Abstract
Although there is a growing interest in using polymer lipid-nanodiscs, polymer charge pose limitations for studies on membrane proteins. Here, we demonstrate the functional reconstitution of large soluble-domain containing positively-charged ~57-kDa cytochrome-P450 and negatively-charged ~16-kDa cytochrome-b5 in lipid-nanodiscs, and the role of polymer charge for high-resolution studies on membrane proteins.
Graphical Abstract
Membrane proteins play a plethora of important roles in cellular function. Despite the need for high-resolution structures to fully understand the function, poor solubility and stability of membrane proteins continue to pose challenges.1 The development of lipid-nanodiscs as a membrane mimetic has enabled structural studies of membrane proteins in their near-native lipid bilayer environment.3 Nanodiscs are discoidal shaped and contain a planar lipid bilayer surrounded by an amphiphilic molecular belt.4 Different types of amphiphilic belts such as membrane scaffold protein (MSP),3, 5, 6 peptides7, 8 and polymers,9, 10 have been used to form lipid-nanodiscs. In recent years, polymer based lipid-nanodiscs have been developed and their advantages over the well-established protein based nanodiscs have been demonstrated.11–14 Polymer based lipid-nanodiscs have been shown to not only form nanodiscs with varying lipid compositions, but have also been used to directly extract membrane proteins from their native cellular membrane environment.15, 16 Styrene maleic anhydride/acid (SMA) polymers were the first amphiphilic polymers shown to form nanodiscs (known as SMALP),9, 17 and since their introduction several other polymers 10, 18 have also been synthesized to improve the nanodiscs properties. Low molecular weight SMA derivatives such as styrene maleic acid – ethanol amine (SMA-EA)19 and styrene maleimide quaternary ammonium (SMA-QA)2 were recently developed to improve the size control and pH stability of nanodiscs. Although polymer nanodiscs are increasingly used for studies on membrane proteins, they have been limited to polytopic transmembrane (TM) proteins.20 On the other hand, unlike the TM proteins, large soluble domain containing membrane proteins (such as single-pass or double-pass TM proteins) exhibit heterogeneous aggregation due to differences between soluble and TM domains. Interestingly, these are the proteins that continue to pose major challenges for structural studies; significant difference in the time scales of motions between the soluble and TM domains makes it very difficult to crystallize them and, functional reconstitution remains to be major a challenge.21 Therefore, it is important to demonstrate the capabilities of polymer nanodiscs to reconstitute such membrane proteins to broaden the applications of polymer nanodiscs.
In this study, we chose the ~57-kDa cytochrome P450 (CytP450) and ~16-kDa cytochrome-b5 (Cyt b5) to demonstrate the feasibility of using polymer nanodiscs because they contain a single TM domain, a large soluble domain, and exhibit their function in a lipid membrane environment.22, 23 The additional complexities due to highly cationic CytP450 and anionic Cyt b5 are also utilized to understand the role of polymer charge on their functional reconstitution. CytP450 and its redox partners are involved in the oxidation of a variety of substrates.24 The active site of CytP450 contains a heme moiety axially coordinated to a cysteine, and this coordination plays a major role in its function.25 When functional CytP450 is reduced and binds to carbon monoxide (CO), it displays a characteristic Soret absorption at 450 nm. CytP450 has an inactive form denoted as P420, which causes a blue shifted Soret absorption peak at 420 nm. Previous studies on using peptide or MSP nanodiscs showed the reconstituted CytP450 to be monomeric, functionally active and more stable as compared to the detergent solubilized protein.7, 26, 27 These studies enabled biophysical and structural characterization of CytP450, whereas in the absence of the nanodiscs the heterogeneous nature of CytP450 makes it difficult.28, 29 In this study, we reconstituted the functional cytochrome P450 2B4 (CYP2B4) in different polymer nanodiscs (negatively charged SMA-EA and SMALP, and positively charged SMA-QA), and demonstrated that CytP450 stability is increased when reconstituted in SMA-QA polymer nanodiscs as compared to that in a lipid-free solution (but in detergent). SMA-EA and SMA polymer nanodiscs were found to inactivate CytP450 during reconstitution. The inactivation of CytP450 was found to be dominated by charge-charge interaction between the negatively charged polymer and cationic CytP450. SMA-EA nanodiscs were found to be effective for Cyt b5 reconstitution, whereas SMA nanodiscs exhibited a strong interaction between Cyt b5 and the polymer belt. On the other hand, SMA-QA nanodiscs successfully reconstituted Cyt b5 only in the presence of a high ionic strength medium. These results demonstrate that the innate chemical properties of the chosen polymer to form nanodiscs can have a large effect on successful functional protein reconstitution.
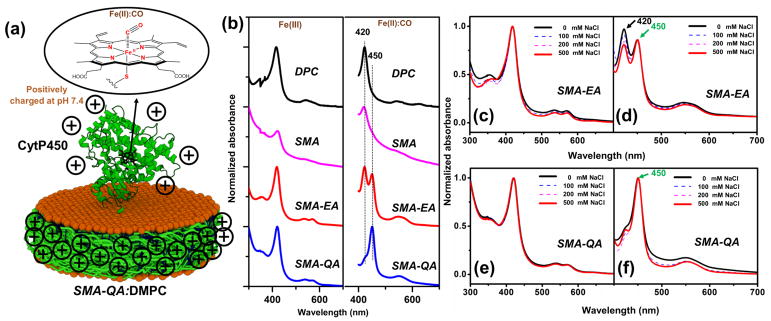
The nanodiscs used in this study were prepared using DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) and a SMA, SMA-EA or SMA-QA (Figure 1a and 1b) as explained in the supporting information. SMA was commercially obtained, whereas both SMA-EA and SMA-QA were synthesized and characterized as previously described.2, 19 The dynamic light scattering (DLS) (Fig.1c–e) and size exclusion chromatography (SEC) (Fig.1f–h) profiles showed that all polymer nanodiscs exhibited a similar size (~10–15 nm diameter). More details on the stability of these polymer nanodiscs can be found in the literature.2, 9, 19 Following SEC, the nanodiscs solutions were incubated with CytP450 at room temperature for 12 hrs. The resulting CytP450-reconstituted nanodiscs were reduced using sodium dithionite and the solution was bubbled with carbon monoxide (Figure 2). Figure 2b shows the absorption spectra of ferric (Fe3+) CytP450 and CO bound ferrous (Fe2+) CytP450. The CytP450 reconstituted in SMALP absorbed at 420 nm showing that the protein is in its inactive form, similar to that observed from DPC micelles (see the experimental data in Figure S1 in the supporting information).30 CytP450 reconstituted in SMA-EA nanodiscs displayed two absorbance peaks at 420 nm and 450 nm indicating the partial inactivation. CytP450 incubated with SMA-QA nanodiscs exhibited a peak maximum at 450 nm demonstrating that the protein is reconstituted in its active form. The overall percentage of inactive CytP450 in SMA-EA nanodiscs decreased when CytP450 was reconstituted in SMA-EA nanodiscs in the presence of 500 mM NaCl (Figure 2c–d). This observation suggests that the positively charged CytP450 (+6.9 at pH 7.4)31 interacts with the negatively charged SMA or SMA-EA via electrostatic charge-charge interactions. The high affinity of positively-charged CytP450 for SMA-EA or SMA can be attributed to the significant charge density of the nanodisc’s polymer-belt that contain negatively-charged carboxylate-groups (see the supporting information).
Figure 1. Characterization of polymer nanodiscs.
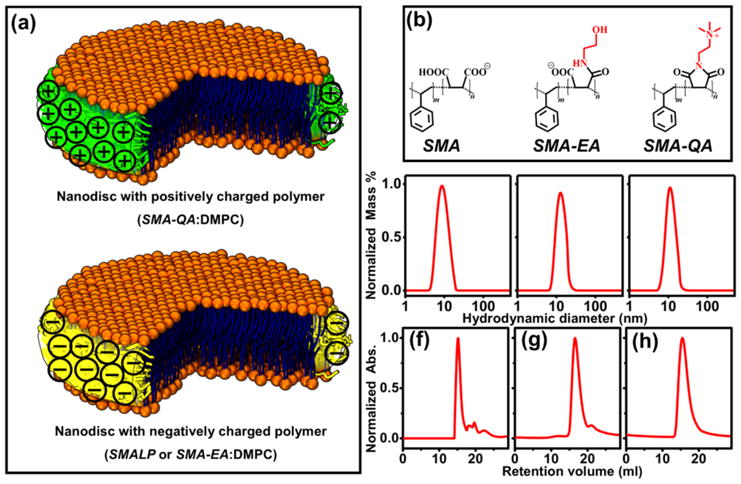
(a) Schematics of nanodiscs with a different charged polymer. (b) Structure of different polymers used. Dynamic light scattering profiles of (c) SMA:DMPC (1.5 :1 w/w), (d) SMA-EA:DMPC (2:1 w/w), (e) SMA-QA:DMPC (1.5:1 w/w). Size exclusion chromatogram profiles of (f) SMA:DMPC (1.5 :1 w/w), (g) SMAEA:DMPC (2:1 w/w), and (h) SMA-QA:DMPC (1.5:1 w/w).2
Figure 2. Reconstitution of CytP450 2B4 in polymer nanodiscs.
(a) Schematic showing CytP450 reconstited in a SMA-QA:DMPC nanodiosc. Heme corination sphere of CO bound state. (b) UV-vis absorption spectra of CytP450 reconstituted in different nanodiscs in its ferric state (left column) and ferrous -carbon monoxide complex (right column). UV-vis absorption spectra of cytP450 reconstituted in SMA-EA nanodiscs: (c) in the presence of indicated NaCl concentrations and (d) ferrous carbon monoxide complex (d). UV-vis spectra of cytP450 reconstituted in SMA-QA nanodiscs and (e) in the presence of NaCl, (f) ferrous carbon monoxide complex.
To further investigate the interaction between the polymer nanodiscs and CytP450, SEC was performed. SEC analysis (Figure S2) of the positively-charged P450 and the negatively-charged polymer (SMA-EA or SMA) exhibited a shift in the retention volume of 4.4 (for SMA-EA) or 4.6 (for SMA) ml. This observation suggests that the formation of large-size particles due to the interaction of the oppositely-charged protein and polymer-belt. On the other hand, when the protein and polymer have the same type of charges, SEC profiles revealed the formation of particles that are larger than that of empty-nanodiscs but smaller than that observed for oppositely-charged protein and polymer-belt (mentioned above). For example, the positively-charged P450 and the positively-charged polymer (SMA-QA) exhibited a shift in the retention volume of 2 ml as compared to that of empty-nanodiscs (Figure S2). This observation indicates a successful reconstitution of P450 in the positively-charged SMA-QA-polymer-nanodiscs. This finding is also in agreement with UV-vis experimental results and demonstrates a successful reconstitution of a functionally active P450.
Reconstitution of functional CytP450 in SMA nanodiscs was not possible as SMA is unstable under high NaCl concentrations; whereas salt is essential to screen the nonspecific electrostatic interactions between the polymer and protein. The complete inactivation of CytP450 by SMA is also partially due to the presence of the hydrophobic domains in SMA polymer as explained in a recently published study.32 Use of salt during SMA-QA reconstitution of CytP450 showed negligible effects on the functional reconstitution efficiency (Figure 2e–f). The ability to monomerize the full-length CytP450 and functionally reconstitute in a planar lipid bilayer using SMA-QA and SMA-EA (in the presence of salt) is remarkable. We believe that this first demonstration should enable high-resolution structural and enzymatic mechanistic studies on the full-length CytP450.
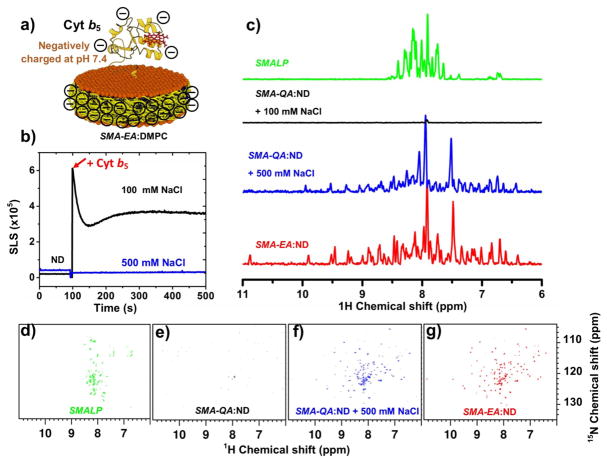
We further examined the effect of polymer charge on the reconstitution of a negatively charged ~16-kDa rabbit Cyt b5 (-8.4 at pH 7.4) which has an isoelectric point of 6.0.31 The incubation of Cyt b5 with SMA-QA nanodiscs was monitored using static light scattering (SLS) (Figure 3b). Increase of light scattering from SMA-QA nanodiscs upon the addition of Cyt b5 indicated the formation of aggregates in the sample. No change in SLS was observed by the addition of Cyt b5 to SMA-QA nanodiscs in the presence of 500 mM of NaCl, suggesting that the positively charged SMA-QA was forming aggregates with negatively charged Cyt b5 via a nonspecific electrostatic interaction; but the addition of salt suppressed the electrostatic interaction between polymer and protein to avoid aggregation. Further insights on the reconstitution of Cyt b5 and stability of resultant nanodiscs were obtained using 2D 1H-15N transverse relaxation optimized spectroscopy heteronuclear single quantum coherence spectroscopy (TROSY-HSQC) NMR experiments on uniformly 15N-labeled Cyt b5 reconstituted nanodiscs (Figure 3). As seen in NMR spectra in Figure 3g, Cyt b5 reconstituted in SMA-EA nanodiscs showed well dispersed resonances indicating that the protein is well folded as reported previously.7, 19 On the other hand, when Cyt b5 is reconstituted in the presence of the SMA polymer nanodiscs, resonances in the 2D TROSY-HSQC NMR spectrum were clustered around in the 7 to 8.5 ppm region signifying a strong interaction between the polymer and Cyt b5 (Figure 3d). The strong intermolecular interaction is most likely due to the hydrophobic domains present in SMA,32 similar to the interactions observed for CytP450 reconstitution as mentioned earlier. It is interesting that SMA-QA polymer nanodiscs containing Cyt b5 produced well dispersed resonances only in the presence of 500 mM NaCl (Figure 3f), while no resonances were observed in the presence of 100 mM NaCl (Figure 3e). This observation suggests that the negatively charged Cyt b5 and positively charged SMA-QA polymer nanodiscs form large aggregates, supporting the above-mentioned SLS results (Figure 3b).
Figure 3. Reconstitution of Cyt b5 in polymer-nanodiscs.
a) Schemtaic representation of a ~16-kDa rabbit Cyt b5 reconstituted in a negatively charged polymer-nanodisc. b) SLS spectra of SMA-QA nanodiscs and Cyt b5 at different NaCl concentrations. c) Projections of 2D TROSY HSQC spectra of a uniformly-15N-Cyt b5 reconstituted in SMALP (d), SMA-QA (e) with 100 mM NaCl, SMA-QA (f) with 500 mM NaCl, and SMA-EA (g) nanodiscs.
In conclusion, we have successfully reconstituted functional CytP450 and Cyt b5 in polymer nanodiscs. We have demonstrated that CytP450’s stability is increased when reconstituted in SMA-QA nanodiscs as compared to that in a lipid-free solution (or in detergent), which is similar to when reconstituted in MSP and peptide nanodiscs. Polymer-protein charge-charge interactions were demonstrated to play an important role in the functional reconstitution of proteins and on the stability of resultant nanodiscs. These interactions are attributed to the presence a high charge density on the polymer-belt of the nanodisc due to the presence of a large number of polymer molecules within a limited surface area. Positively charged SMA-EA polymer nanodiscs were found to be effective for the reconstitution of Cyt b5 which contains a large anionic soluble domain. The negatively charged SMA based polymer nanodiscs, however, were incapable of functionally reconstituting CytP450 or Cyt b5 due to the presence of hydrophobic domains in the SMA polymer, even when the charge-charge interactions were removed as in the case of Cyt b5. Thus, our study reveals that the innate chemical properties of the chosen polymer to form lipid-nanodiscs can have a large effect on the successful functional reconstitution of a membrane protein. In particular, the presence of charged residues in the large soluble-domain(s) containing single-pass (like cytochromes P450 and b5) and double-pass membrane proteins can pose challenges due to their direct interaction with the charged polymer belt. As demonstrated in this study, with a judicious choice of a polymer to form lipid-nanodiscs and salt concentration, membrane proteins can be reconstituted for studies using NMR. We expect the results reported in this study to be useful to prepare polymer based nanodiscs for studies using a variety of biophysical techniques including SAXS, cryo-EM and crystallography. While SMA based polymers exhibit unique advantages and are increasingly used for the structural studies of membrane proteins, our findings reported in this study indicate the need for neutral molecules, that can form lipid-nanodiscs, for the structural studies on the most challenging large soluble-domain containing membrane proteins. Unlike other types of nanodiscs, tolerance exhibited by SMA-QA nanodiscs over a broad range of pH and divalent metal ions can be further utilized. A complete characterization of lipid bilayer properties of polymer nanodiscs would be useful in the structural studies of reconstituted membrane proteins.
Supplementary Material
Acknowledgments
This study was supported by NIH (GM084018 to A.R.).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflicts of interest: There are no conflicts to declare.
Notes and references
- 1.Bill RM, Henderson PJ, Iwata S, Kunji ER, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Nat Biotechnol. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 2.Ravula T, Hardin NZ, Ramadugu SK, Cox SJ, Ramamoorthy A. Angew Chem Int Ed Engl. 2018;57:1342. doi: 10.1002/anie.201712017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 3.Denisov IG, Sligar SG. Chem Rev. 2017;117:4669. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayburt TH, Grinkova YV, Sligar SG. Nano Letters. 2002;2:853–856. [Google Scholar]
- 5.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 6.Hagn F, Etzkorn M, Raschle T, Wagner G. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A. Angew Chem Int Ed Engl. 2016;55:4497–4499. doi: 10.1002/anie.201600073. [DOI] [PubMed] [Google Scholar]
- 8.Ravula T, Barnaba C, Mahajan M, Anantharamaiah GM, Im SC, Waskell L, Ramamoorthy A. Chem Commun (Camb) 2017;53:12798–12801. doi: 10.1039/c7cc07520k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Grobner G, Watts A. Angew Chem Int Ed Engl. 2012;51:4653–4657. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 10.Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi JI, Ramamoorthy A. J Am Chem Soc. 2017;139:18657–18663. doi: 10.1021/jacs.7b10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, Killian JA. Eur Biophys J. 2016;45:3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorr JM, van Coevorden-Hameete MH, Hoogenraad CC, Killian JA. Biochim Biophys Acta. 2017;1859:2155. doi: 10.1016/j.bbamem.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Swainsbury DJK, Scheidelaar S, Foster N, van Grondelle R, Killian JA, Jones MR. Biochim Biophys Acta. 2017;1859:2133–2143. doi: 10.1016/j.bbamem.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grethen A, Oluwole AO, Danielczak B, Vargas C, Keller S. Sci Rep. 2017;7:11517. doi: 10.1038/s41598-017-11616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorr JM, Koorengevel MC, Schafer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, Dafforn TR, Baldus M, Killian JA. Proc Natl Acad Sci U S A. 2014;111:18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR. Nat Protoc. 2016;11:1149. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 17.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M. J Am Chem Soc. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 18.Oluwole AO, Klingler J, Danielczak B, Babalola JO, Vargas C, Pabst G, Keller S. Langmuir. 2017;33:14378. doi: 10.1021/acs.langmuir.7b03742. [DOI] [PubMed] [Google Scholar]
- 19.Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A. Angew Chem Int Ed Engl. 2017;56:11466. doi: 10.1002/anie.201705569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 20.Pollock NL, Lee SC, Patel JH, Gulamhussein AA, Rothnie AJ. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbamem.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Durr UH, Waskell L, Ramamoorthy A. Biochim Biophys Acta. 2007;1768:3235–3259. doi: 10.1016/j.bbamem.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. J Am Chem Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnaba C, Gentry K, Sumangala N, Ramamoorthy A. F1000Research. 2017;6 doi: 10.12688/f1000research.11015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meunier B, de Visser SP, Shaik S. Chem Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 25.Krest CM, Silakov A, Rittle J, Yosca TH, Onderko EL, Calixto JC, Green MT. Nat Chem. 2015;7:696–702. doi: 10.1038/nchem.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luthra A, Gregory M, Grinkova YV, Denisov IG, Sligar SG. In: Cytochrome P450 Protocols. Phillips IR, Shephard EA, Ortiz de Montellano PR, editors. Humana Press; Totowa, NJ: 2013. pp. 115–127. [DOI] [Google Scholar]
- 27.Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im SC, Anantharamaiah GM, Waskell L, Ramamoorthy A. ngew Chem Int Ed Engl. 2018;57:3391–3395. doi: 10.1002/anie.201713167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denisov IG, Sligar SG. Biochim Biophys Acta. 2011;1814:223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath A, Koo PK, Rhoades E, Atkins WM. J Am Chem Soc. 2008;130:15746–15747. doi: 10.1021/ja805772r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Huang R, Im SC, Waskell L, Ramamoorthy A. J Biol Chem. 2015;290:12705–12718. doi: 10.1074/jbc.M114.597096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M, Le Clair SV, Huang R, Ahuja S, Im SC, Waskell L, Ramamoorthy A. Sci Rep. 2015;5:8392. doi: 10.1038/srep08392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheidelaar S, Koorengevel MC, van Walree CA, Dominguez JJ, Dorr JM, Killian JA. Biophys J. 2016;111:1974–1986. doi: 10.1016/j.bpj.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.