have been developed to filter poor quality spectra from MRSI datasets, including exclusion criteria based on peak linewidths (5), reliability testing (9), Cramer-Rao bounds (10), and machine-learning techniques such as random forest classifiers (11,12). In each of these algorithms, the classification of spectral quality is based on a collection of “engineered” features as input; these features are categorical or ordinal vectors that seek to summarize the information encoded in each spectrum. Some features are derived directly from the raw data, such as the magnitude of each point in the spectrum or statistical metrics of variability (e.g. kurtosis, skewness) (12), or derived from analysis of spectral fitting, such as singlet linewidths and Cramer-Rao bounds. Common to all these approaches is that the definition of features is required prior to performing any analysis. Thus, these features capture criteria that MRS experts explicitly believe are important to spectral quality, and machine learning algorithms built using these engineered features have shown promise as spectral quality filters (11–13). In contrast to systems that model expert beliefs explicitly, deep learning broadly defines a category of machine learning algorithms that can learn underlying features from raw data without the need for any a priori definition of such features, and have been able to shatter benchmarks in natural language processing, medical image segmentation, survival analysis, and identification of pathology in medical images (14–18). Deep convolutional neural networks (CNNs) in particular are well suited to analyzing waveforms similar to raw spectra (19,20), and have only recently been applied in the context of MR spectroscopy (21). CNNs consist of sequential layers of convolution, downsampling, and logistic regression that ultimately learn the underlying features of the data that lead to a desired endpoint, in this case whether or not a spectrum is suitable for analysis. Training a CNN involves updating the coefficients of the convolutional layers to reach the endpoint. However, designing the architecture of a CNN – e.g. the number of layers and the size of the convolution kernels – often relies on trial and error. A new framework to determine optimal architecture parameters is known as Bayesian optimization, which performs non-linear optimization without the need for defined derivatives (22).
In this work, a CNN was developed to learn whether or not a spectrum has sufficient quality to be used for clinical assessment. A state-of-the-art Bayesian optimization technique was used to automatically tune the network architecture and train the network on data from patients with glioblastoma. A framework is provided that enables users to visualize the rationale behind the CNN’s classification of individual spectra. Finally, a software pipeline was implemented that enables real-time filtering of whole-brain EPSI data.
Methods
Image Acquisition and Processing
EPSI data were available from a database of patients with glioblastoma previously enrolled in a phase II clinical trial (23) who received post-surgical scans. All scans were conducted in a 3T MRI scanner with a 32-channel head coil (Siemens Medical) and were obtained following surgical resection but prior to the start of radiation therapy and chemotherapy. Anatomic volumes obtained used a T1-weighted (T1w) magnetization-prepared rapid gradient echo pulse sequence (TR = 1900 ms, TE = 3.52 ms, 256 × 256 matrix, flip angle (FA) = 9°). A whole-brain 3D EPSI sequence (TR = 1551 ms, TE = 17.6 ms, FA = 71°, final matrix size of 64 × 64 × 32) was obtained during the same scanning session, as previously reported (3). Both sequences were obtained at a +15 degree tilt in the sagittal plane from the anterior commissure-posterior commissure line in order to capture the entire cerebrum while minimizing acquisition in the clivus, sinuses, and retro-orbital fat. An oblique saturation band was placed in the sagittal plane from the optic chiasm to the cerebellum to suppress signal from those regions. Image reconstruction and formation of metabolite images were carried out using the Metabolite Imaging and Data Analysis System (MIDAS) package (5,6). Briefly, this processing includes spatial reconstruction, frequency alignment, B0 field correction, co-registration of the T1w and EPSI volumes, registration of the T1w volume to an anatomic atlas, lipid suppression, spectral fitting, and normalization with internal water signal to produce relative concentrations of metabolites. Additionally, pre-filtering of data using algorithms built into MIDAS was applied to all data to replicate the workflow currently used in clinical studies. First, a mask based on an anatomic atlas was applied to exclude voxels outside of the brain, which drastically reduces the number of voxels to be analyzed. Voxels with a water linewidth > 18 Hz, as calculated from T2 decay, were removed prior to spectral fitting to save computation time. After fitting, voxels with a metabolite linewidth > 18 Hz were also removed; this step served as an initial filter to remove spectra known to be of poor quality prior to visual review. A representative volume contained 10,298 voxels after filtering, however, since the data is also interpolated and smoothed in each dimension during reconstruction it is estimated that approximately 1,280 independent spectra remain within the brain volume. Spectra were then randomly sampled from a grid with a skip factor of 2, including both regions of tumor and healthy tissue, and exported for analysis. A total of 8,894 spectra collected from 9 patients with glioblastoma were collected.
Data Labeling and Consensus
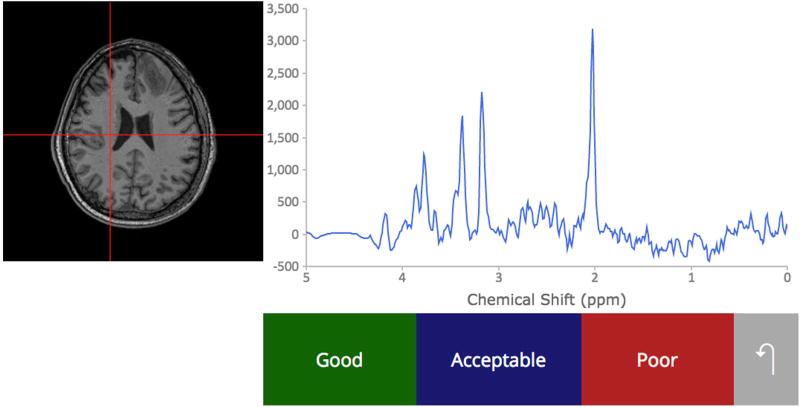
A custom web platform for multi-rater annotation of spectral quality was developed and used to label the exported spectra for training and testing the artifact detection network. Online Spectroscopic Classification and Review (OSCAR) was written in PHP – a language for server-side scripting of webpages – and enables multiple MRS experts to visualize and review spectra, as seen in Figure 2. Each expert is presented with a random subset of spectra and shown the chemical-shift spectrum along with a cross-haired T1-weighted MR image depicting the spectrum’s anatomic location. The user classifies the spectrum’s quality as “Good”, “Acceptable”, or “Poor”, based on their expert opinion.
Figure 2.
To collect ground truth data for machine learning classifiers based on spectral quality, we developed a web-based interface for MR experts to use.
Each spectrum was presented to three randomly chosen users from a pool of experts (authors AAM, JSC, BJS, HP, GV, PBB, and HS) so that an expert consensus could be measured; a total of 8,894 spectra were classified in triplicate. Consensus was based on a majority vote decision rule; if all three voted differently or if there was a large discrepancy in votes (e.g. 2 “Good” and 1 “Poor” or vice versa), the spectrum was discarded. Table 1 includes the results of labeling and consensus; a total of 427 spectra (4.8%) were discarded due to large discrepancy amongst experts. The consensus labels of “Good” and “Acceptable” were merged together into a singular “Good” class, yielding a class proportion of 72% “Good” and 28% “Poor”. The final set of 8,467 spectra were then randomly split 80:10:10 for training, validation, and testing subsets, with class proportions maintained.
Table 1.
Results of the consensus rating of spectral quality by a panel of MR spectroscopy experts. Each spectrum was reviewed independently by three experts, who gave it a label of “Good” (G), “Acceptable” (A), or “Poor” (P) quality. After discarding spectra with strong disagreement between reviewers, the final data set was split into 80:10:10 partitions of training, validation, and testing data sets.
| Reviewer Labels | Consensus Label | # of Spectra | % of Total | |||
|---|---|---|---|---|---|---|
| G | G | G | Good | 360 | 4.05 | |
| G | G | A | Good | 914 | 10.28 | |
|
| ||||||
| G | A | A | Acceptable | 1597 | 17.96 | |
| A | A | A | Acceptable | 1672 | 18.80 | |
| A | A | P | Acceptable | 1448 | 16.28 | |
|
| ||||||
| A | P | P | Poor | 1120 | 12.59 | |
| P | P | P | Poor | 1356 | 15.25 | |
|
| ||||||
| G | G | P | Discarded | 23 | 0.26 | |
| G | A | P | Discarded | 355 | 3.99 | |
| G | P | P | Discarded | 49 | 0.55 | |
|
| ||||||
| Total Spectra: | 8894 | |||||
| Spectra discarded: | 427 | 4.80 | ||||
| Final data set: | 8467 | 95.20 | ||||
|
| ||||||
| Training set: | 6767 | |||||
| Validation set: | 850 | |||||
| Testing set: | 850 | |||||
Network Architecture
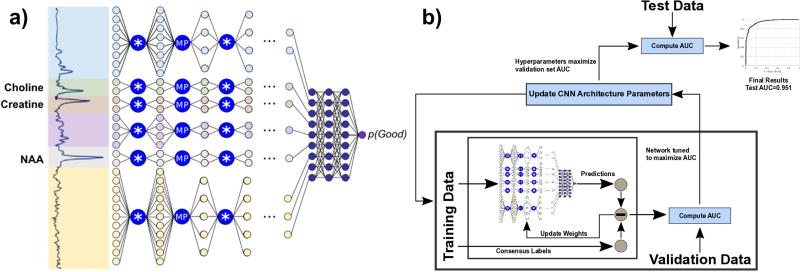
A CNN for spectral quality was developed using the TensorFlow (Google, Inc.) framework on a Windows workstation with two Titan × graphical processing units (GPUs). GPUs are highly optimized for parallel computations and enable rapid development of neural networks. A high-level overview schematic of the CNN is displayed in Figure 3a. It takes the 512-point real component of a spectrum as input. First, the spectrum is normalized to values between 0.0 and 1.0; for this step, a histogram of amplitudes of all spectra in the training data set was computed, and the 1st and 99th percentile values were used as the normalizing bounds. Using these bounds, all spectra are normalized to the same scale. The normalized spectrum is then split into six regions, called “tiles”. Three of these tiles were based on the known location of resonance peaks for the three largest metabolite peaks present: Cho (3.2 ppm), Cr (3.0 ppm), and NAA (2.0 ppm). Given the short TE that the data were acquired at, these three tiles are primarily composted of a Lorentzian-Gaussian singlet as opposed to the slowly-changing spectral baseline and smaller overlaid macromolecule singlets found in other tiles (8). Since these tiles have different shapes, the tiled architecture effectively enables six parallel networks to be trained synchronously. Each tile is passed through a series of convolutional and max-pooling layers. Layers 1 and 2 each perform 32 convolutions on the data, and layers 3–7 each perform 64 convolutions. Since CNNs operate best on low-amplitude data, outputs from each convolution are passed through a parametric rectified linear unit (PReLU), which restricts output range (24). Max-pooling downsamples the output of each convolution by a factor of two, reducing the size of the data going into the next layer. After these 7 layers, outputs from each of the six tiles are concatenated together and passed through two fully-connected layers of 128 nodes. Finally, logistic regression and softmax (25) operations are performed to yield a single scalar output within the range [0.0, 1.0]. The CNN is trained so that this output is the probability of the input spectrum being classified as “Good”.
Figure 3.
(a) A high-level overview of the convolutional neural network for spectral quality analysis. Input spectra are split into six tiles and passed through a series of convolution (*) and max-pooling (MP) layers, then concatenated and passed through fully connected layers to generate a scalar output of spectral quality. (b) Bayesian optimization is used to iteratively optimize architecture hyperparameters.
Training the CNN involves optimizing the coefficients of all the convolution kernels and fully-connected nodes so that the output matches the consensus of the training data generated in OSCAR. If the consensus output was “Good”, the spectrum is given a class label of 1, and if the consensus output was “Poor”, the spectrum is given a class label of 0. The cost function to be minimized by a first-order optimizer is defined as the cross-entropy error of the class label and the output probability of the CNN (26). Spectra are passed in batches of 250 spectra through the network and the cost function is computed; gradient backpropagation then adjusts the network coefficients. For each batch, spectral from the training data set are randomly sampled without replacement; an epoch is defined as one full pass-through of the training set. After each epoch, a receiver-operator characteristic (ROC) curve is computed on the validation (n=850) sets, and the area under the curve (AUC) is reported. Training continued through many epochs until the AUC of the validation set converges.
Bayesian Optimization
CNN architecture hyperparameters, e.g. the number of layers in the CNN and the size of the convolution kernels, can have a large impact on performance. These hyperparameters cannot be defined by linear mathematical functions and therefore do not have explicit derivatives to be used for gradient backpropagation. Because of this, they are outside the scope of neural network training and are often defined by manual trial and error, which can be both time-consuming and highly subjective. Bayesian optimization is a statistical technique that models the performance of an algorithm using Gaussian processes and iteratively seeks to optimize performance over the space of possible algorithm designs (22,27,28). CNNs can be treated as a complex mathematical function with some unknown underlying statistical model. Bayesian optimization assumes a series of Gaussian distributions can reproduce the underlying model and uses those to indirectly optimize the CNN; this technique has previously been successfully applied to tune architecture hyperparameters (18,22). A Bayesian optimization approach using the Spearmint framework is used in this work to tune the size of the convolution kernels and the dropout fraction of the fully connected layers (22). The variable to be minimized for Bayesian optimization is defined as the converged AUC of the validation data set as described in the previous section (Fig. 3b).
Gradient-weighted Class Activation Mapping
While deep learning algorithms such as CNNs extract predictive features from input data, these features do not have any semantic meaning, and it is not readily apparent how to interpret them in a manner similar to feature ranking by standard statistical techniques. A recent method known as gradient-weighted class activation mapping (GradCAM) attempts to visualize which components of a specific input contribute to that input’s classification. GradCAM performs a gradient backpropagation from the output score to the final layer of convolution, which contains the highest level features detected by the network (29). These can then be visualized as heat maps over the original input, highlighting which components contributed most to the CNN’s decision. While this cannot be generalized to all input, it enables insight into what the network is doing. We implemented GradCAM for spectral data in TensorFlow.
Results
Training and Validation
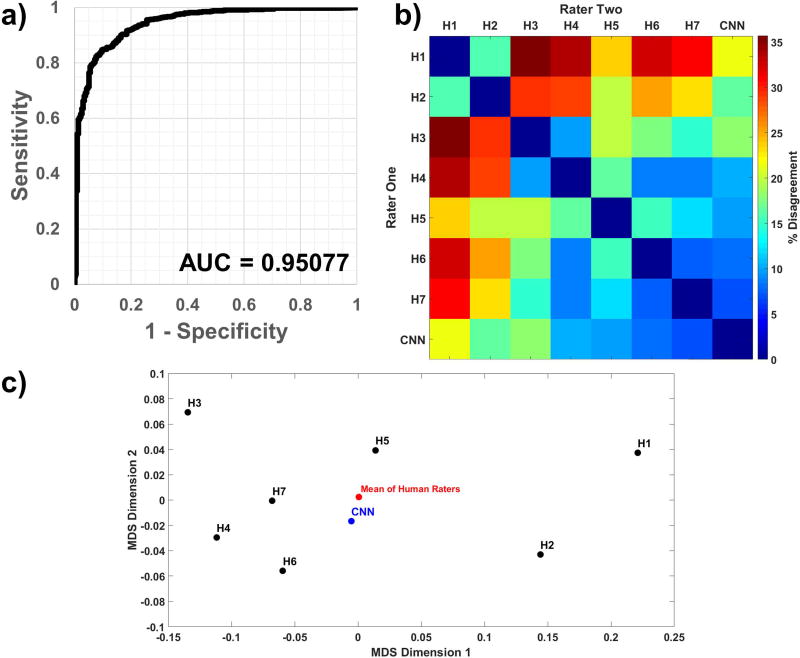
The CNN was trained over multiple epochs (complete passes through the training set) until the validation AUC achieved its peak value, an indication that additional training would result in overfitting. The Bayesian optimization framework was programmed to maximize the peak validation AUC, and this along with a 2×2 contingency table (30) was reported for each parameter set. The final tuned parameters reported by Spearmint were the CNN learning rate and convolution kernel size. After Bayesian optimization was complete, the unused test set (n=850 spectra) was run through inference and probabilities were compared with reviewer labels to generate the final ROC curve shown in Figure 4a, with an AUC of 0.95.
Figure 4.
(a) An unused test data set (n=850 spectra), with class proportions matching that of the full data set, was run through the CNN; comparing the output probabilities to ground truth resulted in a receiver-operator characteristic curve with an AUC of 0.951. The (b) dissimilarity heatmap and (c) a multidimensional scaling plot comparing sets of pairwise inter-rater agreement show that the CNN’s similarity with any given human rater is within the ranges of inter-human rater similarity.
To compare the classifications of the CNN against each human expert’s classifications, an inter-rater agreement analysis was performed. A dissimilarity matrix, representing the percent disagreement between each pair of observers was calculated (Figure 4b). Multidimensional scaling a technique that transforms these pairwise distances into a two-dimensional map to further visualize the relative agreement between pairs of raters, was performed (Figure 4c) for the seven human raters (H1–H7) and the CNN on the full data set. In a multidimensional scaling plot, points that are closer together represent higher agreement between the two raters’ decision on spectral quality. The geometric center of the human raters is also displayed on the plot. After training and tuning of the CNN were complete, it was run on the 427 spectra which the experts disagreed on, using a threshold of 0.7 on the CNN’s output probability for classification. A summary of the results is presented in Supporting Table S2; of note, the majority of these spectra were classified by the CNN as having good quality.
To evaluate the relative performance of the CNN against traditional machine learning with engineered features, a random forest was generated using the same input and ground truth data using the Statistics and Machine Learning Toolbox in MATLAB R2016b (The MathWorks, Inc.). Twenty features generated by MIDAS during the fitting process were used (Supporting Table S1), and the model achieved an AUC of 0.948 after optimizing random forest parameters. The ranking of features is shown in Supporting Figure S1.
Gradient-weighted Class Activation Mapping
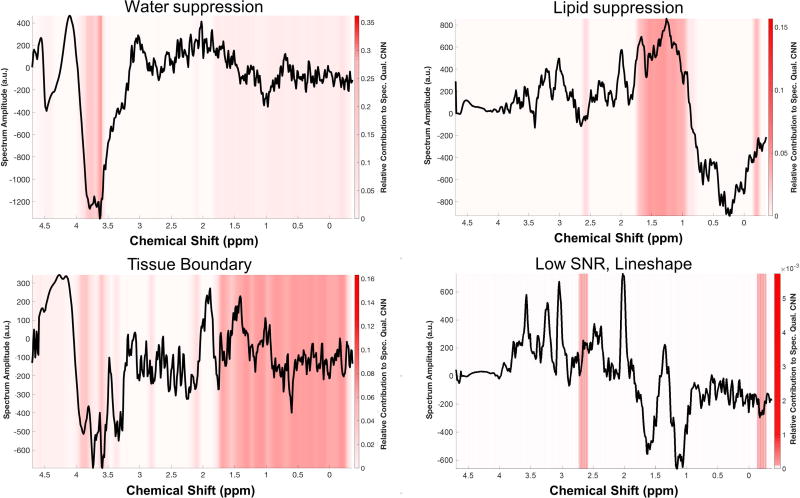
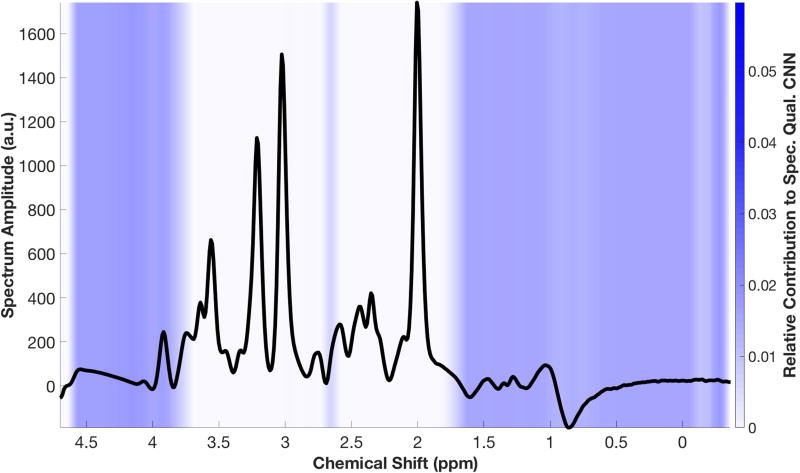
Representative spectra are shown in Figures 5–6 with overlaid heat maps generated by GradCAM, revealing regions of the spectrum that contributed most to the CNN’s classification decision. In Figure 5, four representative spectra are shown, each with different causes leading to poor quality per the opinion of two of the MRS experts. GradCAM highlights the regions of spectrum that correspond to those causes. To assess how the CNN makes a classification decision on good quality spectrum, an idealized “Good” spectrum is generated by taking the average of the amplitude all spectra classified as “Good” by the five readers (Fig. 6), which increases the signal-to-noise ratio and reduces baseline variations. When GradCAM is run on this idealized spectrum, the heat map reveals that it is the regions outside of the metabolite resonance frequencies, e.g. the region of lipid and water signal, that contribute most to the CNN’s classification.
Figure 5.
Four spectra representative of the various phenomena that lead to artifacts were analyzed using GradCAM, a technique which produces a heat map of which portions of a specific input spectrum contributed most to the CNN’s final decision. The results show that the CNN is focusing on appropriate regions for each of these scenarios. Of note, when there is low signal-to-noise ratio due to partial volume effects at a tissue boundary (bottom left), nearly the entire spectrum is detected.
Figure 6.
An idealized “Good” spectrum, created by averaging all spectra classified as “Good,” shows that the CNN focuses most on the regions outside of the metabolic peaks for decision making.
Whole-brain Pipeline
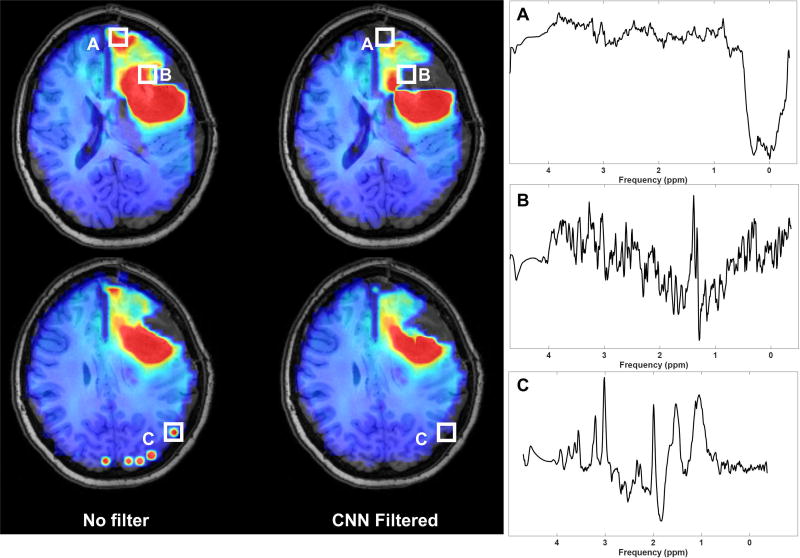
A pipeline was developed for filtering spectral artifacts on whole-brain volumes. Briefly, all spectra were exported to a binary format able to be read by TensorFlow and tagged with their voxel locations for reconstruction. The trained CNN was then loaded and inference is performed on all voxels in a whole-brain volume; a probability of “Good” quality is exported and reconstructed into a 3D volume representing probability. A threshold on this probability can then be selected and applied to all metabolite voxels, eliminating those below it from the map; a threshold of 0.70 was selected for the whole-brain images depicted here. Figure 7 shows a sample pre-radiation therapy Cho/NAA volume from a patient with a newly diagnosed left frontal glioblastoma following surgical resection of the tumor. Voxels with broad linewidths were already removed during data processing in MIDAS with an initial filter based on water and metabolite linewidths. Cho/NAA elevation is observed posterior and medial to the surgical cavity, suggesting residual tumor. Additionally, in the unfiltered Cho/NAA maps, there appears to be residual tumor in the anterior tip of the left frontal lobe. However, inspection of the spectra in these voxels reveals that the spectra are of insufficient quality to make an accurate assessment of pathology. Examples of spectra from eliminated voxels are shown, taken from regions of cellular necrosis, low signal-to-noise-ratio, and insufficient lipid suppression. This whole-brain pipeline is implemented using the Python version of TensorFlow, and is portable across multiple computer operating systems and hardware, including low and mid-tier GPUs. On a system with a low-end GPU (Nvidia GTX 1050 Ti), processing takes less than 2 minutes. Removing the GPU and running the pipeline only on a multicore CPU (Intel i7-6900K, 8 cores) also takes 2 minutes to process a whole-brain volume.
Figure 7.
The spectral quality CNN can be applied to whole-brain volumes in real-time to assist clinicians in making accurate decisions based on MRS, such as the Cho/NAA volume. In a pre-treatment assessment, the CNN filters out voxels in the necrotic tumor core, anterior frontal lobe, and multiple voxels in the occipital lobe which have poor quality.
Discussion
The CNN-based spectral quality filter developed in this study was found to perform well for detection of inadequate quality spectra, as labeled by a consensus decision of MRS experts, with an AUC of 0.951, indicating the network has been tuned for both high sensitivity and specificity. Furthermore, the network is wrapped into a framework that enables rapid deployment of the filter into the clinical research workflow, and can be applied in under 2 minutes to high resolution EPSI data with whole brain coverage. The accuracy achieved is similar to that reported in previous studies using machine learning for MR spectral quality analysis, such as those utilizing random forests with engineered spectral features (11,13,31). It is difficult to compare results across studies, due to variation in study design: how data was collected, the biases of the raters generating ground truth, and which parameters were chosen as features. To enable direct comparison of performance between the CNN and a machine learning algorithm with engineered features, a random forest was built on the same training and testing data collected in OSCAR. While the CNN demonstrates slightly better performance, a key advantage of the deep learning approach over the random forest is that does not require any engineered features generated by spectral fitting. As a result, it is independent of specific features derived from fitting algorithms and is compatible with a broader set of pipelines and workflows. Of note, the spectra in this work do undergo preliminary linewidth filtering in MIDAS prior to being processed by the CNN, which reduced the spectra needed to undergo spectral fitting and excluded spectra with broadened metabolite linewidths known to be of poor quality. This decision was made to use the status quo of spectral quality filtering as a starting point and to develop an algorithm that makes additional improvements to it. In future workflows, applying filtering prior to spectral fitting could reduce the computation time for spectral fitting, a time-intensive process, especially since poor quality spectra may require considerably more processing during fitting, due to their abnormal shapes.
While artifacts can occur anywhere in the tissue parenchyma, they are more common at air/tissue boundaries due to local magnetic field inhomogeneities and near the periphery due to partial volume effects with strong lipid signals in scalp. It is well known that magnetic field inhomogeneity occurs in the inferior frontal lobe, anterior temporal lobe, and superior to the mastoid air spaces due to magnetic susceptibility differences between air and tissue. In patients who have undergone surgery, additional artifacts in EPSI data can occur due to magnetic susceptibility from craniotomy staples and possibly from hemorrhage. In this first iteration of the spectral quality CNN, this information is not taken into account insofar as it is not encoded into the spectrum itself. Future work could include the registration of metabolite or metabolite ratio (e.g. Cho/NAA) maps onto a common anatomical atlas and input the anatomical location of each spectrum into the CNN, either in terms of absolute location or relative distance from the nearest tissue boundary (i.e. ventricle, dura).
Additionally, a label collection scheme was implemented to take into account the inter-reader variability of spectral evaluation by multiple MRS experts. This is a key step in improving the generalizability of our algorithm, since each reader is independently looking for particular spectral patterns based on his or her own expertise. As shown in Table 1, there often was disagreement amongst the experts, including approximately 5% of data with complete discordance. As such, the performance of classifiers reflects subjectivity of the human raters that define the ground truth. Multi-center and multi-expert analyses have previously been conducted for brain tumor classification, and have shown the necessity of such data in establishing quality control norms (32). Inter-rater reliability was assessed by computing the disagreement on spectra for every pair of raters, including the seven MRS experts and the CNN. The multidimensional scaling plot in Figure 4c shows that the distance between the CNN and any of the human raters is not more extreme than the distance between any pair of human reviewers, suggesting that the CNN algorithm agrees with humans about as well as humans agree with each other. Furthermore, the CNN is close to the geometric center of the human raters, which is in accordance with the methodology used to train the CNN; the spatial distance can be attributed to consensus scheme used during data labeling, which deviates from the mean of the three user labels. The OSCAR platform designed for this work is readily available to be used for future experiments requiring multi-user input. For this work, MRS experts were asked to judge the quality of spectra only based on the metabolite spectrum and its location in a 2D slice. In reality, experts use additional information when assessing MRS data: the strength of the water signal; comparison of the fitted and unfitted spectra; and the pathology of the spectrum’s location, e.g. from tumor or healthy tissue. These will be added to OSCAR to supplement data collected in future studies.
Another challenge in developing algorithms for spectral quality filtering is the low percentage of poor quality voxels present in a whole-brain volume compared to good quality voxels, which yields an imbalance in class proportions and consequently can hinder algorithm performance (13). In the dataset collected in this work, 72% of spectra were of good quality and 28% were of poor quality, which is similar to proportions (65 – 84% acceptable spectra) observed in other works (11,13,31). To assess whether balanced class proportions would affect CNN performance, a random minority oversampling scheme was implemented, wherein data from the minority class (poor spectra) are randomly sampled multiple times to artificially increase the number of samples. A new CNN was trained using the same network architecture on this oversampled dataset, with 5991 good quality and 5991 poor quality, and tuned using Bayesian optimization as described in Methods. The resulting AUC was slightly improved compared to the original CNN, at 0.960 (Supporting Figure S2). This suggests that the CNN is robust to the class imbalance of the ground truth dataset, but that a more balanced data set could potentially improve outcomes. Of note, the imbalance is relatively small, being only a factor of ~2.6, whereas in other domains where deep learning has been applied the imbalance can be several orders of magnitude (33).
Based on the results of GradCAM, the user can glean some insight into how the network arrives at a decision. An ideal “good” spectrum is simulated by averaging all spectra labeled as “Good”, creating a high signal-to-noise ratio spectrum with all peaks visible (Fig. 6). The GradCAM evaluation of this simulated spectrum suggests that the network appears to focus on regions outside of the main metabolite peaks, specifically on unsuppressed lipids, unsuppressed water, and the overall baseline waveform. Because all spectra evaluated by spectroscopy experts were already passed through a linewidth filter, they were known to have metabolite peaks with narrow linewidths and low Cramer-Rao bounds. As such, the artifacts are arising from other aspects of the spectrum, and the CNN is focusing on these other spectral features. GradCAM visualization also provides a benefit over traditional machine learning methods, in that it can localize the band(s) of an individual spectrum that explain a CNN classification result. In contrast, interpretation of random forests can only be performed on a “population” level, explaining what features are critical in performing classification in the entire dataset, and cannot provide insights into the classification of individual spectra.
In order to incorporate the spectral quality CNN into a clinical research workflow, a Python application programming interface to query the CNN from other software was developed. The CNN model can perform inference of an entire whole-brain volume on an affordable GPU or on a multicore CPU, and has been deployed to an in-house spectroscopic MRI web app so that it can be easily applied to clinical studies. While training a CNN is computationally intensive and requires powerful GPU hardware, applying the CNN is much less intensive and can be done on commodity hardware, either a low-end GPU or a high-end CPU like the configurations described in the Results section.
Conclusion
A deep learning algorithm was developed to automatically detect poor quality spectra in whole-brain EPSI data that otherwise would lead to incorrect classification of voxel pathology. This approach achieved high accuracy when compared to a consensus decision from a panel of MRS experts, and achieved comparable accuracy to classifiers based on engineered features developed in this work and by others. The key advantage of the CNN developed here is its ability to operate directly on spectra, enabling quality filtering independent of the fitting algorithm and requiring no engineered features. The CNN identifies poor quality spectra and artifacts with a high degree of sensitivity and specificity, and has been integrated into a whole-brain spectroscopy processing pipeline. Frameworks such as convolutional neural networks are well suited to the high dimensionality of EPSI data and can extract information that contributes to spectra quality from the spectral waveform beyond what is extracted using engineered features. To provide feedback to spectroscopy experts, we also implemented a GradCAM-based visualization approach that localizes artifacts in spectra, and that can provide a rationale for classification decisions to the user on individual spectra. In this first iteration, the CNN was trained to identify spectral quality based only on spectral waveforms with no regard to other factors that play a role in experts’ assessments of quality, such as location of the voxel or any known pathologies in the brain. Future work will focus on collecting training data and developing a CNN that can take these into account, by incorporating an anatomic atlas and assessing local pathology. Ultimately, the implementation of an automated spectral quality filter will mitigate errors due to incorrect classification of pathology, and assist in pushing whole brain 3D EPSI technology into clinical decision-making.
Supplementary Material
Supporting Table S1. Features generated by MIDAS used for building a random forest on the same data as used for the CNN.
Supporting Table S2. CNN classifications of the discordant data set, which was discarded prior to training the network. The same threshold of 0.7 used in the whole-brain pipeline is used here.
Supporting Figure S1. The MIDAS features used in the random forest classifier are ranked by their relative strengths in determining the forest’s classification decision. The random forest achieved an area-under-the-curve (AUC) of 0.94 when comparing with the consensus decision of an MRS expert panel.
Supporting Figure S2. An oversampled data set with 50% good and 50% poor quality spectra was generated to assess the effect of class imbalance on CNN performance. A new CNN was trained on this balanced data set and yielded a AUC of 0.960, similar to the CNN trained on the original imbalanced data set.
Acknowledgments
The work described herein was funded by the following grants from the National Institutes of Health: U01CA172027, R21NS100244, and F30CA206291. The authors would like to thank the following individuals for their support in collecting and analyzing data: Sulaiman Sheriff, Michael K. Larche, Robert L. Smith III, Samira Yeboah, and Pooya Mobadersany.
References
- 1.Law M, Cha S, Knopp EA, Johnson G, Arnett J, Litt AW. High-grade gliomas and solitary metastases: differentiation by using perfusion and proton spectroscopic MR imaging. Radiology. 2002;222(3):715–721. doi: 10.1148/radiol.2223010558. [DOI] [PubMed] [Google Scholar]
- 2.Soares DP, Law M. Magnetic resonance spectroscopy of the brain: review of metabolites and clinical applications. Clin Radiol. 2009;64(1):12–21. doi: 10.1016/j.crad.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Cordova JS, Shu H-KG, Liang Z, Gurbani SS, Cooper LAD, Holder CA, Olson JJ, Kairdolf B, Schreibmann E, Neill SG, Hadjipanayis CG, Shim H. Whole-brain spectroscopic MRI biomarkers identify infiltrating margins in glioblastoma patients. Neuro Oncol. 2016;18(8):1180–1189. doi: 10.1093/neuonc/now036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Jin H, Lu Y, Oh J, Chang S, Nelson SJ. Identification of MRI and 1H MRSI parameters that may predict survival for patients with malignant gliomas. NMR Biomed. 2004;17(1):10–20. doi: 10.1002/nbm.858. [DOI] [PubMed] [Google Scholar]
- 5.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn Reson Med. 2009;61(3):548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabati M, Sheriff S, Gu M, Wei J, Zhu H, Barker PB, Spielman DM, Alger JR, Maudsley AA. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn Reson Med. 2015;74(5):1209–1220. doi: 10.1002/mrm.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreis R. Issues of spectral quality in clinical 1H magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004;17(6):361–381. doi: 10.1002/nbm.891. [DOI] [PubMed] [Google Scholar]
- 8.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40(6):822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 9.Slotboom J, Nirkko A, Brekenfeld C, Ormondt Dv. Reliability testing of in vivo magnetic resonance spectroscopy (MRS) signals and signal artifact reduction by order statistic filtering. Meas Sci Technol. 2009;20(10):104030. [Google Scholar]
- 10.Jiru F, Skoch A, Klose U, Grodd W, Hajek M. Error images for spectroscopic imaging by LCModel using Cramer-Rao bounds. MAGMA. 2006;19(1):1–14. doi: 10.1007/s10334-005-0018-7. [DOI] [PubMed] [Google Scholar]
- 11.Pedrosa de Barros N, McKinley R, Knecht U, Wiest R, Slotboom J. Automatic quality control in clinical (1)H MRSI of brain cancer. NMR Biomed. 2016;29(5):563–575. doi: 10.1002/nbm.3470. [DOI] [PubMed] [Google Scholar]
- 12.Menze BH, Kelm BM, Weber MA, Bachert P, Hamprecht FA. Mimicking the human expert: pattern recognition for an automated assessment of data quality in MR spectroscopic images. Magn Reson Med. 2008;59(6):1457–1466. doi: 10.1002/mrm.21519. [DOI] [PubMed] [Google Scholar]
- 13.Kyathanahally SP, Mocioiu V, Pedrosa de Barros N, Slotboom J, Wright AJ, Julià-Sapé M, Arus C, Kreis R. Quality of clinical brain tumor MR spectra judged by humans and machine learning tools. Magn Reson Med. 2017 doi: 10.1002/mrm.26948. [DOI] [PubMed] [Google Scholar]
- 14.Carneiro G, Nascimento JC, Freitas A. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans Image Process. 2012;21(3):968–982. doi: 10.1109/TIP.2011.2169273. [DOI] [PubMed] [Google Scholar]
- 15.Dawes TJW, de Marvao A, Shi W, Fletcher T, Watson GMJ, Wharton J, Rhodes CJ, Howard LSGE, Gibbs JSR, Rueckert D, Cook SA, Wilkins MR, O'Regan DP. Machine Learning of Three-dimensional Right Ventricular Motion Enables Outcome Prediction in Pulmonary Hypertension: A Cardiac MR Imaging Study. Radiology. 2017;283(2):381–390. doi: 10.1148/radiol.2016161315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakhani P, Sundaram B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 17.Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate MR segmentation. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):254–261. doi: 10.1007/978-3-642-40763-5_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousefi S, Amrollahi F, Amgad M, Dong C, Lewis JE, Song C, Gutman DA, Halani SH, Velazquez Vega JE, Brat DJ, Cooper LAD. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Scientific Reports. 2017;7(1):11707. doi: 10.1038/s41598-017-11817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sainath TN, Weiss RJ, Senior A, Wilson KW, Vinyals O. Learning the speech front-end with raw waveform CLDNNs; Sixteenth Annual Conference of the International Speech Communication Association: ee.columbia.edu; 2015. [Google Scholar]
- 20.Wei Dai CDSQJLSD. Very Deep Convolutional Neural Networks for Raw Waveforms. 2016 [Google Scholar]
- 21.Kyathanahally SP, Doering A, Kreis R. Ghostbusters for MRS: Automatic Detection of Ghosting Artifacts using Deep Learning. Honolulu, HI: 2017. p Abstract #5479. [Google Scholar]
- 22.Snoek J, Larochelle H, Adams RP. Practical Bayesian Optimization of Machine Learning Algorithms. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in Neural Information Processing Systems 25. Curran Associates, Inc; 2012. pp. 2951–2959. [Google Scholar]
- 23.Cordova JS, Gurbani SS, Holder CA, Olson JJ, Schreibmann E, Shi R, Guo Y, Shu H-KG, Shim H, Hadjipanayis CG. Semi-Automated Volumetric and Morphological Assessment of Glioblastoma Resection with Fluorescence-Guided Surgery. Mol Imaging Biol. 2016;18(3):454–462. doi: 10.1007/s11307-015-0900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He K, Zhang X, Ren S, Sun J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. 2015 [Google Scholar]
- 25.Bishop C. In: Pattern Recognition and Machine Learning. Jordan MIN, Robert, Schoelkopf, Bernhard, editors. New York, NY: Springer-Verlag; 2006. p. 738. [Google Scholar]
- 26.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization; 3rd International Conference for Learning Representaitons; San Diego, CA, USA. 2015. [Google Scholar]
- 27.Bergstra JS, Bardenet R, Bengio Y, Kégl B. Algorithms for Hyper-Parameter Optimization. In: Shawe-Taylor J, Zemel RS, Bartlett PL, Pereira F, Weinberger KQ, editors. Adv Neur. Curran Associates, Inc; 2011. pp. 2546–2554. [Google Scholar]
- 28.Jones DR. A Taxonomy of Global Optimization Methods Based on Response Surfaces. J Global Optimiz. 2001;21(4):345–383. [Google Scholar]
- 29.Selaraju R, Das A, Vedantam R, Cogswell M, Parikh D, Batra D. Grad-CAM: Why did you say that? Visual Explanations from Deep Networks via Gradient-based Localization. CoRR. 2016 abs/1610.02391. [Google Scholar]
- 30.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27(8):861–874. [Google Scholar]
- 31.Pedrosa de Barros N, McKinley R, Wiest R, Slotboom J. Improving labeling efficiency in automatic quality control of MRSI data. Magn Reson Med. 2017 doi: 10.1002/mrm.26618. [DOI] [PubMed] [Google Scholar]
- 32.García-Gómez JM, Luts J, Julià-Sapé M, Krooshof P, Tortajada S, Robledo JV, Melssen W, Fuster-García E, Olier I, Postma G, Monleón D, Moreno-Torres À, Pujol J, Candiota A-P, Martínez-Bisbal MC, Suykens J, Buydens L, Celda B, Van Huffel S, Arús C, Robles M. Multiproject–multicenter evaluation of automatic brain tumor classification by magnetic resonance spectroscopy. Magn Reson Mater Phy. 2008;22(1):5. doi: 10.1007/s10334-008-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buda M, Maki A, Mazurowski MA. A systematic study of the class imbalance problem in convolutional neural networks. arXiv preprint arXiv:171005381. 2017 doi: 10.1016/j.neunet.2018.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table S1. Features generated by MIDAS used for building a random forest on the same data as used for the CNN.
Supporting Table S2. CNN classifications of the discordant data set, which was discarded prior to training the network. The same threshold of 0.7 used in the whole-brain pipeline is used here.
Supporting Figure S1. The MIDAS features used in the random forest classifier are ranked by their relative strengths in determining the forest’s classification decision. The random forest achieved an area-under-the-curve (AUC) of 0.94 when comparing with the consensus decision of an MRS expert panel.
Supporting Figure S2. An oversampled data set with 50% good and 50% poor quality spectra was generated to assess the effect of class imbalance on CNN performance. A new CNN was trained on this balanced data set and yielded a AUC of 0.960, similar to the CNN trained on the original imbalanced data set.