Abstract
Scientific Abstract
The molecular pathogenesis of autism spectrum disorder (ASD), a neurodevelopmental disorder, is still elusive. In the present study, we investigated the possible roles of endoplasmic reticulum (ER) stress, oxidative stress, and apoptosis as molecular mechanisms underlying autism. This study compared the activation of ER stress signals (PERK, ATF6, IRE1α) in different brain regions (prefrontal cortex, hippocampus, cerebellum) in subjects with autism and in age-matched controls. Our data showed that the activation of three signals of ER stress varies in different regions of the autistic brain. Inositol-requiring enzyme 1 alpha (IRE1α) was activated in cerebellum and prefrontal cortex but activating transcription factor 6 (ATF6) was activated in hippocampus. Protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) was not activated in the three regions. Furthermore, the activation of ER stress was confirmed because the expression of C/EBP-homologous protein (CHOP), which is the common downstream indicators of ER stress signals, and most of ER chaperones were up-regulated in the three regions. Consistent with the induction of ER stress, apoptosis was found in the three regions by detecting the cleavage of caspase 8 and PARP as well as using the TUNEL assay. Moreover, our data showed that oxidative stress was responsible for ER stress and apoptosis because the levels of 4-Hydroxynonenal (4-HNE) and nitrotyrosine-modified proteins were significantly increased in the three regions. In conclusion, these data indicate that cellular stress and apoptosis may play important roles in the pathogenesis of autism.
Lay Abstract
Autism results in significant morbidity and mortality in children. The functional and molecular changes in the autistic brains are unclear. The present study utilized autistic brain tissues from the National Institute of Child Health and Human Development’s Brain Tissue Bank for the analysis of cellular and molecular changes in autistic brains. Three key brain regions, the hippocampus, the cerebellum and the frontal cortex, in six cases of autistic brains and six cases of non-autistic brains from 6-16 years old deceased children, were analyzed. The current study investigated the possible roles of endoplasmic reticulum (ER) stress, oxidative stress, and apoptosis as molecular mechanisms underlying autism. The activation of three signals of ER stress (PERK, ATF6, IRE1α) varies in different regions. The occurrence of ER stress leads to apoptosis in autistic brains. ER stress may result from oxidative stress because of elevated levels of the oxidative stress markers: 4-Hydroxynonenal (4-HNE) and nitrotyrosine-modified proteins in autistic brains. These findings suggest that cellular stress and apoptosis may contribute to the autistic phenotype. Pharmaceuticals and/or dietary supplements, which can alleviate ER stress, oxidative stress and apoptosis, may be effective in ameliorating adverse phenotypes associated with autism.
Keywords: human autism, cerebellum, prefrontal cortex, hippocampus, endoplasmic reticulum stress, apoptosis, oxidative stress
Introduction
Autism is considered to be a part of broader disorder known as autism spectrum disorder (ASD). ASD is characterized by problems in social skills and communication, as well as by a restricted range of interests and activities and repetitive behaviors (DSM-IV criteria, American Psychiatric Association, 1994). Approximately 1 in 68 children has been identified with autism spectrum disorder according to data from the Autism and Developmental Disabilities Monitoring (ADDM) Network of the CDC (CDC, USA, 2010). Approximately 1 in 6 children in the United States had a developmental disability in 2006–2008, ranging from mild disabilities (e.g., speech and language impairments) to serious developmental disabilities (e.g., intellectual disabilities, cerebral palsy, autism) (CDC, USA, 2006–2008). The autistic brain has abnormalities, including overgrowth of axons(Courchesne et al., 2007) and dendritic spines(Hutsler and Zhang, 2010), loss of granule cells(Bauman and Kemper, 2005), decreased Purkinje cell counts in cerebella(Ritvo et al., 1986), and Purkinje cell atrophy(Fatemi et al., 2000). Although numerous structural and functional neuro-imaging studies have been conducted, the underlying molecular etiology of autism is still unknown.
The endoplasmic reticulum (ER) is a membranous network that is primarily recognized as the site of synthesis, folding and translocation of secreted, membrane-bound, and some organelle-targeted proteins(Braakman and Bulleid, 2011). ER homeostasis is very sensitive to perturbations in cellular homeostasis, including nutrient deprivation, hypoxia, Ca+ concentration and the redox state(Szegezdi et al., 2006). Perturbations of ER function, known as ER stress, trigger the unfolded protein response (UPR)(Logue et al., 2013). There are three ER localized transmembrane receptors that act as stress sensors that constantly monitor the condition of the ER: inositol-requiring enzyme 1 alpha (IRE1α), protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6)(Sano and Reed, 2013). Following ER stress, the activation of PERK, ATF6 and IRE1α is a pro-survival response that reduces the accumulation of misfolded or unfolded proteins and restores normal ER function(Sano and Reed, 2013). During conditions when ER stress is chronically prolonged and cannot be resolved, the UPR signal switches from pro-survival to pro-apoptotic(Sano and Reed, 2013). Although the relations between ER stress and neurodegenerative diseases have been well studied(Kaufman, 2002), the associations between ER stress and autism have not been sufficiently investigated.
In the present study, we compared the activation of ER stress signals (PERK, ATF6, IRE1α) in different brain regions (prefrontal cortex, hippocampus, cerebellum) in subjects with autism and in age-matched controls. Our studies showed that the activation of three signals of ER stress (PERK, ATF6, IRE1α) varies in different regions. The occurrence of ER stress leads to apoptosis in autistic brains because the levels of cleaved caspase 8 and PARP were significantly increased in autistic brains. ER stress may result from oxidative stress because of elevated levels of 4-Hydroxynonenal (4-HNE) and nitrotyrosine-modified proteins in autistic brains. Our results suggest that cellular stress and apoptosis may contribute to the autistic phenotype.
Materials and methods
Brain tissue
Brain tissue samples from autistic subjects and non-neurological control cases were obtained from the Brain and Tissue Bank at the University of Maryland, Baltimore (a brain and tissue repository of the NIH NeuroBioBank). The collection protocol at the University of Maryland, Baltimore was reviewed and approved by its Institutional Review Board. All of the autistic cases fit the diagnostic criteria established in the Autism Diagnostic Interview-Revised (ADI-R). The clinical characteristics of all autistic subjects and controls included in this study are described in Table 1. The samples of the prefrontal cortex, hippocampus and cerebellum were obtained from six autistic subjects and six controls that were matched for age, postmortem interval (PMI) and sex. This study was approved by the Institutional Review Board at the University of Maryland School of Medicine.
Table 1.
Clinical characteristics of the autistic subjects and controls
| UMB# | Group | Brain Region | Age (yrs) | Gender | PMI (hr) | Storage (yr) | Cause of death | Clinical Brain Diagnosis | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 4849 | ASD | P | H | C | 7.46 | Male | 20 | 8.66 | Drowning | ASD - Autism, Language Delay, Lead Poisoning |
| 5408 | CTRL | P | H | C | 6.84 | Male | 18 | 4.39 | Drowning | Unaffected Control |
| 5144 | ASD | P | H | C | 7.20 | Male | 3 | 6.86 | Unknown | ASD - Autism |
| 5391 | CTRL | P | H | C | 8.78 | Male | 12 | 4.71 | Drowning | Unaffected Control |
| 4334 | ASD | P | H | C | 11.04 | Male | 27 | 5.95 | Acute hemorrhagic tracheobronchitis and bronchiolitis (Probable viral etiology) | ASD - Autism |
| 5334 | CTRL | P | H | C | 12.68 | Male | 15 | 5.67 | Asphyxiation | Unaffected Control |
| 5308 | ASD | P | --- | C | 4.49 | Male | 21 | 5.36 | Head injury | ASD - Autism |
| 4670 | CTRL | P | --- | C | 4.64 | Male | 17 | 9.63 | Cardiac arrest | Unaffected Control |
| 5403 | ASD | P | H | C | 16.72 | Male | 35 | 4.64 | Cardiac arrhythmia | ASD - Autism |
| 5407 | CTRL | P | H | C | 16.53 | Male | 33 | 4.41 | Acute bronchopneumonia | Unaffected Control |
| 5565 | ASD | P | H | C | 12.92 | Male | 22 | 6.17 | Complications of seizure disorder | ASD - Autism, Epilepsy |
| 1714 | CTRL | P | H | C | 12.44 | Male | 22 | 11.89 | Cardiac arrhythmia | Unaffected Control |
P: prefrontal cortex; H: hippocampus; C: cerebellum; yrs: years; hrs: hours; PMI: postmortem interval
Immunoblotting
Frozen brain tissue samples were homogenized in Neuronal Protein Extraction Reagent (Thermo Scientific) supplemented with protease inhibitors (Sigma) and phosphatase inhibitors (New England BioLabs). The supernatants were collected and assayed for total protein using the DC™ protein assay (Bio-Rad). Samples were aliquoted into labeled vials and stored at −80°C until use. Equal amounts of protein (50 µg) were resolved by SDS-PAGE and transferred onto Immobilon-P membranes (Millipore). Five micrograms of Precision Plus Protein Standards (Bio-Rad) were loaded into one lane of the SDS-PAGE gel. The Immobilon-P membranes were incubated in 5% nonfat milk for 1 hour and were then incubated for overnight at 4°C with the following primary antibodies at dilutions of 1:1000: PERK (Cell Signaling Technology), phosphorylated PERK (p-PERK) (Cell Signaling Technology), ATF6, and CHOP (Cell Signaling Technology), IRE1α (Cell Signaling Technology), phosphorylated IRE1α (p-IRE1α) (Abcam), caspase 8 (Enzo Life Sciences), PARP (Cell Signaling Technology), 4-HNE (Millipore) and nitrotyrosine (Millipore). The membranes were then exposed to goat anti-rabbit or anti-mouse secondary antibodies (Kirkegaard & Perry Laboratories). To ensure that equivalent amounts of protein were loaded, the membranes were stripped and probed with a mouse antibody against β-actin (Abcam, 1:5000 dilution). Signals were detected using a SuperSignal West Femto Maximum Sensitivity Substrate kit (Thermo Scientific). Quantitative analysis of blots was performed using VisionWorksLS software (UVP). Fold changes were calculated by the ratio of interest proteins to β-actin.
RNA extraction and Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from brain tissue using the Trizol reagent (Ambion) and reverse transcribed using the QuantiTect Reverse Trancription Kit (Qiagen). RT-qPCR for ER chaperone genes and GAPDH was performed using the Maxima SYBR Green/ROX qPCR Master Mix assay (Thermo Scientific). The primers for RT-qPCR are listed in Table 2. RT-qPCR and subsequent calculations were performed by a StepOnePlus™ Real-Time PCR System (Applied Biosystem).
Table 2.
The sequences of primers used in RT-qPCR.
| Primers name | Primer sources | Primer sequences |
|---|---|---|
| CHOP F | Own design | GCACCTCCCAGAGCCCTCACTCTCC |
| CHOP R | Own design | GTCTACTCCAAGCCTTCCCCCTGCG |
| BiP F | Own design | CGAGGAGGAGGACAAGAAGG |
| BiP R | Own design | CACCTTGAACGGCAAGAACT |
| calnexin F | Own design | CCTTCCTGTGTTCCTGGTTATC |
| calnexin R | Own design | TCCTTCTCTTCTTCCTCTTCCT |
| GRP94 F | Own design | GCTGACGATGAAGTTGATGTGG |
| GRP94 R | Own design | CATCCGTCCTTGATCCTTCTCTA |
| PDIA2 F | Own design | GGACACTGCAAAAACCTAGAGC |
| PDIA2 R | Own design | CCAGAACCTGATTGACTGTAGCA |
| Calreticulin F | Primerbank ID: 209862753c1 | CCTGCCGTCTACTTCAAGGAG |
| Calreticulin R | GAACTTGCCGGAACTGAGAAC | |
| GRP58 F | Primerbank ID: 67083697c1 | GCCTCCGACGTGCTAGAAC |
| GRP58 R | GCGAAGAACTCGACGAGCAT | |
| GRP170 F | Primerbank ID: 169234641c3 | CGAGCTGACTTTCGACCCAC |
| GRp170 R | TGAGAACCATGCCCAACACTT | |
| SIL1 F | Primerbank ID: 83641898c2 | AGGCAAAACTCCAATATGAGGAC |
| SIL1 F | GATGTGTAGGTGTTGGTGTTGAT | |
| P58IPK F | Primerbank ID: 300192905c3 | GGATGCAGAACTACGGGAACT |
| P58IPK R | TCTTCAACTTTGACGCAGCTT | |
| Herp F | Primerbank ID: 58530858c2 | TGCTGGTTCTAATCGGGGACA |
| Herp R | CCAGGGGAAGAAAGGTTCCG | |
| EDEM F | Primerbank ID: 197304767c1 | CGGACGAGTACGAGAAGCG |
| EDEM R | CGTAGCCAAAGACGAACATGC | |
| ERp72 F | Primerbank ID: 157427676c1 | GGCAGGCTGTAGACTACGAG |
| ERp72 R | TTGGTCAACACAAGCGTGACT | |
| GAPDH F | Primerbank ID: 378404907c2 | ACAACTTTGGTATCGTGGAAGG |
| GAPDH R | GCCATCACGCCACAGTTTC |
F: forward; R: reverse.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
The ApopTag Red In Situ Apoptosis Detection Kit (Millipore) was used to detect apoptotic cells as previously described(Wang et al., 2017). Briefly, formalin-fixed brain tissues were dehydrated by series of 50%–100% ethanol and then embedded in paraffin for sectioning. Ten-µm paraffin-embedded sections were rehydrated, washed with PBS, and incubated with the TUNEL reaction agents. The percentage of apoptotic cells was obtained by dividing the number of TUNEL positive cells with the total number of cells in a microscopic field and then multiplying by 100 from six autistic brains and six control brains.
Statistics
Data are presented as the means ± SEM. SPSS 20.0 was used for statistical analysis. The Mann-Whitney U test was used to compare group differences because the datasets are not normally distributed. P < 0.05 was considered statistical significant.
Results
Activation of ER stress signals in human autistic brain
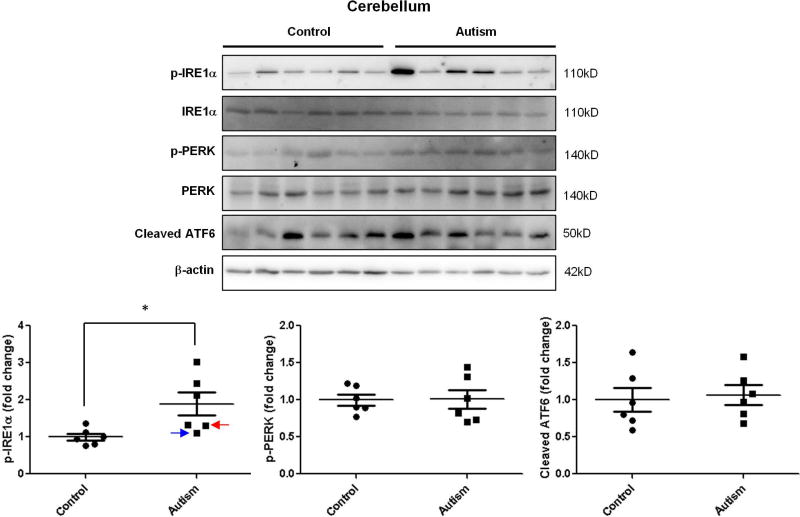
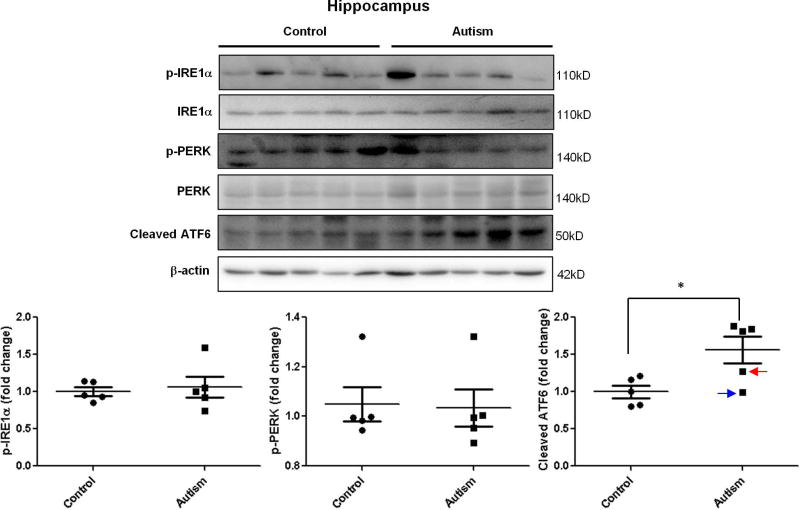
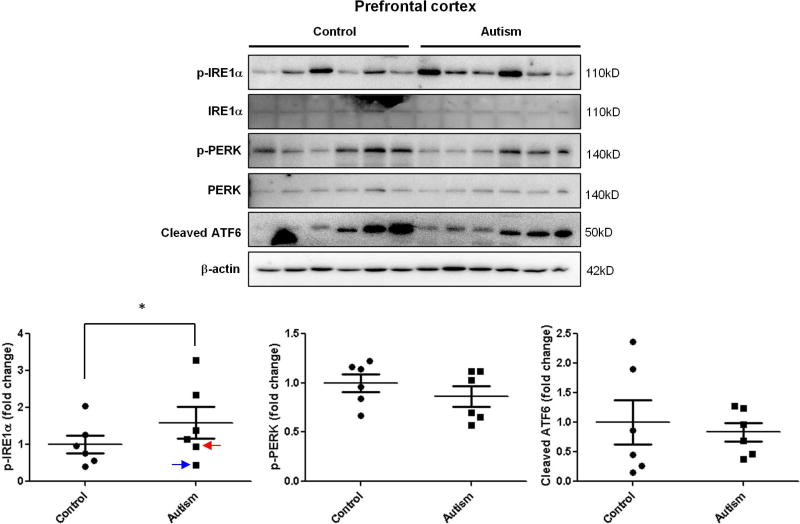
To investigate the possible role of ER stress in the pathogenesis of autism, we determined the activation of UPR signals in autistic brains. The levels of three ER stress signals were measured by immunoblotting. In the cerebellum, phosphorylated IRE1α was significantly increased in autistic subjects compared with age-matched normal controls (Figure 1.). There were no changes in the phosphorylated PERK and cleaved ATF6 levels in the cerebellum (Figure 1.). However, the cleaved ATF6 was significantly up-regulated in the hippocampus of autistic subjects (Figure 2.). Phosphorylated IRE1α and PERK were comparable in the hippocampus between autistic subjects and controls (Figure 2.). Similarly with cerebellum, phosphorylated IRE1α was significantly elevated in prefrontal cortex of autistic subjects compared to controls (Figure 3.). The levels of phosphorylated PERK and cleaved ATF6 in the prefrontal cortex were similar between autistic subjects and controls (Figure 3.). The ER stress-induced signals were differentially activated in three main regions of the autistic brain. The overall findings indicate that ER stress signals were activated in human autistic brains.
Figure 1.
Immunoblot analysis of ER stress signals in the autistic cerebellum. Immunoblot analysis of the cerebellum homogenate was performed using p-IRE1α, p-PERK and total ATF6 antibodies. Representative images of the immunoblots are shown in the top panel. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM (n=6). * indicates a significant difference in autistic brains compared to controls (P < 0.05).
Figure 2.
Immunoblot analysis of ER stress signals in the autistic hippocampus. Immunoblot analysis of the hippocampus homogenate was performed using p-IRE1α, p-PERK and total ATF6 antibodies. Representative images of the immunoblots are shown in the top panel. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM (n=5). * indicates a significant difference in autistic brains compared to controls (P < 0.05).
Figure 3.
Immunoblot analysis of ER stress signals in the autistic prefrontal cortex. Immunoblot analysis of the prefrontal cortex homogenate was performed using p-IRE1α, p-PERK and total ATF6 antibodies. Representative images of the immunoblots are shown in the top panel. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM (n=6). * indicates a significant difference in autistic brains compared to controls (P < 0.05).
Induction of ER stress downstream gene CHOP and ER chaperones in human autistic brain
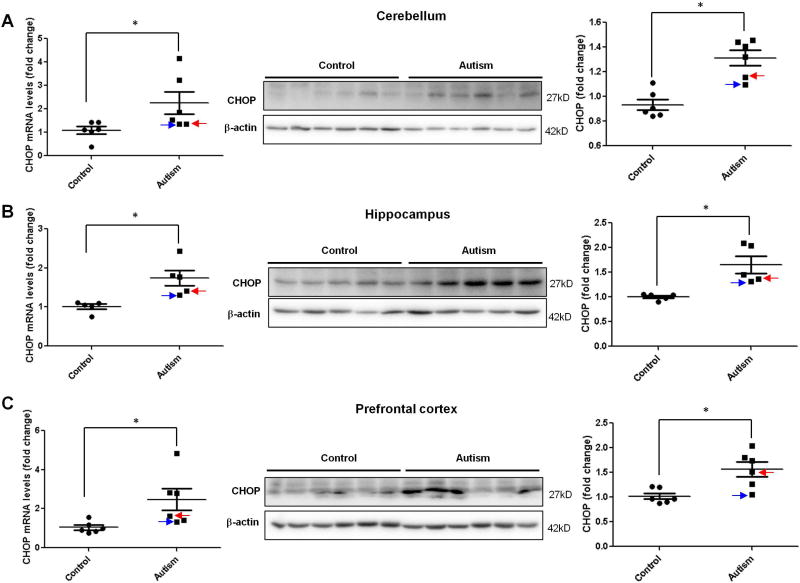
CHOP, also known as growth arrest and DNA damage inducible gene 153 (GADD153), is up-regulated by ER stress(Szegezdi, et al., 2006). All three signals of ER stress can induce the transcription of CHOP through transcription factors: ATF4, ATF6, and XBP-1, respectively(Szegezdi, et al., 2006). To further confirm the activation of ER stress signals, we examined CHOP mRNA and protein levels in human autistic brain using RT-qPCR and immunoblotting. Consistent with the activation of ER stress signals, CHOP mRNA and protein levels in all three regions were increased in autistic subjects compared to controls (Figure 4.).
Figure 4.
Induction of CHOP expression in the autistic brain. RT-qPCR (left) and Immunoblot (right) analysis of CHOP in the autistic: (A) cerebellum (n=6), (B) hippocampus (n=5) and (C) prefrontal cortex (n=6). Representative images of the immunoblots are shown in the left panel. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM. * indicates a significant difference in autistic brains compared to controls (P < 0.05).
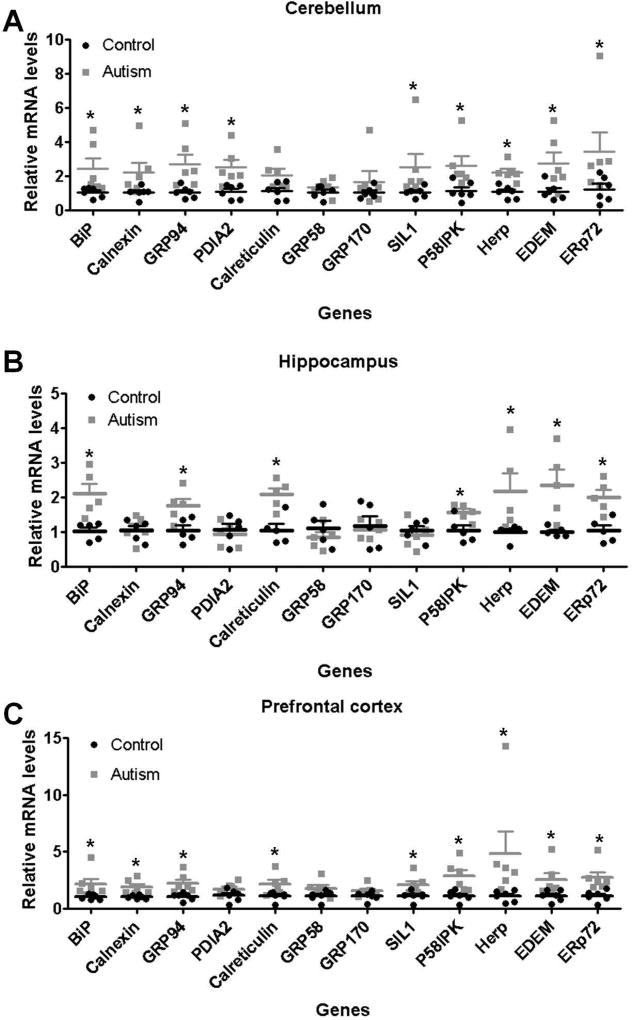
The ER contains a number of molecular chaperones physiologically involved in the posttranscriptional modifications, disulfide bond formation, folding, assembly and quality-control of newly synthesized proteins(Ni and Lee, 2007). The transcriptional up-regulation of ER chaperones is the hallmark of the ER stress response(Ma and Hendershot, 2004). In the present study, we determined the expressions of ER chaperones including GRP78/BiP, calnexin, GRP94/gp96, PDIA2, calreticulin, GRP58/ERp57, GRP170/ORP150, SIL1, P58IPK, Herp, EDEM, and ERp72(Ni and Lee, 2007). As shown in Figure 5., most of ER chaperones were up-regulated in autistic brain compared to controls, whereas there were only slight differences among three brain regions. GRP78/BiP, GRP94/gp96, P58IPK, Herp, EDEM, and ERp72 were increased in three brain regions of autistic subjects. GRP58/ERp57 and GRP170/ORP150 had similar levels between autistic brain and controls in three brain regions (Figure 5). Calreticulin was increased in both hippocampus and prefrontal cortex of autistic subjects, whereas SIL1 was induced in both cerebellum and prefrontal cortex of autistic subjects (Figure 5). PDIA2 was induced only in cerebellum of autistic subjects (Figure 5.).
Figure 5.
Induction of ER chaperones expression in the autistic brain. RT-qPCR analysis of ER chaperones expression in the autistic: (A) cerebellum (n=6), (B) hippocampus (n=5) and (C) prefrontal cortex (n=6). Values are the means ± SEM. * indicates a significant difference in autistic brains compared to controls (P < 0.05).
The combined data from the CHOP expression and ER chaperones consistently showed that ER stress was elevated in human autistic brain. Thus, these findings suggest a possible association between the activation of ER stress signals and pathological changes in the autistic brain.
Activation of caspase 8 and PARP in human autistic brain
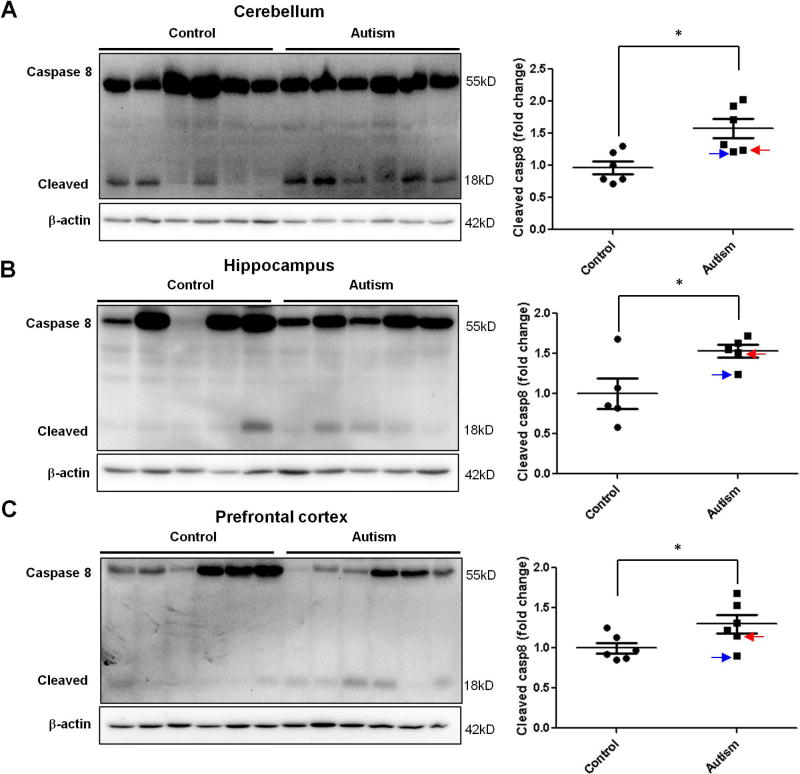
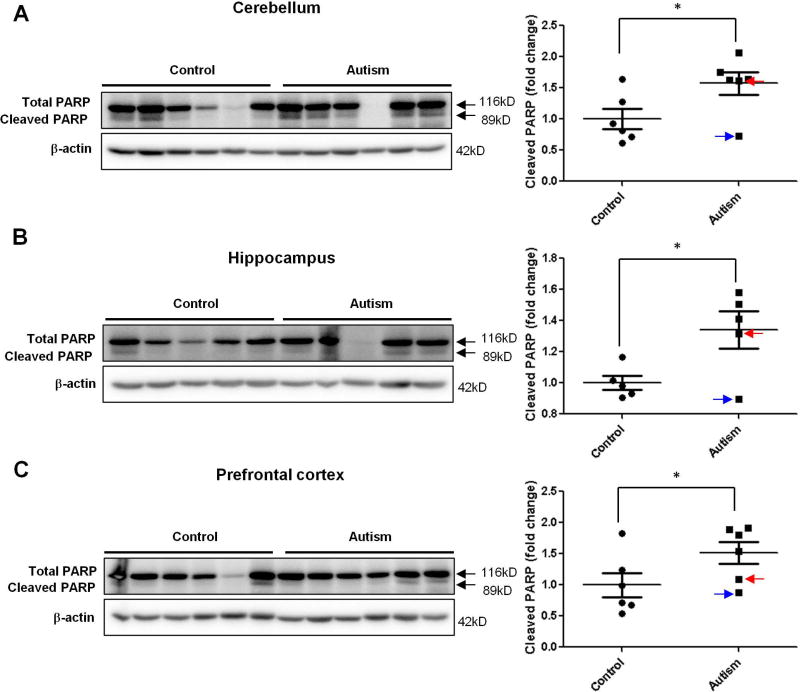
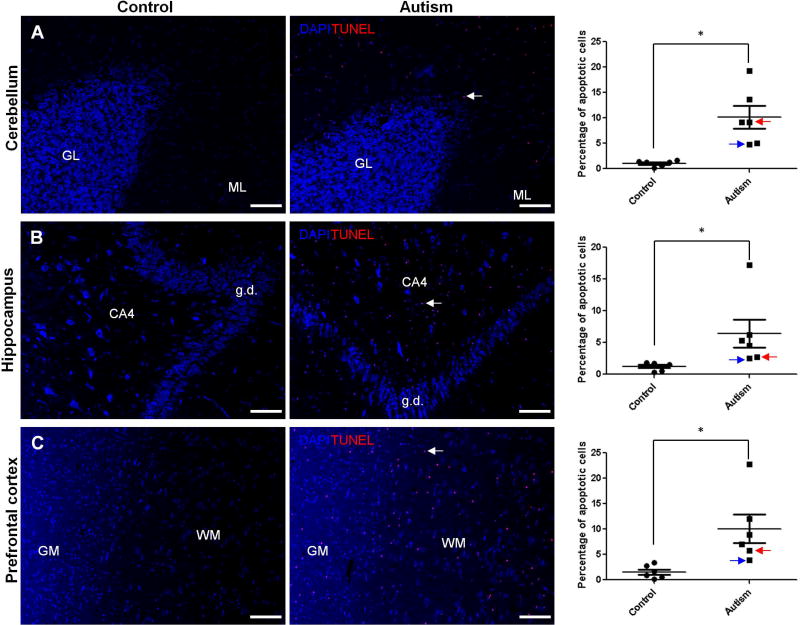
Finding increased levels of CHOP in the autistic brain would suggest a possible increase of apoptosis in the autistic brain because CHOP is primarily considered to be a pro-apoptotic factor that mediates ER stress-induced cell death(Szegezdi, et al., 2006). To further investigate whether there is increased apoptosis in the autistic brain, we determined the cleavage of caspase 8, an initiator of caspases for extrinsic pathway of apoptosis(Ichim and Tait, 2016), as the marker of apoptosis because it has been shown that ER stress can mediate cleavage of caspase 8 via CHOP-death receptor 4 (DR4, also named TRAILR1)/ death receptor 5 (DR5, also named TRAILR2) pathway(Iurlaro and Munoz-Pinedo, 2016; Lu et al., 2014). At the same time, we measured the cleavage of poly(ADP-ribose) polymerase (PARP) as another marker of apoptosis based on that PARP is one of several known cellular substrates of caspases and considered to be a hallmark of apoptosis(Boulares et al., 1999; Chaitanya et al., 2010). Almost all caspases are known to cleave PARP(Boulares, et al., 1999; Chaitanya, et al., 2010). The results of immunoblotting analysis showed that the cleavage of caspase 8 and PARP was significantly increased in human autistic brain in all three regions with the same pattern of activation of ER stress signals and increased CHOP expression was found in every region (Figure 6 and 7.). The TUNEL assay is a direct method for detecting in situ apoptotic cells by measuring DNA fragmentation generated during apoptosis. Our data revealed the occurrence of apoptosis in the molecular layer of the autistic cerebellum (Fig. 8A), cornu ammonis 4 (CA4) of the autistic hippocampus (Fig. 8B), and both of grey and white matters of the autistic prefrontal cortex (Fig. 8C). In contrast, there were none to negligible apoptotic cells presented in the control brains (Fig. 8A, B, C). Taken together, our results suggest that the activation of ER stress signals and increase of CHOP expression lead to apoptosis in human autistic brain.
Figure 6.
The cleavage of caspase 8 in the autistic brain. Immunoblot analysis of cleaved caspase 8 in the autistic: (A) cerebellum (n=6), (B) hippocampus (n=5) and (C) prefrontal cortex (n=6). Representative images of the immunoblots are shown in the left panel. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM. * indicates a significant difference in autistic brains compared to controls (P < 0.05).
Figure 7.
The cleavage of PARP in the autistic brain. Immunoblot analysis of cleaved PARP in the autistic: (A) cerebellum (n=6), (B) hippocampus (n=5) and (C) prefrontal cortex (n=6). Representative images of the immunoblots are shown in the left panel. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM. * indicates a significant difference in autistic brains compared to controls (P < 0.05).
Figure 8.
Apoptotic cells in the autistic brain. TUNEL assay was performed in the autistic: (A) cerebellum (n=6), (B) hippocampus (n=5) and (C) prefrontal cortex (n=6). Representative images of the TUNEL staining were shown in the left panel. The quantification of apoptotic cells was shown in the bar graphs. Blue: DAPI cell nuclear staining; Red: TUNEL signal. The typical apoptotic cells labeled by Red were indicated by arrows. GL: granular layer; ML: molecular layer; CA: cornu ammonis; g.d.: gyrus dentatus; GM: grey matter; WM: white matter. Scale bars: 100 µm. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. Values are the means ± SEM. * indicate a significant difference in autistic brains compared to the controls (P < 0.05).
Elevation of oxidative stress in human autistic brain
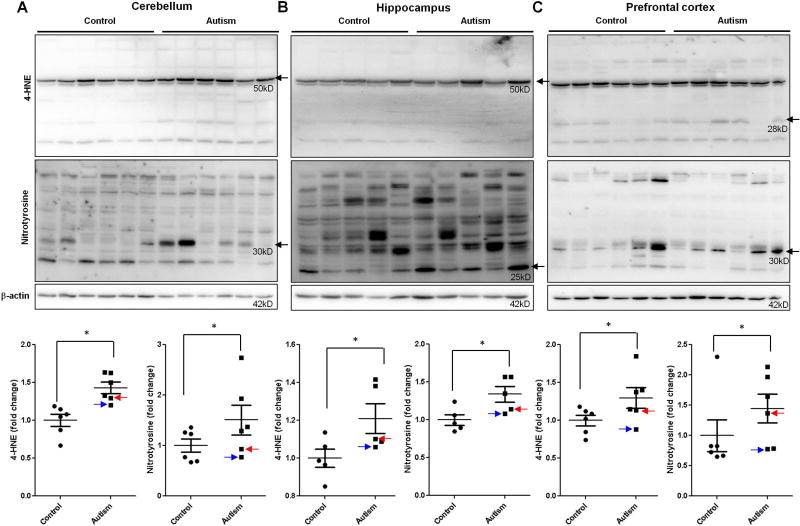
To investigate the causal factors that result in the activation of ER stress signals and subsequent apoptosis, we examined oxidative stress in human autistic brain. Oxidative stress is induced by reactive oxygen species (ROS) and reactive nitrogen species (RNS), such as superoxide, hydroxyl, peroxyl and nitric oxide (NO) free radicals(Chauhan and Chauhan, 2006). 4-Hydroxynonenal (4-HNE) and nitrotyrosine are two important markers for oxidative stress. 4-HNE is one of the most studied products of phospholipid peroxidation, and it efficiently reacts with sulfhydryl groups or histidine and lysine groups of proteins to form stable 4-HNE-protein adducts. Nitrotyrosine is formed by the nitration of tyrosine resides of proteins by RNS, such as peroxynitrite, nitrogen dioxide and nitryl chloride. In the present study, we determined the levels of 4-HNE- and nitrotyrosine-modified proteins in human autistic brain by immunoblotting. We calculated all blots in the membrane and found that part of blots had significant differences between autistic and control groups. These data suggest that 4-HNE or nitrotyrosine modifies part of proteins in autistic brains and 4-HNE or nitrotyrosine-modified proteins were different in different regions of brain (Figure 9.). For example, in hippocampus and cerebellum, proteins with the molecular weight (MW) 50kD were differentially modified by 4-HNE (Figure 9A and B). However, in prefrontal cortex, proteins with the MW 28kD were differentially modified by 4-HNE (Figure 9C). Similarly, in cerebellum and prefrontal cortex, proteins with the MW 30kD were differentially modified by nitrotyrosine (Figure 9A and C). However, in hippocampus, proteins with the MW 25kD were differentially modified by nitrotyrosine (Figure 9B). Because it has been shown that oxidative stress induces ER stress(Dong et al., 2016; Gu et al., 2016; Li et al., 2012; Li et al., 2013), these results suggest that oxidative stress may be responsible for the induction of ER stress in human autistic brain.
Figure 9.
Lipid peroxidation and nitrotyrosine in the autistic brain. Immunoblot analysis of 4-HNE and nitrotyrosine in the autistic: (A) cerebellum (n=6), (B) hippocampus (n=5) and (C) prefrontal cortex (n=6). Representative images of the immunoblots are shown in top panel. The modified blots were indicated with arrow. The blue arrow indicates sample #4849 and the red arrow indicates sample # 5565. The quantification of immunoblots normalized to β-actin is shown by bar graphs. Values are the means ± SEM. * indicates a significant difference in autistic brains compared to controls (P < 0.05).
Discussion
In the current study, we showed that a marked increase in ER stress, characterized by the activation of ER stress signals (IRE1α, ATF6 and PERK), occurred in three regions (cerebellum, hippocampus and prefrontal cortex) of autistic brains. A common downstream gene of ER stress signals CHOP and most of ER chaperones were also up-regulated in three regions of autistic brain, which confirmed the activation of ER stress signals. Increased ER stress is likely to be associated with oxidative stress in which the levels of 4-HNE and nitrotyrosine-modified proteins significantly increased in three regions of autistic brain. In addition, elevated ER stress may lead to apoptosis because the cleavage of caspase 8 and PARP was increased in the autistic brain. These results from the present studies support the findings of the increase of oxidative stress, ER stress and apoptosis in the autistic brain.
The emerging evidence shows that ER stress is associated with the pathogenesis of autism(Fujita-Jimbo et al., 2015; Fujita et al., 2010; Momoi et al., 2010; Ulbrich et al., 2015). Genetic testing in individuals with autism has identified mutations in several genes encoding synaptic cell adhesion molecules, such as neuroligin-3 and -4 (NLGN-3 and -4)(Jamain et al., 2003) and cell adhesion molecule-1 (CADM-1)(Zhiling et al., 2008). It has been shown that mutated NLGN-3 and CADM-1 induce local misfolding and cause partial retention of proteins in the ER that lead to ER stress in PC12 and C2C5 cell models(Fujita, et al., 2010; Ulbrich, et al., 2015). Overexpression of mutated NLGN-3 leads to the activation of three ER stress signals, as well as their downstream genes, such as BiP and CHOP in PC12 cells(Ulbrich, et al., 2015). In addition to mutations in genes encoding synaptic adhesion molecules, mutations in the GPR85 gene that is located in the AD linkage locus 9 (AUTS9) on chromosome 7q31 are associated with autism(Fujita-Jimbo, et al., 2015). Mutated GPR85 protein is more preferentially accumulated in the ER, causing ER stress characterized by induction of CHOP in C2C5 cells(Fujita-Jimbo, et al., 2015). In the present study, we further showed that ER stress indeed occurred in human autistic brain. Our data also revealed that the activation of ER stress signals varied in different regions of the autistic brain. IRE1α were activated in cerebellum and prefrontal cortex, but ATF6 was activated in the hippocampus. There was no activation of PERK pathway. However, the expression of CHOP, which is the common downstream indicators of ER stress signals, and most of ER chaperones were up-regulated in all three regions of autistic brain. It is suggested the differential pattern of ER stress signals activation but with the same downstream genes up-regulation in different brain regions. It has been demonstrated that three ER stress signals are differentially activated under different stimulation. For example, a plant metabolite andrographolide activates IRE1α but not PERK and ATF6 in cancer cells(Banerjee et al., 2016). However, bortezomib, an selective inhibitor of the 26S proteasome, induces the activation of PERK and ATF6 but not IRE1α in myeloma cells(Davenport et al., 2007).
It is well-known that ER stress can induce apoptosis(Sano and Reed, 2013). CHOP is primarily considered to be a pro-apoptotic transcription factor that mediates ER stress-induced apoptosis(Szegezdi, et al., 2006). Our data indicate that the protein levels of CHOP were increased in three regions of the autistic brain. Therefore, we determined whether apoptosis occurred in the autistic brain. Our results showed that apoptosis increased in three regions of the autistic brain, as characterized by increased cleavage of caspase 8 and PARP. It was shown that CHOP interacts with the phosphorylated form of the transcription factor JUN in a complex that binds to the promoter regions of DR4 and activates DR4 expression(Iurlaro and Munoz-Pinedo, 2016). It was also observed that CHOP interacts with GCN5, a histone acetyltransferase, to form a complex regulating both DR4 and DR5 expressions(Iurlaro and Munoz-Pinedo, 2016). DR4 and DR5 lead to widespread cleavage of caspases including caspase 8 and rapid cell death(Ichim and Tait, 2016). In addition, CHOP-mediated apoptosis manifests through the repression of Bcl2 expression, which then increases the proportion of pro-apoptotic Bcl-2 proteins that lead to the release of cytochrome c(Szegezdi, et al., 2006). In actuality, Bcl-2 expression is significantly reduced in the autistic brain(Fatemi and Halt, 2001; Fatemi et al., 2001; Sheikh et al., 2010a; Sheikh et al., 2010b; Wei et al., 2014). Fatemi et al. showed a significant reduction of 34 to 51% in the Bcl-2 protein levels in the autistic cerebellum(Fatemi, et al., 2001) and a 32% reduction in the autistic parietal cortex(Fatemi and Halt, 2001). Sheikh et al. also found that Bcl-2 expression was significantly reduced in the frontal cerebral cortex and cerebellum of autistic subjects(Sheikh, et al., 2010a; Sheikh, et al., 2010b). Taken together, it is possible that ER stress mediates apoptosis through the CHOP-DR4/5-caspase 8 or CHOP-Bcl2-cytochrome c pathway in the autistic brain.
Despite the possibility that the mutation of autism-related genes and the aggregation of their encoded proteins in the ER may be responsible for ER stress, oxidative stress may be another important cause for ER stress in the autistic brain. Indeed, metabolic markers of oxidative stress were previously detected in plasma from autistic subjects(Frye and James, 2014; Howsmon et al., 2017; James et al., 2004; Melnyk et al., 2012; Rose et al., 2012b). It has been demonstrated that micronutrient supplementation targeting redox metabolism exerts beneficial effects on some children with autism, implicating the presence of oxidative stress in autistic brains(Blossom et al., 2012; Frye et al., 2013; Hamlin et al., 2013; Hendren et al., 2016). Oxidative stress-induced modifications of proteins, such as the nitration of tyrosine residues, may lead to the retention of proteins in the ER that may contribute to ER stress in the autistic brain because our data showed marked elevation of nitrotyrosine-modified proteins in three regions of the autistic brain. Nitrotyrosine is formed primarily from peroxynitrite, a free radical generated from superoxide and nitric oxide (NO)(Bishop and Anderson, 2005). On the one hand, the decreased activities of glutathione peroxidase, catalase and superoxide dismutase (SOD) lead to the overproduction of superoxide in the autistic brain(Chauhan and Chauhan, 2006). The mitochondrion may be the source of superoxide because several studies have revealed mitochondrial overactivities or abnormalities in the autistic brain(Griffiths and Levy, 2017; Palmieri et al., 2010; Tang et al., 2013). On the other hand, elevated plasma levels of NO in autism have been reported(Sogut et al., 2003; Sweeten et al., 2004; Zoroglu et al., 2003), suggesting that NO synthase (NOS) may be activated in autism(Chauhan and Chauhan, 2006). In actuality, neuroglial cells can express inducible NOS (iNOS)(Bolanos et al., 2007; Brown et al., 1995). Rose et al have found 3-nitrotyrosine in the autistic brain(Rose et al., 2012a). Therefore, it is suggested that excess superoxide and NO may contribute to peroxynitrite formation and the increased nitrotyrosine protein damage observed in the present study.
The 4-HNE results from the cleavage of the oxidized fatty acyl chain, including ω-6 polyunsaturated fatty acid(Spickett, 2013). Some studies have reported oxidative damage to brain lipids in autism(Chauhan and Chauhan, 2006; Rossignol and Frye, 2014). Previous studies have reported a significant increase in lipofuscin-containing cells in language areas of the autistic brain(López-Hurtado and Prieto, 2008; Rossignol and Frye, 2014). Another study demonstrated the increase of lipid hydroperoxides (LOOH) in the temporal cortex and cerebellum in the autism group(Chauhan et al., 2011). In addition, plasma malonyldialdehyde (MDA), which is an end product of peroxidation of polyunsaturated fatty acids, as measured by thiobarbituric acid (TBA), had higher levels in 13/15 (87%) of autistic subjects(Chauhan et al., 2004). Taken together, oxidative damage to brain lipids occurs in autism, which is consistent with our findings in human autistic brain with elevation of 4-HNE-modified proteins.
Data from the Centers for Disease Control and Prevention have shown that ASD is approximately 4.5 times more common in boys (1 in 42) than in girls (1 in 189)(Christensen et al., 2016). The data is the average from multiple US and worldwide studies (Halladay et al., 2015). The difference in ascertainment procedures may contribute to the sex difference in ASD (Frazier et al., 2014; Idring et al., 2015; Ozonoff et al., 2011). The intellectual quotient measurement may also contribute to this variability (Autism et al., 2012; Sullivan et al., 2007). In addition, there is evidence showing that diagnostic biases lead to an overestimation of sex bias(Autism, et al., 2012; Zwaigenbaum et al., 2012). Excluding the variability in ascertainment and IQ measurement, it is believed that there is still a magnitude of at least 2:1–3:1 ASD prevalence difference between boys and girls(Halladay, et al., 2015). However, One study did not observe sex difference in autistic symptoms(Mandic-Maravic et al., 2015). Thus, our study used male ASD subjects from the brain tissue bank. Our current finding in boys may represent common pathways for autism because there is so far no report showing any sex difference in oxidative stress, ER stress and apoptosis. Future studies need validate our hypothesis using a large sample size including both sexes.
The limitation of this study is that all data were obtained from small sample size. In the current study, we observed significantly occurrence of ER stress, oxidative stress, and apoptosis in three regions of autistic brains in 6 of autistic samples compared to 6 of matched control samples. This results need to be verified in large sample size in the future. In addition, the cause of death may be a parameter that leads to changes observed in the present study. Overall, the causes of death for these patients showing in Table 1 are varied and in majority they are not related to brain injury, indicating that the causes of death may not affect our conclusions. Sample #4849 had lead poisoning and sample #5565 had epilepsy. Lead, a systemic toxicant affecting virtually every organ system, primarily affects the central nervous system, particularly the developing brain. However, the cause of death for #4849 was drowning. Epilepsy is a central nervous system (neurological) disorder in which brain activity becomes abnormal, causing seizures or periods of unusual behavior, sensations, and sometimes loss of awareness. Epilepsy can sometimes be associated with developmental disorders, such as autism and neurofibromatosis. However, when we labeled the values of these two samples in the graphs of Figures showing significant difference between the control group and the autism group, we found that the values from these two samples are at the low end in the autism group and comparable to the values in the control group, suggesting that lead poisoning and epilepsy in these two samples may not contribute to the observed cellular stress and apoptosis in autistic brains. Nevertheless, our findings need to be verified in a large sample size.
Moreover, we observed apoptosis in three regions of autistic brains but did not identify which kind of cell type undergoing apoptosis. Our next work will focus on which cell type such as neurons or glial cells undergoing apoptosis as well as co-localization of ER stress markers and apoptosis markers for identifying the cause-effect relationship between ER stress and apoptosis.
In summary, we showed the elevation of ER stress signals, oxidative stress and apoptosis in three regions of autistic brains. Based on these findings, we reason that increased cellular stress and apoptosis in the autistic brain may be associated with the pathogenesis of autism. Because autism is affected by multiple genetic and environmental factors that are case-specific and there are inherent limitations in the post-mortem brain, the present observations will need further confirmation in future studies. Further research with larger sample sizes is needed to investigate the association of cellular stress and apoptotic events with the severity and clinical phenotypes of autism.
Acknowledgments
This research is supported by NIH R01DK083243, R01DK101972, R01HL131737, R01HL134368, R01HL139060 and R01DK103024. We are grateful to the Brain and Tissue Bank at the University of Maryland Baltimore (Director Dr. Tomas Blanchard, https://neurobiobank.nih.gov) for providing brain tissues.
Footnotes
Disclosure: None of the authors have a conflict of interest.
References
- Autism, Developmental Disabilities Monitoring Network Surveillance Year Principal, I., Centers for Disease, C., and Prevention. Prevalence of autism spectrum disorders--Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. Morbidity and mortality weekly report. Surveillance summaries. 2012;61(3):1–19. [PubMed] [Google Scholar]
- Banerjee A, Ahmed H, Yang P, Czinn SJ, Blanchard TG. Endoplasmic reticulum stress and IRE-1 signaling cause apoptosis in colon cancer cells in response to andrographolide treatment. Oncotarget. 2016 doi: 10.18632/oncotarget.9180. 10.18632/oncotarget.9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2005;23(2–3):183–7. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Bishop A, Anderson JE. NO signaling in the CNS: from the physiological to the pathological. Toxicology. 2005;208(2):193–205. doi: 10.1016/j.tox.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Blossom SJ, Melnyk S, Cooney CA, Gilbert KM, James SJ. Postnatal exposure to trichloroethylene alters glutathione redox homeostasis, methylation potential, and neurotrophin expression in the mouse hippocampus. Neurotoxicology. 2012;33(6):1518–27. doi: 10.1016/j.neuro.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos JP, Herrero-Mendez A, Fernandez-Fernandez S, Almeida A. Linking glycolysis with oxidative stress in neural cells: a regulatory role for nitric oxide. Biochemical Society transactions. 2007;35(Pt 5):1224–7. doi: 10.1042/BST0351224. [DOI] [PubMed] [Google Scholar]
- Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. The Journal of biological chemistry. 1999;274(33):22932–40. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annual review of biochemistry. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- Brown GC, Bolanos JP, Heales SJ, Clark JB. Nitric oxide produced by activated astrocytes rapidly and reversibly inhibits cellular respiration. Neuroscience letters. 1995;193(3):201–4. doi: 10.1016/0304-3940(95)11703-y. [DOI] [PubMed] [Google Scholar]
- Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell communication and signaling : CCS. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology : the official journal of the International Society for Pathophysiology / ISP. 2006;13(3):171–81. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V, Brown WT, Cohen I. Oxidative stress in autism: increased lipid peroxidation and reduced serum levels of ceruloplasmin and transferrin--the antioxidant proteins. Life sciences. 2004;75(21):2539–49. doi: 10.1016/j.lfs.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Gu F, Essa MM, Wegiel J, Kaur K, Brown WT, Chauhan V. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. Journal of neurochemistry. 2011;117(2):209–20. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DL, Bilder DA, Zahorodny W, Pettygrove S, Durkin MS, Fitzgerald RT, Rice C, Kurzius-Spencer M, Baio J, Yeargin-Allsopp M. Prevalence and Characteristics of Autism Spectrum Disorder Among 4-Year-Old Children in the Autism and Developmental Disabilities Monitoring Network. Journal of Developmental & Behavioral Pediatrics. 2016;37(1):1–8. doi: 10.1097/dbp.0000000000000235. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. Mapping early brain development in autism. Neuron. 2007;56(2):399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Davenport EL, Moore HE, Dunlop AS, Sharp SY, Workman P, Morgan GJ, Davies FE. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110(7):2641–9. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- Dong D, Reece EA, Yang P. The Nrf2 Activator Vinylsulfone Reduces High Glucose-Induced Neural Tube Defects by Suppressing Cellular Stress and Apoptosis. Reproductive sciences. 2016;23(8):993–1000. doi: 10.1177/1933719115625846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Earle JA, McMenomy T. Hippocampal CA4 Reelin-positive neurons. Molecular psychiatry. 2000;5(6):571. doi: 10.1038/sj.mp.4000794. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR. Altered levels of Bcl2 and p53 proteins in parietal cortex reflect deranged apoptotic regulation in autism. Synapse. 2001;42(4):281–4. doi: 10.1002/syn.10002. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Realmuto GM, Jalali-Mousavi M. Reduction in anti-apoptotic protein Bcl-2 in autistic cerebellum. Neuroreport. 2001;12(5):929–33. doi: 10.1097/00001756-200104170-00013. [DOI] [PubMed] [Google Scholar]
- Frazier TW, Georgiades S, Bishop SL, Hardan AY. Behavioral and cognitive characteristics of females and males with autism in the Simons Simplex Collection. Journal of the American Academy of Child and Adolescent Psychiatry. 2014;53(3):329–40 e1. 3. doi: 10.1016/j.jaac.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RE, James SJ. Metabolic pathology of autism in relation to redox metabolism. Biomarkers in medicine. 2014;8(3):321–30. doi: 10.2217/bmm.13.158. [DOI] [PubMed] [Google Scholar]
- Frye RE, Melnyk S, Fuchs G, Reid T, Jernigan S, Pavliv O, Hubanks A, Gaylor DW, Walters L, James SJ. Effectiveness of methylcobalamin and folinic Acid treatment on adaptive behavior in children with autistic disorder is related to glutathione redox status. Autism research and treatment. 2013;2013:609705. doi: 10.1155/2013/609705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita-Jimbo E, Tanabe Y, Yu Z, Kojima K, Mori M, Li H, Iwamoto S, Yamagata T, Momoi MY, Momoi T. The association of GPR85 with PSD-95-neuroligin complex and autism spectrum disorder: a molecular analysis. Molecular autism. 2015;6:17. doi: 10.1186/s13229-015-0012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita E, Dai H, Tanabe Y, Zhiling Y, Yamagata T, Miyakawa T, Tanokura M, Momoi MY, Momoi T. Autism spectrum disorder is related to endoplasmic reticulum stress induced by mutations in the synaptic cell adhesion molecule, CADM1. Cell death & disease. 2010;1:e47. doi: 10.1038/cddis.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths KK, Levy RJ. Evidence of Mitochondrial Dysfunction in Autism: Biochemical Links, Genetic-Based Associations, and Non-Energy-Related Mechanisms. Oxidative medicine and cellular longevity. 2017;2017:4314025. doi: 10.1155/2017/4314025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Yu J, Dong D, Zhou Q, Wang JY, Fang S, Yang P. High Glucose-Repressed CITED2 Expression Through miR-200b Triggers the Unfolded Protein Response and Endoplasmic Reticulum Stress. Diabetes. 2016;65(1):149–63. doi: 10.2337/db15-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halladay AK, Bishop S, Constantino JN, Daniels AM, Koenig K, Palmer K, Messinger D, Pelphrey K, Sanders SJ, Singer AT, Taylor JL, Szatmari P. Sex and gender differences in autism spectrum disorder: summarizing evidence gaps and identifying emerging areas of priority. Molecular autism. 2015;6:36. doi: 10.1186/s13229-015-0019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JC, Pauly M, Melnyk S, Pavliv O, Starrett W, Crook TA, James SJ. Dietary intake and plasma levels of choline and betaine in children with autism spectrum disorders. Autism research and treatment. 2013;2013:578429. doi: 10.1155/2013/578429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendren RL, James SJ, Widjaja F, Lawton B, Rosenblatt A, Bent S. Randomized, Placebo-Controlled Trial of Methyl B12 for Children with Autism. Journal of child and adolescent psychopharmacology. 2016;26(9):774–783. doi: 10.1089/cap.2015.0159. [DOI] [PubMed] [Google Scholar]
- Howsmon DP, Kruger U, Melnyk S, James SJ, Hahn J. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS computational biology. 2017;13(3):e1005385. doi: 10.1371/journal.pcbi.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain research. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- Ichim G, Tait SW. A fate worse than death: apoptosis as an oncogenic process. Nature reviews. Cancer. 2016;16(8):539–48. doi: 10.1038/nrc.2016.58. [DOI] [PubMed] [Google Scholar]
- Idring S, Lundberg M, Sturm H, Dalman C, Gumpert C, Rai D, Lee BK, Magnusson C. Changes in prevalence of autism spectrum disorders in 2001–2011: findings from the Stockholm youth cohort. Journal of autism and developmental disorders. 2015;45(6):1766–73. doi: 10.1007/s10803-014-2336-y. [DOI] [PubMed] [Google Scholar]
- Iurlaro R, Munoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. The FEBS journal. 2016;283(14):2640–52. doi: 10.1111/febs.13598. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T Paris Autism Research International Sibpair, S. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature genetics. 2003;34(1):27–9. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, Neubrander JA. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. The American journal of clinical nutrition. 2004;80(6):1611–7. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Orchestrating the unfolded protein response in health and disease. The Journal of clinical investigation. 2002;110(10):1389–98. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Weng H, Xu C, Reece EA, Yang P. Oxidative stress-induced JNK1/2 activation triggers proapoptotic signaling and apoptosis that leads to diabetic embryopathy. Diabetes. 2012;61(8):2084–92. doi: 10.2337/db11-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xu C, Yang P. c-Jun NH2-terminal kinase 1/2 and endoplasmic reticulum stress as interdependent and reciprocal causation in diabetic embryopathy. Diabetes. 2013;62(2):599–608. doi: 10.2337/db12-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue SE, Cleary P, Saveljeva S, Samali A. New directions in ER stress-induced cell death. Apoptosis : an international journal on programmed cell death. 2013;18(5):537–46. doi: 10.1007/s10495-013-0818-6. [DOI] [PubMed] [Google Scholar]
- López-Hurtado E, Prieto JJ. A microscopic study of language-related cortex in autism. American Journal of Biochemistry and Biotechnology. 2008;4(2):130. doi: 10.3844/ajbbsp.2008.130.145. [DOI] [Google Scholar]
- Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345(6192):98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. ER chaperone functions during normal and stress conditions. Journal of chemical neuroanatomy. 2004;28(1–2):51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Mandic-Maravic V, Pejovic-Milovancevic M, Mitkovic-Voncina M, Kostic M, Aleksic-Hil O, Radosavljev-Kircanski J, Mincic T, Lecic-Tosevski D. Sex differences in autism spectrum disorders: does sex moderate the pathway from clinical symptoms to adaptive behavior? Scientific reports. 2015;5:10418. doi: 10.1038/srep10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk S, Fuchs GJ, Schulz E, Lopez M, Kahler SG, Fussell JJ, Bellando J, Pavliv O, Rose S, Seidel L, Gaylor DW, James SJ. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. Journal of autism and developmental disorders. 2012;42(3):367–77. doi: 10.1007/s10803-011-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momoi T, Fujita E, Senoo H, Momoi M. Genetic factors and epigenetic factors for autism: endoplasmic reticulum stress and impaired synaptic function. Cell biology international. 2010;34(1):13–9. doi: 10.1042/CBI20090250. [DOI] [PubMed] [Google Scholar]
- Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS letters. 2007;581(19):3641–51. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, Bryson S, Carver LJ, Constantino JN, Dobkins K, Hutman T, Iverson JM, Landa R, Rogers SJ, Sigman M, Stone WL. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128(3):e488–95. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, Sacco R, Hager J, Rousseau F, Curatolo P, Manzi B, Militerni R, Bravaccio C, Trillo S, Schneider C, Melmed R, Elia M, Lenti C, Saccani M, Pascucci T, Puglisi-Allegra S, Reichelt KL, Persico AM. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Molecular psychiatry. 2010;15(1):38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, Guthrie D, Ritvo A. Lower Purkinje cell counts in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research Report. The American journal of psychiatry. 1986;143(7):862–6. doi: 10.1176/ajp.143.7.862. [DOI] [PubMed] [Google Scholar]
- Rose S, Melnyk S, Pavliv O, Bai S, Nick TG, Frye RE, James SJ. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Translational psychiatry. 2012a;2:e134. doi: 10.1038/tp.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S, Melnyk S, Trusty TA, Pavliv O, Seidel L, Li J, Nick T, James SJ. Intracellular and extracellular redox status and free radical generation in primary immune cells from children with autism. Autism research and treatment. 2012b;2012:986519. doi: 10.1155/2012/986519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol DA, Frye RE. Evidence linking oxidative stress, mitochondrial dysfunction, and inflammation in the brain of individuals with autism. Frontiers in physiology. 2014;5:150. doi: 10.3389/fphys.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochimica et biophysica acta. 2013;1833(12):3460–70. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh AM, Li X, Wen G, Tauqeer Z, Brown WT, Malik M. Cathepsin D and apoptosis related proteins are elevated in the brain of autistic subjects. Neuroscience. 2010a;165(2):363–70. doi: 10.1016/j.neuroscience.2009.10.035. [DOI] [PubMed] [Google Scholar]
- Sheikh AM, Malik M, Wen G, Chauhan A, Chauhan V, Gong CX, Liu F, Brown WT, Li X. BDNF-Akt-Bcl2 antiapoptotic signaling pathway is compromised in the brain of autistic subjects. Journal of neuroscience research. 2010b;88(12):2641–7. doi: 10.1002/jnr.22416. [DOI] [PubMed] [Google Scholar]
- Sogut S, Zoroglu SS, Ozyurt H, Yilmaz HR, Ozugurlu F, Sivasli E, Yetkin O, Yanik M, Tutkun H, Savas HA, Tarakcioglu M, Akyol O. Changes in nitric oxide levels and antioxidant enzyme activities may have a role in the pathophysiological mechanisms involved in autism. Clinica chimica acta; international journal of clinical chemistry. 2003;331(1–2):111–7. doi: 10.1016/s0009-8981(03)00119-0. [DOI] [PubMed] [Google Scholar]
- Spickett CM. The lipid peroxidation product 4-hydroxy-2-nonenal: Advances in chemistry and analysis. Redox biology. 2013;1:145–52. doi: 10.1016/j.redox.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Finelli J, Marvin A, Garrett-Mayer E, Bauman M, Landa R. Response to joint attention in toddlers at risk for autism spectrum disorder: a prospective study. Journal of autism and developmental disorders. 2007;37(1):37–48. doi: 10.1007/s10803-006-0335-3. [DOI] [PubMed] [Google Scholar]
- Sweeten TL, Posey DJ, Shankar S, McDougle CJ. High nitric oxide production in autistic disorder: a possible role for interferon-gamma. Biological psychiatry. 2004;55(4):434–7. doi: 10.1016/j.biopsych.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO reports. 2006;7(9):880–5. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gutierrez Rios P, Kuo SH, Akman HO, Rosoklija G, Tanji K, Dwork A, Schon EA, Dimauro S, Goldman J, Sulzer D. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiology of disease. 2013;54:349–61. doi: 10.1016/j.nbd.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich L, Favaloro FL, Trobiani L, Marchetti V, Patel V, Pascucci T, Comoletti D, Marciniak SJ, De Jaco A. Autism associated R451C mutation in Neuroligin3 leads to the activation of the unfolded protein response in a PC12 Tet-On inducible system. The Biochemical journal. 2015 doi: 10.1042/BJ20150274. 10.1042/BJ20150274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu C, Reece EA, Li X, Wu Y, Harman C, Yu J, Dong D, Wang C, Yang P, Zhong J, Yang P. Protein kinase C-alpha suppresses autophagy and induces neural tube defects via miR-129-2 in diabetic pregnancy. Nature communications. 2017;8:15182. doi: 10.1038/ncomms15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Alberts I, Li X. The apoptotic perspective of autism. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2014;36:13–8. doi: 10.1016/j.ijdevneu.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Zhiling Y, Fujita E, Tanabe Y, Yamagata T, Momoi T, Momoi MY. Mutations in the gene encoding CADM1 are associated with autism spectrum disorder. Biochemical and biophysical research communications. 2008;377(3):926–9. doi: 10.1016/j.bbrc.2008.10.107. [DOI] [PubMed] [Google Scholar]
- Zoroglu SS, Yurekli M, Meram I, Sogut S, Tutkun H, Yetkin O, Sivasli E, Savas HA, Yanik M, Herken H, Akyol O. Pathophysiological role of nitric oxide and adrenomedullin in autism. Cell biochemistry and function. 2003;21(1):55–60. doi: 10.1002/cbf.989. [DOI] [PubMed] [Google Scholar]
- Zwaigenbaum L, Bryson SE, Szatmari P, Brian J, Smith IM, Roberts W, Vaillancourt T, Roncadin C. Sex differences in children with autism spectrum disorder identified within a high-risk infant cohort. Journal of autism and developmental disorders. 2012;42(12):2585–96. doi: 10.1007/s10803-012-1515-y. [DOI] [PubMed] [Google Scholar]