Abstract
Purpose
To develop a fast and automated volume-of-interest (VOI) prescription pipeline (AutoVOI) for single-voxel MR spectroscopy (MRS) that removes the need for manual VOI placement, allows flexible VOI planning in any brain region, and enables high inter- and intra-subject consistency of VOI prescription.
Methods
AutoVOI was designed to transfer pre-defined VOIs from an atlas to the 3D anatomical data of the subject during the scan. The AutoVOI pipeline was optimized for consistency in VOI placement (precision), enhanced coverage of the targeted tissue (accuracy) and fast computation speed. The tool was evaluated against manual VOI placement using existing T1-weighted datasets and corresponding VOI prescriptions. Finally, it was implemented on two scanner platforms to acquire MRS data from clinically-relevant VOIs that span the cerebrum, cerebellum and the brainstem.
Results
The AutoVOI pipeline includes skull stripping, non-linear registration of the atlas to the subject’s brain, and computation of the VOI coordinates and angulations using a minimum oriented bounding box algorithm. When compared against manual prescription, AutoVOI showed higher intra- and inter-subject spatial consistency, as quantified by generalized Dice coefficients (GDC), lower intra- and inter-subject variability in tissue composition (gray matter, white matter and cerebrospinal fluid) and higher or equal accuracy, as quantified by GDC of prescribed VOI with targeted tissues. High quality spectra were obtained on Siemens and Philips 3T systems from 6 automatically prescribed VOIs by the tool.
Conclusion
Robust automatic VOI prescription is feasible and can help facilitate clinical adoption of MRS by avoiding operator dependence of manual selection.
Keywords: MRS, Automation, Registration, Prescription, Generalized Dice Coefficient
Introduction
The acquisition of single-voxel Magnetic Resonance Spectroscopy (MRS) data in the human brain currently involves the manual selection of the volume-of-interest (VOI) by MR technologists, clinicians such as radiologists, or scientists. This process requires an understanding of neuroanatomy as well as familiarity with MRS acquisition processes. Consistent VOI prescription within and across subjects is crucial to minimize variability in metabolite quantification that results from inconsistencies in VOI position and tissue composition. Achieving high consistency in VOI prescriptions across subjects is particularly challenging due to anatomical variations between subjects (1). The issue can be exacerbated when the data are acquired at multiple sites and/or by different technologists. Hence, the use of automated VOI prescription is highly desirable to reduce the operator-induced variability in volume selection for MRS.
Existing automated prescription methods for MR imaging on clinical scanners, such as the Siemens (Erlangen, Germany) AutoAlign (2), align scout images of a subject to predefined landmarks (3) or an average atlas (4), and guarantee that the subsequent images are acquired in a consistent orientation. Such methods facilitate consistent VOI prescription in follow-up scans of the same subject in both single-voxel (5,6) and multi-voxel MR spectroscopic imaging (MRSI) applications (7). However, they do not guarantee that any particular brain structure is aligned across subjects due to inter-individual anatomical variability. They also require manual VOI prescription during the initial reference scan. Another automated VOI prescription method for single-voxel MRS computes VOIs by registering pre-segmented brain regions and calculating the tightest fitting oriented bounding box (8) around the segmented region-of-interest (ROI). However, this approach requires lengthy off-line processing of brain segmentation and thereby is unlikely to be feasible within the clinical workflow. Additionally, it only allows prescription of VOIs in structures that can be segmented.
The feasibility of automated prescription of MRSI volumes and outer volume suppression (OVS) slices was shown using affine registration of an atlas to the subject while the subject is in the scanner (9–13). Affine registration was preferable for these applications due to its speed and the time limitations during clinical scans. However, affine registration may not sufficiently capture local anatomical variations within the brain. Such variations are more critical for VOI selection in single-voxel spectroscopy than for MRSI that allows retrospective selection of ROI within the investigated volume. There is currently no method to automatically prescribe VOIs for single voxel MRS without reference to manual VOI prescription during a baseline acquisition. To eliminate the need for manual VOI prescription, we set out to develop an on-line automated VOI prescription tool, named AutoVOI, for single-voxel MRS that can capture inter-individual anatomical variability by leveraging recent improvements in computational efficiency of non-linear registration methods.
To allow flexibility in VOI planning, we designed the tool to transfer VOIs defined on an atlas to subject space during the scanning session. Design criteria included inter-subject consistency of VOI placement (precision), optimal coverage of the targeted tissue (accuracy) and fast computation speed. To address precision, we compared inter-subject consistency of VOI prescription using affine (linear with translation) and non-linear (b-spline) registrations of the atlas to the subject’s brain. We hypothesized that non-linear registration would be necessary to accommodate variations in anatomical features. We further investigated inter-subject consistency of VOI content (gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF)). To address accuracy, we investigated overlap of automatically placed VOIs with the targeted tissue obtained from state-of-the-art segmentation tools. To address speed, we tested the feasibility of completing the AutoVOI pipeline, including registration, computation of the VOI coordinates and angulation, and transfer between the scanner and the computation server, within the constraints of a clinical scan (under one minute). Once the components of the AutoVOI pipeline were optimized based on these criteria, we determined if AutoVOI improves precision and accuracy of VOI prescription over manual placement by expert MRS investigators and MR technologists, using 100 existing T1-weighted MPRAGE datasets and corresponding VOI prescriptions from prior publications (14–17). Finally, we implemented the AutoVOI pipeline on two widely used clinical scanner platforms (Siemens and Philips) and acquired in vivo MRS data from 6 clinically-relevant VOIs that span the cerebrum, cerebellum and the brainstem.
Methods
Design and Optimization of the AutoVOI
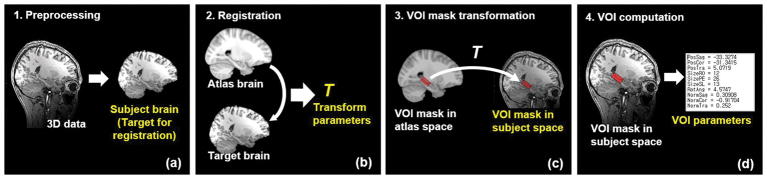
The proposed AutoVOI pipeline, shown in Figure 1, utilizes registration-based automatic prescription that aligns an atlas to the 3D T1-weighted MPRAGE structural scan of a subject in the scanner. AutoVOI uses 3D T1-weighted data that are typically taken for VOI prescription in MRS acquisitions. However, it can be extended to any other MR contrasts (e.g. T2-weighted) as long as good registration performance can be achieved. Prior to running registration, the T1-weighted data undergo preprocessing steps (Figure 1a). Then, a brain atlas is registered to the subject’s brain data to estimate the transformation parameters from atlas (generic or custom) to the subject’s space (Figure 1b). The VOI mask(s), defined once on the atlas by an expert, are transformed to the subject space using the parameters obtained from the registration step (Figure 1c). Multiple VOIs can be computed in a single run as the same registration parameters can be applied to different VOI definitions. After transformation, each VOI mask is converted into a 3D point cloud, and the coordinates, angulations and size of the VOIs are computed using a minimal oriented bounding box (OBB) algorithm (18) (Figure 1d). Finally, the position, dimension and orientation of the cuboid-shaped OBB are exported to the MR scanner. For the VOI dimensions, users can choose between values computed from the OBB estimation or a predetermined value. In this study, the dimensions of the VOIs were set to predetermined values, as is common in MRS studies.
Figure 1.
Overview of automatic VOI prescription for single-voxel spectroscopy. (a) DICOM images of the subject’s head are used to create a NIFTI file of the skull-stripped brain data. (b) Running the registration of the atlas to the skull-stripped subject’s brain yields transformation parameters from atlas space to subject space. (c) Transformation of VOI masks from atlas space to subject space using the transformation obtained from the registration. (d) Computation of VOI coordinates, angulation and size and writing of this information to the file that enables MRI scanners to retrieve and use any of the computed VOI in the pulse sequence.
To build the pipeline, we first investigated if any preprocessing of the head MRI data was necessary (Step 1, Figure 1a) and evaluated existing brain extraction routines for this purpose (19–21). For the registration step (Step 2, Figure 1b), we evaluated registration algorithms for speed, and thereby for feasibility to utilize them online during a clinical scan. Next, we tested the effect of registration algorithm on inter-subject VOI prescription consistency (precision) by comparing inter-subject variability in prescription of three VOIs of high relevance for neurodegenerative diseases with affine and non-linear (b-spline) registration. VOIs covering the posterior cingulate cortex (PCC, 20×20×20mm3), left hippocampus (LHC, 13×12×26mm3) and cerebellar vermis (Vermis, 10×25×25mm3) were defined on the T1-weighted MNI152 brain atlas with 1 mm3 spatial resolution (22,23), using anatomical landmarks employed by previous studies (14,15,24). The MNI ICBM 152 6th generation atlas is an average of 152 T1-weighted MRI scans, and is available from the MNI website and FSL (25). T1-weighted MPRAGE data of 19 subjects (8 females, age 31±8) from three different 3T scanners were utilized: 7 from a Siemens Trio 3T at the University of Minnesota - Center for Magnetic Resonance Research (CMRR), 5 from a Siemens Verio 3T at the Korea Advanced Institute of Science and Technology (KAIST), and 7 from a Philips Achieva 3T at Johns Hopkins University (JHU). All datasets had a spatial resolution of 1 mm3, and were acquired with TR/TI/TE = 2530/1100/3.65ms (Trio), 1900/900/2.48ms (Verio), and 3000/1100/3.78ms (Achieva). All subjects were enrolled after giving informed consent approved by the Institutional Review Board at each participating institution. All shared datasets were de-identified to meet HIPAA compliance.
The final steps for computing VOI coordinates involve transformation of the VOI mask from the atlas to the subject space (Step3, Figure 1c), followed by conversion of the transformed mask to the point cloud format, and OBB computation (Step 4, Figure 1d). We compared singular value decomposition (SVD) and polygonal surface-based (18) minimization method (brute-force method) for OBB computation.
The precision and accuracy metrics that were used during the design and evaluation of the tool are described below, under “Metrics for evaluating VOI prescription performance”.
Comparison of AutoVOI Performance with Manual VOI prescription
For an objective performance assessment of AutoVOI against manual placement, 100 3D T1-weighted datasets and respective manual VOI prescription results from previously published studies (14–17) were used, where the VOIs were prescribed without the knowledge that the manual placement was going to be compared to an automated method.
First, datasets from two studies on test-retest reproducibility of MRS data acquisitions were used to measure inter- and intra-subject spatial consistency and accuracy of AutoVOI vs. manual placement by expert MRS investigators. The first study involved MRS scans of LHC on 10 subjects with each subject scanned twice (14). The second study involved MRS scans of PCC and Vermis with six subjects each scanned four times (15).
Next, MRI data from healthy elderly subjects and VOI prescriptions by MR technologists were used to evaluate the performance of AutoVOI in a clinical setting. The VOIs were prescribed by one of three technologists for each MRS acquisition in this existing dataset (16,17). As the elderly brain usually presents atrophy, this analysis utilized both the MNI152 atlas, which was generated from a younger cohort, and a custom atlas generated from elderly subject images. 3D T1-weighted MPRAGE datasets for 62 healthy elderly subjects (age 80±6) from two previously published studies (16,17) were utilized for this analysis. Of these, 30 datasets were used to generate the custom atlas, and the remaining 32 were used for quantifying the performance of AutoVOI with custom and MNI152 atlas against manual VOI prescriptions by MR technologists. The Advanced Normalization Tools (ANTS) (26) was used to construct the elderly brain atlas, by spatial normalization of the 30 elderly datasets. The elderly atlas then underwent skull stripping with ROBEX (21).
System integration and multi-vendor in vivo testing
In vivo MRS data were acquired using the pipeline on a Siemens 3T Prisma scanner (CMRR), and a Philips 3T Achieva scanner (JHU). A dedicated processing server installed at CMRR (Intel Xeon E5-2650@2.6 Ghz, 64 GB-RAM, NVIDIA Tesla K80 24 GB graphics card) ran AutoVOI. The AutoVOI pipeline was integrated on these systems such that once AutoVOI is initiated, a batch script running on the MR console PC securely transmits compressed NIFTI files to the processing server. NIFTI files without patient information were chosen as the format for file transfer in order to meet HIPAA compliance. The batch script periodically checks the status of computation (on the server) after transmission, and retrieves the computed VOI coordinates to the MR console once the VOI calculation is finished. The pulse sequence then reads the text file containing the VOI coordinates, and allows the MR operator to select and review the VOI before deciding whether to proceed or to adjust the automatically prescribed VOI.
Healthy participants (N=5; all male, age= 39±13 years) were scanned to acquire MR spectra from clinically relevant VOI on the Siemens (CMRR, N=2) and Philips (JHU, N=3) platforms. Subjects were scanned after giving informed consent approved by the Institutional Review Board. MRS data were acquired from VOIs automatically prescribed by AutoVOI in PCC, LHC, Vermis, right cerebellar white matter (CBWM, 17×17×17 mm3), pons (16×16×16 mm3) and right putamen (Putamen, 10×25×11 mm3). Following first and second order B0 shimming, MR spectra were acquired using the modified semi-LASER (sLASER) sequence (27) (TR/TE = 5000/30ms, 64 transients), as described previously (17). Unsuppressed water scans were obtained for eddy current correction (24). Single-shot spectra were corrected for eddy-current, frequency and phase variations prior to summation using MRspa (28). These data were acquired with the purpose of demonstrating feasibility of utilizing the AutoVOI on different scanner platforms and spectra are therefore provided for visual comparisons only rather than for a quantitative analysis.
Metrics for evaluating VOI prescription performance
The goal of automated VOI prescription is to achieve superior inter- and intra-subject consistency, and greater accuracy over manual prescriptions. To reflect this, we employed the following quantitative metrics to evaluate spatial consistency, consistency in VOI composition and accuracy of VOI prescription.
Spatial consistency
Normalization
In order to evaluate the spatial consistency of VOI prescription, VOIs prescribed either by AutoVOI or human experts have to be normalized to a standard space to measure their overlap. Elastix (29), based on the Insight Toolkit library (30), was chosen as an independent normalization technique for the purpose of this evaluation, in order to avoid normalizing masks with BROCCOLI (31), since AutoVOI utilizes BROCCOLI within the pipeline.
For inter-subject VOI spatial consistency analysis, non-linear registration was used for normalization of head data and transformation of VOI masks to account for structural variations in different subjects. Rigid registration was used for intra-subject analyses as no anatomical changes are expected between scan sessions of the same subject. All head data underwent skull stripping with ROBEX (21) prior to registration. ROBEX was chosen as an independent skull stripping technique for the purpose of this evaluation as it was less prone to errors on atrophied brains than BET2 (19) used in the AutoVOI pipeline.
Generalized Dice Coefficient
The Dice coefficient (32) is a metric commonly used to evaluate the overlap between binary masks, and can be calculated from the Tannimoto (TC, also known as Jaccard) coefficient (33). However, AutoVOI requires spatial resampling (e.g. interpolation) of the atlas VOI masks to adapt them to each subject’s anatomy and the resulting masks contain fractional values (non-binary), which the Dice and TC cannot handle. Hence, we used the modified forms of Dice and TC that consider fractional labels, and are respectively referred to as the generalized Dice coefficient (GDC) (34) and the generalized Tannimoto coefficient (GTC), as defined in Equations 1 and 2, to quantify the spatial overlap between a large number of fractional masks. The GDC and GTC values are computed as follows,
| [1] |
and
| [2] |
where GTC is computed as the sum of fuzzy intersections (MIN) over the sum of fuzzy unions (MAX) for every pair (α, β) from the set of N normalized masks. Equation 2 is a special case of Equation 2 in Crum et al (34). Like Dice and TC, GDC and GTC are intended to have the minimum value of 0 when none of the masks overlap, and the maximum value of 1 when there is a complete overlap for all masks.
Consistency of VOI composition
The tissue composition of the prescribed VOI provided another metric for consistency of VOI selection. Contributions of WM, GM and CSF to each VOI were analyzed using the tissue segmentation module of SPM12 (35). Standard deviation (SD) in the tissue fraction estimation across subjects was calculated to assess the compositional consistency of the selected VOIs.
Accuracy of VOI prescription
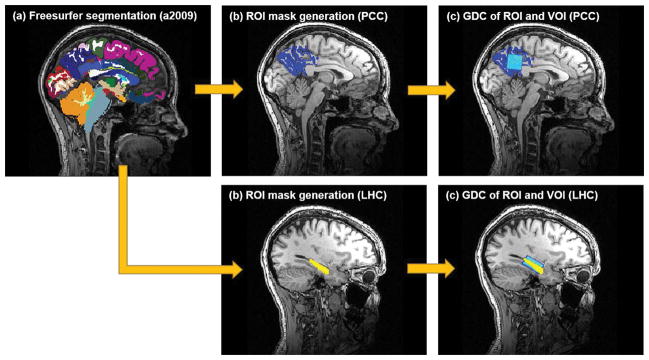
In addition to inter- and intra-subject spatial/compositional consistency analysis, VOI’s GDC with targeted ROI was measured to assess the accuracy of the VOI prescription (Figure 2). Each 3D T1-weighted MPRAGE dataset underwent automated cortical segmentation using FreeSurfer (36) to obtain anatomical labels. Using the cortical parcellation results with Destrieux atlas (a2009) (37), binary masks of PCC with Precuneus and LHC on each subject were generated. Then, GDC overlap between the binary ROI mask and VOI mask generated from either AutoVOI or manual prescriptions were computed for each subject. Mean, SD and coefficient of variance (CV) were computed. Note that unlike spatial consistency, ROI overlap and tissue fraction analyses do not require any spatial normalization, and are performed in each individual subject’s native space.
Figure 2.
Visual description of ROI coverage analysis shows how the FreeSurfer segmentation result with Destrieux atlas (a2009) (a) is used to create a binary ROI mask (b). Once the binary ROI mask is obtained, GDC between the ROI and VOI mask is computed for the PCC and LHC prescription results.
Results
Design and Optimization of the AutoVOI
To build the AutoVOI pipeline, we first tested the need for preprocessing of subject’s T1-weighted MRI data to minimize registration errors. We found that registration errors were frequent if the images contained unwanted anatomical regions such as neck and shoulders. Therefore, skull stripping was deemed necessary in the pipeline (Step 1, Figure 1a). Advanced brain extraction routines, such as BET (which includes BET2 and BETsurf) (19), BEaST (20) and ROBEX (21) were tested but ultimately not considered suitable for the AutoVOI pipeline due to time constraints. BET2 with robust option (38) was chosen as the routine to remove any unnecessary regions outside the brain. Although BET skull stripping method satisfied the design time constraints, it was vulnerable to incorrect center-of-mass estimations. Therefore, an automated routine using FSLROI, a tool within FSL (25), was added prior to skull stripping to automatically remove additional problematic areas such as the lower neck, shoulders or phase-wrap artifact of the nose (Supporting Figure S1).
For the atlas-to-brain registration module (step 2, Figure 1b), computation speed was a critical consideration. To execute AutoVOI within a time comparable to manual VOI prescriptions, the target computation time was set to be within one minute on a regular desktop PC. Therefore standard registration programs such as FLIRT (39) and FNIRT (40) of the FSL (25) suite, SPM12 (35) or AFNI (41) were not suitable. Instead, GPU-based registration methods Elastix (29) and BROCCOLI (31), respectively based on the Insight Toolkit (ITK) library (30) and OpenCL (42), were considered as they offer fast non-linear registration. Ultimately, BROCCOLI was used with 5 iterations of affine and 3 iterations of non-linear registrations, as it offered a good compromise between speed and VOI prescription performance.
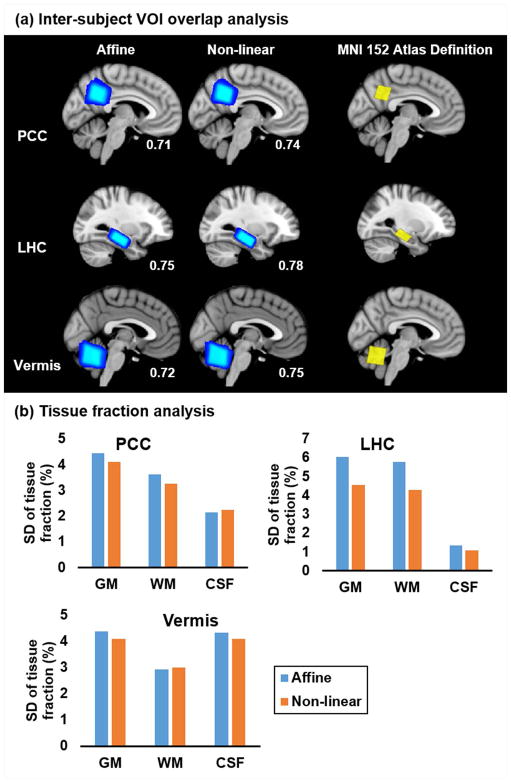
We then compared linear and non-linear registration methods to evaluate if non-linear registration provided improvement over affine registration in precision and accuracy of VOI prescription, justifying the extra time needed for non-linear registration. The time spent for registration, using BROCCOLI, was around 1.2 seconds for affine registration only and 1.7 seconds for full registration including non-linear registration. Non-linear (b-spline) registration provided higher GDC values, thus better inter-subject overlap, over affine registrations for three clinically-relevant VOIs (PCC, LHC and Vermis) tested with 19 T1-weighted datasets obtained from a healthy adult cohort (Figure 3a). For WM/GM/CSF tissue fraction estimations, lower SD values were obtained with non-linear (compared to affine) registration in 2 of the 3 VOIs (PCC and LHC) with the remaining third VOI (Vermis) having a comparable SD, confirming lower or equivalent inter-subject variance in VOI tissue content of select regions (Figure 3b). ROI overlap with targeted tissue showed no significant differences between affine and non-linear registrations in PCC (0.24±0.03 for both affine and non-linear, p=0.84 paired, two-tailed Student’s t-test) and LHC (0.4±0.03 for both affine and non-linear, p=0.95 paired, two-tailed Student’s t-test). Because of the improvement in spatial and compositional consistency provided by non-linear over affine registration and because GPU-based non-linear registration could be executed within our time constraint, we chose to proceed with non-linear registration for step 2 of the AutoVOI pipeline.
Figure 3.
Spatial consistency and GDC values of the normalized (a) posterior cingulate (PCC), left hippocampus (LHC) and cerebellar vermis (Vermis) prescriptions for affine and non-linear registration show that non-linear registration leads to higher inter-subject consistency over affine registration, as indicated by minimal areas of dark blue pixels and higher GDC values than those with affine registration. SD of VOI tissue fraction estimations (b) show that non-linear registration has lower variability than affine registration, confirming greater inter-subject consistency.
For the OBB computation, the brute-force method (18) was chosen over SVD as it consistently yielded a tighter fitted volume. Furthermore, it was practical to use the brute-force method because the polygonal representation of the warped VOI mask contains a relatively small number of triangles. Nonetheless, the convex hull using the quickhull algorithm (43) was used to decrease the number of triangles used in the OBB calculation. The overall time for computing OBB was between 1 to 2 seconds.
The average processing time for the entire pipeline, including read and write processes of NIFTI files, on 19 datasets was 32.5±4.2 sec with NVIDIA GTX650 graphics card and 20.9±5.6 seconds with NVIDIA GTX1060 graphics card, with the BROCCOLI affine and non-linear registration step taking less than two seconds to complete.
Performance evaluation against manual prescription
We next evaluated if VOI prescription with AutoVOI improved inter- and intra-subject consistency (precision) and accuracy in PCC, LHC and Vermis over expert manual VOI prescriptions (Table 1) using data from two test-retest reproducibility studies (14,15). Intra-subject spatial consistency was higher, as indicated by higher GDC values, with AutoVOI vs. manual prescription for all 3 VOIs (Table 1a). Inter-subject spatial consistency was higher with AutoVOI for 2 of 3 VOIs, specifically for PCC and LHC, likely because of the ease of manual placement of VOIs in the vermis based on anatomical landmarks. Consistently, inter-subject analysis of tissue content showed very low SD in GM and WM fractions (1–2%) in the vermis, leaving little room for improvement with AutoVOI (Table 1b). On the other hand, consistent improvement in the SD of GM and WM content was apparent in PCC and LHC, where SD in GM and WM fractions were higher (3–5%) with manual prescription. A similar improvement in SDs of CSF fractions was not apparent with AutoVOI vs. manual prescription. To evaluate accuracy of prescription, VOI overlap with the PCC and LHC masks generated by FreeSurfer was computed; vermis was not included in this analysis because it is not generated by FreeSurfer. This analysis showed higher coverage of PCC ROI across subjects with AutoVOI than manual prescription, as well as lower inter-subject CV of GDC values for both PCC and LHC (Table 1c).
Table 1. Quantitative comparison of AutoVOI and manual VOI prescription.
Comparison of AutoVOI against manual VOI prescriptions by expert MRS investigators shows improved inter- and intra-subject spatial consistency (a), and inter-subject tissue content variations (b) by AutoVOI, indicating improvement of the precision of VOI prescriptions. In the inter-subject overlap, normalized VOI masks from each head data is used to compute a single inter-subject GDC. In contrast, intra-subject overlap first computes the GDC value among the VOI prescriptions of different scans of each subject, then computes the average and SD of the GDC values across subjects. ROI overlap analysis (c) confirms improvement of accuracy in VOI placement with AutoVOI over manual placement, based on higher GDC values and lower variability (standard deviation, SD and coefficient of variance, CV) among subjects.
| (a) Inter and intra-subject VOI overlap analysis
| ||||
|---|---|---|---|---|
| VOI overlap analysis | Inter-subject overlap GDC | Intra-subject overlap Mean (SD) of GDC |
||
| AutoVOI | Manual | AutoVOI | Manual | |
| PCC | 0.79 | 0.45 | 0.90 (0.05) | 0.85 (0.06) |
| LHC | 0.77 | 0.72 | 0.89 (0.07) | 0.82 (0.06) |
| Vermis | 0.80 | 0.80 | 0.89 (0.04) | 0.85 (0.05) |
| (b) Tissue fraction analysis
| ||||||
|---|---|---|---|---|---|---|
| Tissue % Mean (SD) |
GM | WM | CSF | |||
| AutoVOI | Manual | AutoVOI | Manual | AutoVOI | Manual | |
| PCC | 70.24 (3.85) | 68.61 (3.81) | 19.06 (1.54) | 20.18 (3.22) | 10.70 (3.39) | 11.21 (1.65) |
| LHC | 61.52 (4.08) | 64.40 (5.55) | 33.27 (2.44) | 30.96 (5.11) | 5.20 (2.56) | 4.64 (2.19) |
| Vermis | 72.21 (1.50) | 69.49 (2.21) | 19.13 (1.74) | 20.30 (1.60) | 8.66 (2.00) | 10.21 (3.00) |
| (c) ROI overlap analysis
| ||||
|---|---|---|---|---|
| ROI overlap (GDC) | PCC | LHC | ||
| AutoVOI | Manual | AutoVOI | Manual | |
| Mean (SD) | 0.23 (0.02) | 0.17 (0.03) | 0.46 (0.03) | 0.47 (0.04) |
| CV | 11.01 | 19.41 | 6.97 | 8.35 |
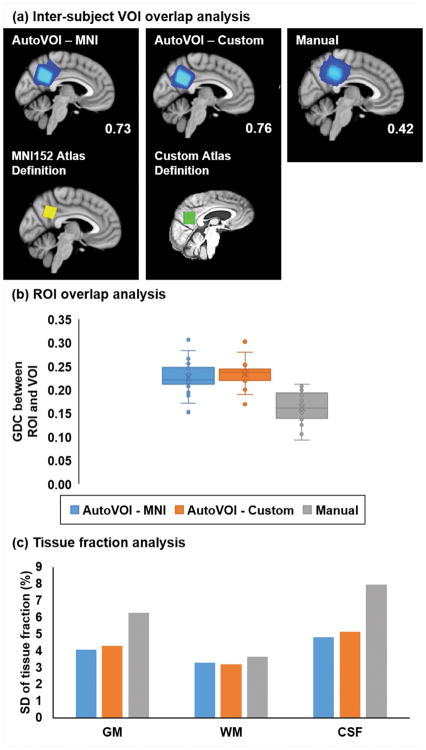
An existing dataset from an elderly cohort (16,17) allowed evaluation of the AutoVOI tool against manual prescription by multiple MR technologists, the ultimate targeted user base for the tool. AutoVOI using either MNI152 or a custom elderly atlas showed substantially improved GDC values (Figure 4a), higher overlap with the targeted tissue in PCC (Figure 4b) and lower variability in tissue content (Figure 4c) over manual prescription by MR technologists. The custom atlas showed marginal advantages over MNI152 atlas with higher GDC and smaller variability in ROI overlap. Note that automatic segmentation in FreeSurfer failed in 6 datasets, hence these were not included in the ROI overlap analysis. In addition, BET skull stripping failed in one dataset, omitting the cerebellum, and therefore was not included in the analysis. Finally, registration of one dataset to the MNI152 atlas failed (outlier VOI mask in Figure 4a), however that particular dataset was left in the analysis since the registration was successful with the custom atlas.
Figure 4.
Comparison of AutoVOI against manual prescriptions of a posterior cingulate cortex voxel by three rotating MR technologists in an elderly cohort (N=32). AutoVOI was run both with the MNI atlas and a custom elderly atlas generated from 30 independent elderly datasets. The precision of VOI prescription was improved with AutoVOI vs. manual prescription based on greater spatial overlap, as shown by higher GDC values (shown in the corner of each panel) (a) in AutoVOI. Improved accuracy was shown by higher mean overlap with intended ROI (b) using AutoVOI. Precision of VOI prescription was also enhanced with lower tissue content variability (c) between subjects. The custom atlas shows features commonly seen among elderly subjects including enlarged ventricles and head posture that is tilted backwards. The custom atlas allowed small improvement in inter-subject consistency (slightly higher inter-subject GDC and smaller SD in ROI overlap) compared to standard MNI152 atlas.
System integration and multi-vendor in vivo testing
AutoVOI was implemented on 3T Siemens and Philips systems for in vivo data acquisition at two sites (CMRR, JHU). The processing time of AutoVOI for a single VOI prescription was ~50 seconds with ~10 seconds spent for AutoVOI computation and ~40 seconds for file preparations and transmission between MR consoles at the two institutions and the processing server at CMRR. If more than one VOI were requested, ~10 seconds were added to the total computation time for each additional VOI. This was comparable to time required for manual prescriptions, which involves additional steps such as 3D re-slicing of image data before performing manual prescriptions of VOIs.
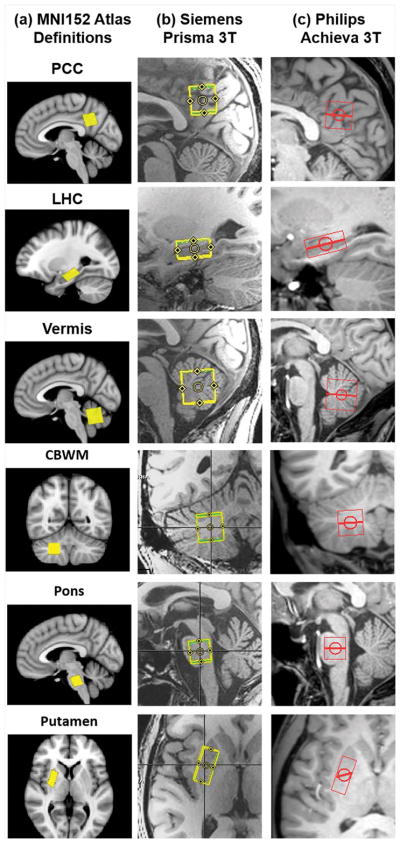
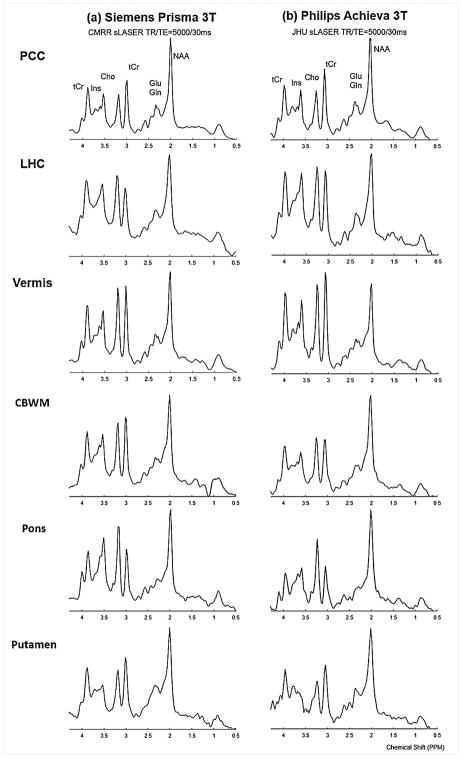
The six clinically-relevant VOIs that were defined on the MNI atlas for these in vivo acquisitions were prescribed consistently on both systems (Figure 5) and spectra of comparable quality were obtained from these automatically prescribed VOIs (Figure 6).
Figure 5.
Atlas definitions for six clinically-relevant VOIs (PCC - posterior cingulate cortex/LHC - left hippocampus/Vermis - cerebellar vermis/CBWM - right cerebellar white matter/Pons/Putamen - right putamen) over the MNI152 atlas (a), and AutoVOI prescriptions on (b) Siemens and (c) Philips systems that were based on these definitions.
Figure 6.
MR Spectra acquired from the six VOIs (PCC - posterior cingulate cortex/LHC - left hippocampus/Vermis: cerebellar vermis/CBWM - right cerebellar white matter/Pons/Putamen - right putamen) shown in Fig. 5 prescribed using AutoVOI on (b) Siemens and (c) Philips 3T systems at the University of Minnesota - Center for Magnetic Resonance Research (CMRR) and Johns Hopkins University (JHU) respectively. Spectra were acquired with a semi-LASER (sLASER) pulse sequence at TR/TE = 5000/30ms and 64 transients. Spectra were apodized with line broadening (1 Hz) and Gaussian multiplication (σ=0.12 s) for display purposes.
Discussion
In this study, we have developed a tool that can automatically prescribe VOIs for single voxel MRS data acquisitions within 1 minute during an MRS scan session and with high intra- and inter-subject consistency. The proposed tool was tested for precision and accuracy on existing T1-weighted datasets from published studies with varying imaging parameters. In addition, in vivo testing was performed on Siemens and Philips 3T systems to demonstrate the feasibility of automated prescription of clinically-relevant VOIs. The proposed atlas-based AutoVOI tool with non-linear registration yields higher inter- and intra-subject precision and accuracy compared to manual prescription.
Non-linear registration was chosen for the AutoVOI pipeline because it yielded more consistent results than affine registration across subjects, with higher GDC values for spatial overlap, and lower SD values for the estimated tissue (WM/GM/CSF) fractions (Figure 3). We have shown that advanced methods, such as BROCCOLI registration algorithm, and the use of advanced graphics cards (GPU) make it possible to integrate fast non-linear registration methods into the pipeline, with minimal burden on the overall performance. Hence, for atlas-based automatic VOI prescription in multi-subject MRS studies, non-linear registration must be considered for consistent outcomes.
In the analysis of VOI prescription precision and accuracy against prescriptions by expert MRS investigators, AutoVOI showed superior or equivalent prescription performance in both inter- and intra-subject analyses. The advantages of AutoVOI over manual prescription were more profound in the prescription of PCC compared to LHC and Vermis. This could be due to the spatial spread of the ROI for PCC (Figure 2). As the region for PCC is generally large, manual VOI prescriptions for PCC are often not in oblique orientations, regardless of the position and orientation of the subject’s head. There was little improvement for the vermis VOIs with AutoVOI vs. expert prescription because the corners of the VOIs are well defined by the boundaries of the cerebellum, simplifying manual prescriptions. Therefore, greater benefits in spatial and compositional consistency and accuracy are expected when using AutoVOI for cerebral cortical regions that are more prone to variability in manual prescription, even by experts.
The advantages of AutoVOI against manual prescription were more apparent in the typical clinical setting where different MR technologists were involved in voxel placement depending on clinical schedules. Namely, AutoVOI prescriptions using either the MNI152 or custom elderly atlas showed benefits in all 3 metrics we used, demonstrating improved spatial and compositional consistency and accuracy over manual prescriptions. Importantly, the performance by MR technologists (Figure 4) was comparable to manual prescription on PCC by expert MRS investigators in the previous analysis (Table 1) based on GDC (14,15), because these neuroradiology MR technologists were well-trained to position this voxel (15–17). Therefore, the improvements are likely to be more profound for technologists less familiar with the placement of particular VOIs. Between the atlases, AutoVOI with the custom atlas showed marginal improvement in inter-subject VOI prescription consistency in ROI overlap over the use of the standard MNI152 atlas. As the MNI152 atlas was created using brain data from a younger population, it lacks the characteristic atrophy seen in elderly subjects, which may decrease overall performance with this atlas. While this analysis demonstrated some benefits of using a custom atlas for the target cohort to facilitate robust automatic VOI prescriptions, it also showed that use of the standard MNI atlas allows acceptable automated prescription and that benefits over manual prescription are apparent even without the use of a custom atlas.
In-vivo tests at two institutions used AutoVOI running on a centralized server and demonstrated the potential to use AutoVOI in acquiring datasets with consistent voxel placement from sites with different MR systems. Executing AutoVOI in a central server ensures that all sites in a multi-site trial utilize the same VOI definitions and that any updates to the software are applied consistently and seamlessly across sites. Furthermore, secure data transfer across sites was feasible within the targeted execution time for AutoVOI. However, the tool can also be utilized in single site studies and further developed and modified if desired.
Although this study demonstrated the feasibility of an automated VOI prescription method through off-line evaluations using existing datasets and in vivo data acquisition, some limitations remain. First, the present study lacks testing on disease cohorts with significant brain deformations. Specifically, alterations caused by vascular, structural or mass lesions in the brain could cause substantial alterations in the T1-weighted structural data, and could yield incorrect registration results with MNI152 atlas designed for healthy subjects. Development and application of disease-specific atlases containing anatomical information to the AutoVOI pipeline could help mitigate some of these issues.
Another limitation is that the analysis presented here used fixed VOI dimensions. The evaluations presented in this work have the VOI dimensions fixed to a predetermined value because the manual VOI prescriptions also had the same volume across subjects and it is a common practice to have a fixed volume when acquiring single voxel MRS data in clinical studies. In traditional MRS scan protocols, it is challenging to incorporate manual calculation of VOI volumes based on the subject’s head volume with high consistency during the VOI prescription and any changes in the VOI size that the operator implements are likely to be subjective. Since the registration step yields scaling factors and allows the computation of customized VOI size with ease, incorporation of variable VOI is also feasible in AutoVOI.
Finally, optimization of the AutoVOI code and imaging parameters could further enhance speed and accuracy as the current AutoVOI pipeline utilizes existing registration algorithms intended primarily for functional MRI or voxel-based morphometry analyses. Imaging parameters of the reference structural 3D data, such as resolution, TE, TI and TR, could bring further performance enhancements in both reducing the computation time as well as improving the inter-subject consistency (44). Migration of the aforementioned advanced skull stripping techniques to GPUs may also improve registration speed and accuracy.
Conclusion
Here we demonstrated the feasibility of a fast and consistent inter-subject automatic VOI prescription. We have shown higher intra- and inter-subject spatial consistency and accuracy with AutoVOI over manual VOI prescription, which are expected to be of benefit in future clinical trials with single-voxel MRS as an outcome measure for accurate assessment of treatment efficacy. We expect that automation of VOI prescription will enhance the ability of MR technologists to acquire high quality MRS data by reducing operator dependence, and thus will lead to increased adoption of MRS in clinical settings.
Supplementary Material
Tests showed that BET skull stripping performs better when neck and aliased nose are removed from the 3D data. A dedicated script was integrated to AutoVOI to remove these regions automatically using FSLROI prior to performing BET. To select the head with FSLROI, the head dataset is first reoriented to the standard orientation. Then, the inferior section of the head image, specifically 200mm below the top of the head, is removed based on the average human head size (a). Intensity projection in the IF (inferior/superior)-direction is used to detect the top of the head, and 20cm was chosen as the vertical threshold. To detect nose aliasing, intensity projection in the AP direction is performed (b). Nose aliasing is present if both ends of the projection are above a certain intensity threshold (5% of the peak intensity value in the projection was used in AutoVOI).
Acknowledgments
We would like to thank Dr. Petr Bednařík, Dr. Karim Snoussi and Mr. Ian Cheong for providing data and feedback during our work.
This work was supported in part by NIH grants R01 NS080816, R01 NS070815, P41 EB015894, P30 NS076408 and S10OD017974. This work was also funded by the Ministry of Science and ICT (NRF 2014M3C7033999) and the Ministry of Health & Welfare (KHIDI HI14C1135) of Republic of Korea, and Minnesota Partnership for Biotechnology and Medical Genomics (MNP 12.15).
Footnotes
Code availability statement
The implementation and source code of the AutoVOI will be made freely available at http://www.cmrr.umn.edu/downloads.
References
- 1.Arthofer C, SoYoung K, Draper A, Stephenson MC, Jackson SR, Pitiot A, Morgan PS. How Variable is Manual MRS Volume Positioning within Subjects?. Proc. Org. for Human Brain Mapp. 21; Honolulu, Hawaii, USA. 1903. [Google Scholar]
- 2.van der Kouwe AJ, Benner T, Fischl B, Schmitt F, Salat DH, Harder M, Sorensen AG, Dale AM. On-line automatic slice positioning for brain MR imaging. Neuroimage. 2005;27(1):222–230. doi: 10.1016/j.neuroimage.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 3.Han Y, Park H. Automatic registration of brain magnetic resonance images based on Talairach reference system. J Magn Reson Imaging. 2004;20(4):572–580. doi: 10.1002/jmri.20168. [DOI] [PubMed] [Google Scholar]
- 4.Itti L, Chang L, Ernst T. Automatic scan prescription for brain MRI. Magn Reson Med. 2001;45(3):486–494. doi: 10.1002/1522-2594(200103)45:3<486::aid-mrm1064>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Hancu I, Blezek DJ, Dumoulin MC. Automatic repositioning of single voxels in longitudinal 1H MRS studies. NMR Biomed. 2005;18(6):352–361. doi: 10.1002/nbm.965. [DOI] [PubMed] [Google Scholar]
- 6.Dou W, Speck O, Benner T, Kaufmann J, Li M, Zhong K, Walter M. Automatic voxel positioning for MRS at 7 T. MAGMA. 2015;28(3):259–270. doi: 10.1007/s10334-014-0469-9. [DOI] [PubMed] [Google Scholar]
- 7.Ratai EM, Hancu I, Blezek DJ, Turk KW, Halpern E, Gonzalez RG. Automatic repositioning of MRSI voxels in longitudinal studies: impact on reproducibility of metabolite concentration measurements. J Magn Reson Imaging. 2008;27(5):1188–1193. doi: 10.1002/jmri.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Kouwe AJ, Balasubramanian M, Busa EA, Fischl B. Automatic prospective spectroscopy VOI placement based on brain segmentation. Proc. Intl. Soc. Mag. Reson. Med; Berlin, Germany. 2007. p. 773. [Google Scholar]
- 9.Martinez-Ramon M, Gallardo-Antolin A, Cid-Sueiro J, Heileman GL, Yung KT, Zheng W, Zhao C, Posse S. Automatic placement of outer volume suppression slices in MR spectroscopic imaging of the human brain. Magn Reson Med. 2010;63(3):592–600. doi: 10.1002/mrm.22275. [DOI] [PubMed] [Google Scholar]
- 10.Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer-volume suppression saturation bands. J Magn Reson Imaging. 2011;33(4):792–802. doi: 10.1002/jmri.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yung KT, Zheng W, Zhao C, Martinez-Ramon M, van der Kouwe A, Posse S. Atlas-based automated positioning of outer volume suppression slices in short-echo time 3D MR spectroscopic imaging of the human brain. Magn Reson Med. 2011;66(4):911–922. doi: 10.1002/mrm.22887. [DOI] [PubMed] [Google Scholar]
- 12.Ozhinsky E, Vigneron DB, Chang SM, Nelson SJ. Automated prescription of oblique brain 3D magnetic resonance spectroscopic imaging. Magn Reson Med. 2013;69(4):920–930. doi: 10.1002/mrm.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bian W, Li Y, Crane JC, Nelson SJ. Fully automated atlas-based method for prescribing 3D PRESS MR spectroscopic imaging: Toward robust and reproducible metabolite measurements in human brain. Magn Reson Med. 2017 doi: 10.1002/mrm.26718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bednarik P, Moheet A, Deelchand DK, Emir UE, Eberly LE, Bares M, Seaquist ER, Oz G. Feasibility and reproducibility of neurochemical profile quantification in the human hippocampus at 3 T. NMR Biomed. 2015;28(6):685–693. doi: 10.1002/nbm.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terpstra M, Cheong I, Lyu T, Deelchand DK, Emir UE, Bednarik P, Eberly LE, Oz G. Test-retest reproducibility of neurochemical profiles with short-echo, single-voxel MR spectroscopy at 3T and 7T. Magn Reson Med. 2016;76(4):1083–1091. doi: 10.1002/mrm.26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeydan B, Deelchand DK, Tosakulwong N, Lesnick TG, Kantarci OH, Machulda MM, Knopman DS, Lowe VJ, Jack CR, Jr, Petersen RC, Oz G, Kantarci K. Decreased Glutamate Levels in Patients with Amnestic Mild Cognitive Impairment: An sLASER Proton MR Spectroscopy and PiB-PET Study. J Neuroimaging. 2017 doi: 10.1111/jon.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deelchand DK, Kantarci K, Öz G. Improved localization, spectral quality, and repeatability with advanced MRS methodology in the clinical setting. Magn Reson Med. 2017 doi: 10.1002/mrm.26788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orourke J. Finding Minimal Enclosing Boxes. International Journal of Computer & Information Sciences. 1985;14(3):183–199. [Google Scholar]
- 19.Jenkinson M, Pechaud M, Smith S. BET2: MR-based estimation of brain, skull and scalp surfaces. Proc Org for Human Brain Mapp. 2005;11 [Google Scholar]
- 20.Eskildsen SF, Coupe P, Fonov V, Manjon JV, Leung KK, Guizard N, Wassef SN, Ostergaard LR, Collins DL Alzheimer’s Disease Neuroimaging I. BEaST: brain extraction based on nonlocal segmentation technique. Neuroimage. 2012;59(3):2362–2373. doi: 10.1016/j.neuroimage.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Iglesias JE, Liu CY, Thompson PM, Tu ZW. Robust Brain Extraction Across Datasets and Comparison With Publicly Available Methods. Ieee Transactions on Medical Imaging. 2011;30(9):1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- 22.Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL Brain Development Cooperative G. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric Atlasing and Model Based Segmentation: An Application to the Hippocampus in Older Adults. In: Larsen R, Nielsen M, Sporring J, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2006: 9th International Conference; Copenhagen, Denmark. October 1–6, 2006; Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. pp. 58–66. Proceedings, Part II. [DOI] [PubMed] [Google Scholar]
- 24.Deelchand DK, Adanyeguh IM, Emir UE, Nguyen TM, Valabregue R, Henry PG, Mochel F, Oz G. Two-site reproducibility of cerebellar and brainstem neurochemical profiles with short-echo, single-voxel MRS at 3T. Magn Reson Med. 2015;73(5):1718–1725. doi: 10.1002/mrm.25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Avants BB, Tustison N, Song G. Advanced normalization tools (ANTS) Insight j. 2009;2:1–35. [Google Scholar]
- 27.Öz G, Tkáč I. Short-Echo, Single-Shot, Full-Intensity Proton Magnetic Resonance Spectroscopy for Neurochemical Profiling at 4 T: Validation in the Cerebellum and Brainstem. Magnetic Resonance in Medicine. 2011;65(4):901–910. doi: 10.1002/mrm.22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deelchand DK. MRspa: Magnetic Resonance signal processing and analysis. Minneapolis; Minnesota, U.S.A: 2016. https://www.cmrr.umn.edu/downloads/mrspa/ [Google Scholar]
- 29.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: A Toolbox for Intensity-Based Medical Image Registration. Ieee Transactions on Medical Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 30.Ackerman MJ, Yoo TS. The Visible Human data sets (VHD) and Insight Toolkit (ITk): experiments in open source software. AMIA Annu Symp Proc. 2003:773. [PMC free article] [PubMed] [Google Scholar]
- 31.Eklund A, Dufort P, Villani M, Laconte S. BROCCOLI: Software for fast fMRI analysis on many-core CPUs and GPUs. Front Neuroinform. 2014;8:24. doi: 10.3389/fninf.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dice LR. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 33.Duda RO, Hart PE. Pattern Classification and Scene Analysis. Wiley; 1973. [Google Scholar]
- 34.Crum WR, Camara O, Hill DLG. Generalized overlap measures for evaluation and validation in medical image analysis. Ieee Transactions on Medical Imaging. 2006;25(11):1451–1461. doi: 10.1109/TMI.2006.880587. [DOI] [PubMed] [Google Scholar]
- 35.Ashburner J, Friston KJ. Chapter 4 - Rigid Body Registration and Chapter 5 - Non-linear Registration. In: Friston KJ, Ashburner J, Kiebel SJ, Nichols TE, Penny WD, editors. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Academic Press; 2007. pp. 49–80. [Google Scholar]
- 36.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53(1):1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 40.Andersson JLR, Jenkinson M, Smith SM. Non-linear registration, aka spatial normalisation. FMRIB technical report. 2010 [Google Scholar]
- 41.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 42.Munshi A. OpenCL programming guide. Upper Saddle River, NJ: Addison-Wesley; 2012. p. xliv.p. 603. [Google Scholar]
- 43.Barber CB, Dobkin DP, Huhdanpaa H. The Quickhull algorithm for convex hulls. Acm Transactions on Mathematical Software. 1996;22(4):469–483. [Google Scholar]
- 44.Popescu V, Battaglini M, Hoogstrate WS, Verfaillie SC, Sluimer IC, van Schijndel RA, van Dijk BW, Cover KS, Knol DL, Jenkinson M, Barkhof F, de Stefano N, Vrenken H, Group MS. Optimizing parameter choice for FSL-Brain Extraction Tool (BET) on 3D T1 images in multiple sclerosis. Neuroimage. 2012;61(4):1484–1494. doi: 10.1016/j.neuroimage.2012.03.074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tests showed that BET skull stripping performs better when neck and aliased nose are removed from the 3D data. A dedicated script was integrated to AutoVOI to remove these regions automatically using FSLROI prior to performing BET. To select the head with FSLROI, the head dataset is first reoriented to the standard orientation. Then, the inferior section of the head image, specifically 200mm below the top of the head, is removed based on the average human head size (a). Intensity projection in the IF (inferior/superior)-direction is used to detect the top of the head, and 20cm was chosen as the vertical threshold. To detect nose aliasing, intensity projection in the AP direction is performed (b). Nose aliasing is present if both ends of the projection are above a certain intensity threshold (5% of the peak intensity value in the projection was used in AutoVOI).