Abstract
Gastroesophageal reflux disease (GERD) is a common gastrointestinal disorder with an increasing prevalence. GERD develops when the reflux of stomach contents causes troublesome typical and atypical symptoms and/or complications. Several risk factors of GERD have been identified and evaluated over the years, including a considerable amount of genetic factors. Multiple mechanisms are involved in the pathogenesis of GERD including: (1) motor abnormalities, such as impaired lower esophageal sphincter (LES) resting tone, transient LES relaxations, impaired esophageal acid clearance and delayed gastric emptying; and (2) anatomical factors, such as hiatal hernia and obesity. Genetic contribution seems to play a major role in GERD and GERD- related disorders development such Barrett’s esophagus and esophageal adenocarcinoma. Twin and family studies have revealed an about 31% heritability of the disease. Numerous single-nucleotide polymorphisms in various genes like FOXF1, MHC, CCND1, anti-inflammatory cytokine and DNA repair genes have been strongly associated with increased GERD risk. GERD, Barrett’s esophagus and esophageal adenocarcinoma share several genetic loci. Despite GERD polygenic basis, specific genetic loci such as rs10419226 on chromosome 19, rs2687201 on chromosome 3, rs10852151 on chromosome 15 and rs520525 on the paired related homeobox 1 gene have been mentioned as potential risk factors. Further investigation on the risk genes may elucidate their exact function and role and demonstrate new therapeutic approaches to this increasingly common disease.
Keywords: Single nucleotide polymorphisms, Genetic risk loci, Risk factors, Gastroesophageal reflux disease, Gastroesophageal reflux disease development
Core tip: Gastroesophageal reflux disease (GERD) is a common gastrointestinal disorder, which develops when the reflux of stomach contents causes troublesome symptoms and/or complications. Several risk factors of GERD have been identified and evaluated over the years. Motor esophageal and gastric abnormalities, along with anatomical factors could contribute to GERD development. Genetic contributors seem to play a major role in GERD. Numerous single-nucleotide polymorphisms in various genetic loci have been mentioned as potential risk factors. Further investigation on the risk genes may elucidate their exact function and role and demonstrate new therapeutic approaches to this increasingly common disease.
INTRODUCTION
Gastroesophageal reflux disease (GERD) is a highly frequent gastrointestinal disorder with prevalence up to 20% in Europe and USA. Its prevalence is also increasing in the Far East and other Asian areas[1,2]. According to the Montreal definition and classification system, GERD represents a condition which develops when the reflux of stomach contents causes troublesome symptoms and/or complications[3]. The disease is characterized by a broad spectrum of typical symptoms, such as heartburn and acid regurgitation, some atypical ones, like dysphagia and chest pain, in addition to extra-esophageal manifestations, such as asthma, chronic cough and laryngitis[4-6].
GERD and its complications have a high impact on every day clinical practice, as well as on patients suffering regularly from discomforting symptoms of refluxate[7]. Several risk factors of GERD have been identified and evaluated over the years, including a considerable amount of genetic factors[4,8].
A PubMed search was performed using the key words (“gastroesophageal reflux disease” OR “GERD” OR “chronic reflux disease” OR “reflux disease”) AND (“oesophagus” OR “esophagus” OR “esophageal” OR “oesophageal”) AND (“risk factors” OR “contributors”) AND (“genetic background” OR “genetics” OR “susceptible genetic loci” OR “ SNPs”) AND (“Barrett’s” OR “adenocarcinoma”) AND (“genome wide association study” OR “GWAS”) AND (“pathogenesis” OR “pathogenetic mechanisms”) AND (“epidemiology”) AND (“biomarkers”). The present review aims to report the genetic contributors of GERD, enriched with the pathogenetic mechanisms of the main risk factors, based on current literature.
PATHOGENETIC MECHANISMS OF THE MAIN RISK FACTORS
GERD is developed when detrimental to the esophagus factors transcend protective mechanisms, such as the esophago-gastric junction barrier and esophageal acid clearance, which normally contribute to the maintenance of a physiologically balanced condition. There are multiple mechanisms involved in the pathogenesis of GERD including: (1) motor abnormalities, such as impaired lower esophageal sphincter (LES) resting tone, transient LES relaxations (TLESR), impaired esophageal acid clearance and delayed gastric emptying; and (2) anatomical factors, such as hiatal hernia and obesity[4,9,10]. A valve mechanism exists between the esophagus and the stomach, formed by the LES and its adjacent anatomical structures, including the gastric sling and the crural diaphragm[11]. The main role of this valve mechanism in resting conditions is to create a fine-tuned high-pressure zone (15-30 mmHg above intragastric pressures), preventing gastric contents reflux. A minority of GERD patients experiences extremely low LES resting pressure (< 6 mmHg); every time stomach pressure exceeds the LES pressure, reflux occurs. Such a decreased LES resting tone is ordinarily correlated to severe grade of esophagitis and/or presence of GERD complications, including peptic stricture and Barrett’s esophagus. However, in the majority of GERD patients, a high frequency of inappropriate LES relaxations is the cause of abnormal gastro-esophageal reflux[4]. TLESRs are spontaneous LES relaxations of 10-60 s duration, which are unrelated to swallowing[12,13]. Gastric distension, via stimulation of proximal gastric tension and stretch receptors, is considered the major contributor generating TLESRs. Although TLESRs occur in healthy individuals with a similar frequency to GERD patients, a higher percentage of TLESRs is associated with reflux in GERD patients[4,9,14-17]. Like LES resting pressure, the frequency of TLESRs is influenced by endogenous hormones (cholocystokinin, progesterone etc.)[18], drugs (calcium channel blockers, nitrates, tricyclic antidepressant medications, benzodiazepine, anticholinergic drugs, theophylline etc.)[19], specific foods (fat, chocolate, etc.)[20] and daily habits (alcohol, caffeine, smoking)[21].
Ineffective esophageal motility (IEM) is considered, along with TLESR, another significant contributor to the appearance of GERD, as it leads to impaired esophageal clearance[22]. Esophageal acid clearance is a critical protective process involving primary and secondary peristalsis as well as the swallowing of salivary bicarbonate. Primary peristalsis occurs approximately 60 times per hour just after every swallow, whereas secondary peristalsis is observed in the absence of swallowing as a result of esophageal distension or of the presence of acidic contents into the esophageal lumen. The swallowing of saliva (pH 7.8-8.0) is pivotal in the accomplishment of esophageal acid clearance and in the restitution of esophageal pH. Evidence suggests that GERD patients show 2-3 fold longer acid clearance times compared to normal subjects[4]. The slower the esophageal clearance is, the longest the refluxate (comes into contact) with the esophageal mucosa. Thus, IEM leads to more severe GERD, in terms of both symptoms and mucosal damage[9].
Delayed gastric emptying might contribute to GERD in a small yet significant amount of patients, especially those who do not respond to proton pump inhibitors (PPIs) therapy[4,9]. An increase of the intra-gastric pressure, due to gastric distension, resulting in an overwhelming amount of refluxate, could be a putative mechanism for deteriorating GERD. Gastric distension could also contribute to an increase of the postprandial TLESR’s rate[4].
Hiatal hernia is often found in patients with GERD with a prevalence of 0.8% up to 43.0%[23]. Hiatal hernia is considered to be a significant factor, since it disintegrates the gastro-esophageal sphincter, as the proximal stomach is dislocated into the chest and the crural diaphragm becomes separated from the LES. In patients with severe erosive esophagitis and in those with GERD complications, hiatal hernia is present in most cases. A linear correlation between hernia’s size and the severity of reflux symptoms seems to exist. Hiatal hernia loosens the lower esophageal sphincter and increases the frequency of TLESRs. Moreover, it decreases esophageal clearance and enhances reflux by acting as a reservoir for gastric acid that becomes trapped in its sac[24,25].
Obesity has been considered to be a key risk factor of GERD. The rising rates of obesity (35.5% for men and 35.8% for women estimated by the National Health and Nutrition Examination Survey for the years 2005-2009)[26] are associated with early onset of GERD, as an independent factor (approximately 50% in morbid obesity)[27]. Among the possible mechanisms by which obesity promotes GERD, increased abdominal pressure, delayed gastric emptying, increased frequency of TLESR and reduced LES resting pressure are considered to play a crucial role[2,28,29]. The incidence of reflux symptoms rises progressively with increasing BMI. It is widely accepted that even short-term weight gain is associated with a three- to four-fold higher risk of GERD symptoms. This positive association between increasing BMI and GERD has been confirmed by a recent meta-analysis[29-31].
An interesting potential factor in peptic acid diseases is the gastric acid secretion in the interprandial periods. As suggested (1) by Feldman and Richardson[32] in a study on 8 patients with duodenal ulcer disease versus 7 normal subjects; and (2) by Caboclo et al[33] in a study on rats the possible mechanisms are: (1) increased oxyntic gland sensitivity; (2) hyperplasia of parietal cells, hypercorresponding to the vagal release of gastrin; or (3) cortical-stimulated secretion through methods of learning and memory, for example combing the food intake with specific sounds and emotions.
Additionally, several other factors have been asserted as causes of heartburn symptoms. Metabolic syndrome or its components- and especially hypertriglyceridemia- have been associated with erosive esophagitis or reflux symptoms respectively[34]. Zheng et al[21] found that a dose-dependent smoking was linked to the occurrence of gastroesophageal reflux symptoms with a risk of about 37% risk among women and 53% among men. In the same analysis, coffee intake was considered to be protective factor for GERD in men, contrary to women, probably due to different caffeine metabolic patterns. Exercise at work expedites the appearance of reflux symptoms, whereas leisure-time exercise was protective to the disease.
In a recently published study, Mungan and colleague[19] estimated the correlation between several categories of drugs and GERD. They deduced that non-steroidal anti-inflammatory drugs (especially when combined with acetylsalicylic acid, estrogen replacement therapy, calcium channel blockers (CCBs), nitrates, tricyclic antidepressant medications (particularly amitriptyline and clomipramine), hypnotics and benzodiazepine, anticholinergic drugs and theophylline promote the onset of GERD.
GENETIC RISK FACTORS
For over a decade, the role of genes in the development of gastroesophageal reflux disease and GERD-related disorders [Barrett’s esophagus (BE), esophageal adenocarcinoma (EAC)] has been introduced. This statement was verified through population-based studies on twins. Numerous single-nucleotide polymorphisms (SNPs) have been proposed in genome-wide association studies (GWAS) as potential factors in the appearance of reflux disease [35,36] (Table 1).
Table 1.
Studies performed on measuring the possible heritability of gastroesophageal reflux disease and the severity of reflux symptoms among twins and family members and on identifying the risk genetic loci for gastroesophageal reflux disease
| Studies | |
| Twin and family studies | |
| Cameron A et al[37] | ↑ Heritability of GERD, ↑ symptoms in MZ |
| Mohammed I et al[38] | ↑ Heritability of GERD, ↑ symptoms in MZ |
| Reding-Bernal A et al[39] | ↑GERD symptoms severity in families in Mexico |
| GERD risk genetic loci studies | |
| Ghoshal and Chourasia[8] | > 10 genes, up/down regulating GERD |
| Liu WF et al[40] | C allele in FOX1 rs9936833, A allele in MHC rs9257809 : ↑reflux symptoms |
| Gharahkhani P et al[41] | rs10419226 (chr 19), rs2687201 (chr 3) : ↑GERD symptoms |
| Bonfiglio F et al[35] | > 30 susceptible gene loci for GERD |
GERD: Gastroesophageal reflux disease; MZ: Monozygotic twins; chr: Chromosome.
Twin and family studies
In 2002, Cameron et al[37] examined 8411 pairs of twins [2178 monozygotic twins pairs (MZ) and 6233 dizygotic ones (DZ)] and concluded that the genetic influence on GERD reached 31% (95%CI, 0.23-0.38). After taking several factors into account, such as age, gender and daily habits, GERD was found to be strongly correlated to MZ. A year later, Mohammed et al[38] examined 4480 pairs of twins, which inferred 30% heritability of GERD, while the same symptoms in MZ outnumbered the DZ ones. Another study of 585 individuals concerning 32 families living in Mexico revealed an association between the severity of GERD symptoms, metabolic syndrome components and inflammatory markers due to common genetic back round[39].
Studies on identifying risk genetic loci for GERD
Ghoshal and Chourasia[8] attempted to enumerate the host genetic factors responsible for GERD and to explain their role in the pathogenesis of the disease, as well as its complications. The presence of the pro-inflammatory cytokines interleukin-1beta and IL-1RN (gene encoding for a non-signaling molecule IL-1 receptor antagonist) (IL-1B-511*T/IL-1RN*1) combined with Helicobacter pylori infection, has a protective effect against the development of GERD. Their presence results in extended gastritis and the destruction of parietal cells, leading in hypochlorhydria and thus reducing the risk for GERD (Figure 1). On the contrary, altered expression of Cyclooxygenase-2 (COX-2) (enzyme in prostaglandin biosynthesis), IL-10 (anti-inflammatory cytokine), Glutathione-S-transferases (especially GSTP1*b), Cyclin D1 (CCND1) and DNA repair genes (XRCC1, hMLH1) have been associated with a high risk of GERD, BE or EAC. Additionally, the homozygous G/G variant genotype of epidermal growth factor (A61G), and the -C825T- genetic polymorphism of GNB3 (G protein) also appear to contribute to an elevated risk of these conditions (Figure 2)[8].
Figure 1.
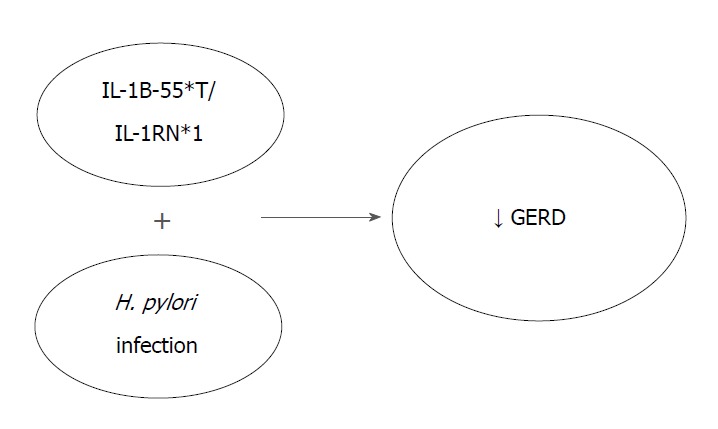
The presence of the genes IL-1B and IL-1RN combined with Helicobacter pylori infection is associated with hypochlorydria and thus reducing the risk for gastroesophageal reflux disease. GERD: Gastroesophageal reflux disease; H. pylori: Helicobacter pylori; IL-1B-511*T: Interleukin-1 beta T allele; IL-1RN: Gene encoding for a non-signaling molecule IL-1 receptor antagonist (IL-1Ra).
Figure 2.
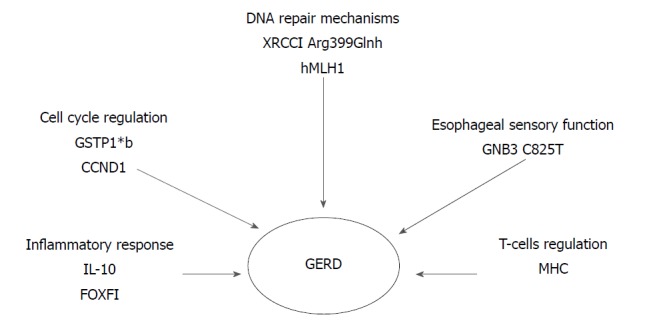
Susceptible risk genes for gastroesophageal reflux disease. Their increased or reduced (DNA repair genes) expression alters different biological pathways. GERD: Gastroesophageal reflux disease; IL-10: Anti-inflammatory cytokine interleukin 10; FOXF1: Forkhead BOX F1; GSTP1*b: Glutathione- S- transferases b allele; CCND1: Cyclin D1 gene; XRCC1: X-ray repair complementing defective repair in Chinese hamster cells 1; Hmlh1: Humal homolog of the E. coli DNA mismatch repair gene mutL; GNB3: Guanine nucleotide binding protein beta polypeptide 3; MHC: Major histocompatibility complex.
In 2014, a study of 182 patients concluded that FOXF1 (C allele in FOXF1 rs9936833) (95%CI: 1.1-3.0; P = 0.02) and MHC (A allele in MHC rs9257809) polymorphisms (95%CI: 2.9-3.0; P < 0.001) were strongly associated with increased GERD risk in patients with reflux symptoms. FOXF1 gene may play a role in the regulation of the contraction of the lower oesophageal sphincter, due to its involvement in the development of the gastrointestinal smooth muscle. Furthermore, the possibility that MHC genes are associated with HLA alleles and, therefore, could influence the activity of T-cells, reveals a T-cell involvement in reflux esophagitis (Figure 2)[40].
Gharahkhani et al[41] investigated the involvement of SNPs in the development of GERD and the shared genetic loci of GERD, BE and EAC.; The variability of SNPs could explain the 7% phenotypic variance present in GERD (BEACON and 23 and ME studies). Despite GERD polygenic basis, they suggested two specific genetic loci with high association with GERD: rs10419226 on chromosome 19 (95%CI: 1.00-1.07; P = 0.038) and rs2687201 on chromosome 3 (95%CI: 1.01-1.09; P = 0.025) (Figure 3).
Figure 3.
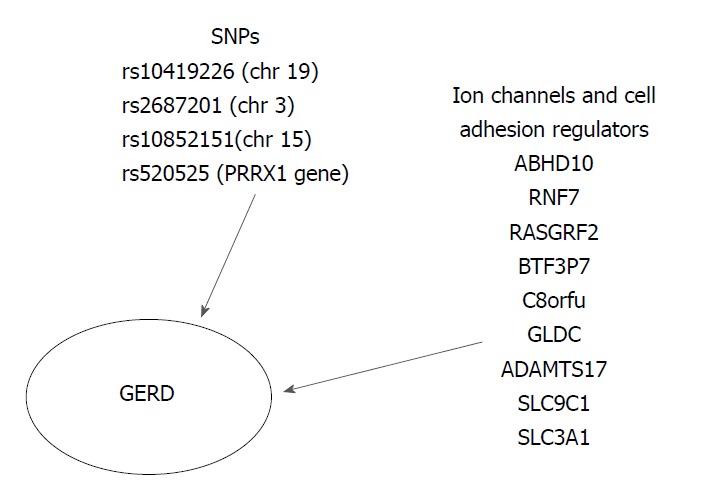
Genetic risk loci associated with the development of gastroesophageal reflux disease[32,38]. GERD: Gastroesophageal reflux disease; SNPs: Single-nucleotide polymorphisms.
Bonfiglio et al[35] recently conducted a GWAS meta-analysis of three independent population-based studies from Sweden, UK (TwinsUK) and Northern Finland (NFBC1966) in order to elucidate the pathogenesis of GERD. A total of 30 susceptible GERD risk loci were identified (P < 0.5 × 10-5). The strongest evidence suggesting a correlation between GERD and the various genetic loci was found at the SNP rs10852151 on chromosome 15 (P = 2.3 × 10-7) and at rs520525 on the paired related homeobox 1 (PRRX1) gene (P = 0.011). Their effect on gene expression in relevant tissues, such as gastroesophageal junction, esophagus muscularis, esophagus mucosa and stomach, was also evaluated. GERD risk genes influence the regulation of several biological pathways, including the ion channel and the cell adhesion. Moreover, expression trait quantitative loci (eQTL) analyses revealed that these risk genetic loci were enriched for significant eQTLs from GERD-related tissues. The following seven genes, ABHD10, RNF7, RASGRF2, BTF3P7, C8orf4, GLDC, and ADAMTS17, located in GERD risk region could be potential GERD risk candidate. Additionally, they pointed at two more ion-channel genes, the SLC9C1 gene (a NA+/H+ exchanger) associated with eQTLs in the gastroesophageal junction and the SLC3A1 (an amino-acid transporter) associated with eQTLs in the esophagus mucosa, which is of great interest in terms of treatment with PPIs. Moreover, the authors suggested that the risk genes ADAMTS17 (rs4965272) and ADAM10 should be investigated in the future, since ADAMTS17 participates in numerous biological processes and ADAM10 controls the e-cadherin proteolytic cleavage in GERD patients. Finally, they attempted to link the identified GERD risk loci with the known therapeutic compounds, by using a computational Connectivity Map analysis. Interesting results were obtained for omeprazole (P = 0.032) and fludroxycortide (P = 1.04 × 10-4). After the Anatomical Therapeutic Chemical index system was taken into account, the class JO4A (anti-tuberculosis drugs) showed high scores after normalization (P ≤ 0.033), as the ion channel genes could be an antituberculosis target. Nevertheless, there was a significant limitation in predicting the total drug regulation effect, despite the fact that it was suggested that drugs affect the expression of these genes (Figure 3)[35].
Genetic factors substantially explain the phenotypic variance of the severity of some GERD symptoms and increase our knowledge of the etiology of the disease. Future genetic studies should define the relation between GERD and its pathophysiological features such as BMI, body fat distribution and hiatal hernia, leading to the identification of biomarkers for GERD prevention and molecular targets for novel treatment. The genetic overlap between GERD, BE and EA may be helpful in future treatments targeting shared molecular pathways involved in pathogenesis of these diseases. Furthermore genetic markers can be discovered to help identify the highest risk individuals for intervention, patients with GERD that will or not progress to EA. That genetic difference could be exploited to determine which patients with GERD are at risk; as such more aggressive screening and treatment could be focused on a clear high risk group[42-44].
CONCLUSION
GERD has proven to be a multivariate disorder, including abnormal anatomical structures and co-morbidities, affected by environmental and genetic factors. The latter has been also found in several studies and in a newly published GWAS, although none of them has established specific genetic loci with certainty. Further investigation on the mentioned risk genes is needed, in order to evaluate their exact function and role, to probably use them as screening tools or biomarkers and to demonstrate new therapeutic approaches to this increasingly common disease.
Footnotes
Conflict-of-interest statement: No conflicts of interest.
Manuscript Source: Invited Manuscript
Peer-review started: May 4, 2018
First decision: May 22, 2018
Article in press: June 8, 2018
Specialty type: Medicine, research and experimental
Country of origin: Greece
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): E
P- Reviewer: Caboclo GF, Garcia-Compean D, Kim GH, Lan C, Lei JJ S- Editor: Wang JL L- Editor: A E- Editor: Tan WW
Contributor Information
Alexandra Argyrou, Department of Basic Medical Sciences, Laboratory of Biology, School of Medicine, National and Kapodistrian University of Athens, Athens 11527, Greece.
Evangelia Legaki, Department of Basic Medical Sciences, Laboratory of Biology, School of Medicine, National and Kapodistrian University of Athens, Athens 11527, Greece.
Christos Koutserimpas, 2nd Department of General Surgery, “Sismanoglion” General Hospital of Athens, Athens 11527, Greece.
Maria Gazouli, Department of Basic Medical Sciences, Laboratory of Biology, School of Medicine, National and Kapodistrian University of Athens, Athens 11527, Greece. mgazouli@med.uoa.gr.
Ioannis Papaconstantinou, 2nd Department of Surgery, School of Medicine, National and Kapodistrian University of Athens, Athens 11527, Greece.
George Gkiokas, 2nd Department of Surgery, School of Medicine, National and Kapodistrian University of Athens, Athens 11527, Greece.
George Karamanolis, Gastroenterology Unit, 2nd Department of Surgery, School of Medicine, National and Kapodistrian University of Athens, Athens 11527, Greece.
References
- 1.El-Serag HB, Sweet S, Winchester CC, Dent J. Update on the epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut. 2014;63:871–880. doi: 10.1136/gutjnl-2012-304269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaheen N, Provenzale D. The epidemiology of gastroesophageal reflux disease. Am J Med Sci. 2003;326:264–273. doi: 10.1097/00000441-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R; Global Consensus Group. The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus. Am J Gastroenterol. 2006;101:1900–1920; quiz 1943. doi: 10.1111/j.1572-0241.2006.00630.x. [DOI] [PubMed] [Google Scholar]
- 4.De Giorgi F, Palmiero M, Esposito I, Mosca F, Cuomo R. Pathophysiology of gastro-oesophageal reflux disease. Acta Otorhinolaryngol Ital. 2006;26:241–246. [PMC free article] [PubMed] [Google Scholar]
- 5.Galmiche JP, Bruley des Varannes S. Symptoms and disease severity in gastro-oesophageal reflux disease. Scand J Gastroenterol Suppl. 1994;201:62–68. doi: 10.3109/00365529409105366. [DOI] [PubMed] [Google Scholar]
- 6.Katz PO, Gerson LB, Vela MF. Guidelines for the diagnosis and management of gastroesophageal reflux disease. Am J Gastroenterol. 2013;108:308–328; quiz 329. doi: 10.1038/ajg.2012.444. [DOI] [PubMed] [Google Scholar]
- 7.Dent J, El-Serag HB, Wallander MA, Johansson S. Epidemiology of gastro-oesophageal reflux disease: a systematic review. Gut. 2005;54:710–717. doi: 10.1136/gut.2004.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghoshal UC, Chourasia D. Genetic factors in the pathogenesis of gastroesophageal reflux disease. Indian J Gastroenterol. 2011;30:55–62. doi: 10.1007/s12664-011-0095-7. [DOI] [PubMed] [Google Scholar]
- 9.Herbella FA, Patti MG. Gastroesophageal reflux disease: From pathophysiology to treatment. World J Gastroenterol. 2010;16:3745–3749. doi: 10.3748/wjg.v16.i30.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhmer AC, Schumacher J. Insights into the genetics of gastroesophageal reflux disease (GERD) and GERD-related disorders. Neurogastroenterol Motil. 2017:29. doi: 10.1111/nmo.13017. [DOI] [PubMed] [Google Scholar]
- 11.Nadaleto BF, Herbella FA, Patti MG. Gastroesophageal reflux disease in the obese: Pathophysiology and treatment. Surgery. 2016;159:475–486. doi: 10.1016/j.surg.2015.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Boeckxstaens GE. The lower oesophageal sphincter. Neurogastroenterol Motil. 2005;17 Suppl 1:13–21. doi: 10.1111/j.1365-2982.2005.00661.x. [DOI] [PubMed] [Google Scholar]
- 13.Spechler SJ, Castell DO. Classification of oesophageal motility abnormalities. Gut. 2001;49:145–151. doi: 10.1136/gut.49.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter J. Do we know the cause of reflux disease? Eur J Gastroenterol Hepatol. 1999;11 Suppl 1:S3–S9. [PubMed] [Google Scholar]
- 15.Liakakos T, Karamanolis G, Patapis P, Misiakos EP. Gastroesophageal reflux disease: medical or surgical treatment? Gastroenterol Res Pract. 2009;2009:371580. doi: 10.1155/2009/371580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karamanolis G, Sifrim D. Developments in pathogenesis and diagnosis of gastroesophageal reflux disease. Curr Opin Gastroenterol. 2007;23:428–433. doi: 10.1097/MOG.0b013e328133f56a. [DOI] [PubMed] [Google Scholar]
- 17.Campos GM, Peters JH, DeMeester TR, Oberg S, Crookes PF, Mason RJ. The pattern of esophageal acid exposure in gastroesophageal reflux disease influences the severity of the disease. Arch Surg. 1999;134:882–887; discussion 887-888. doi: 10.1001/archsurg.134.8.882. [DOI] [PubMed] [Google Scholar]
- 18.Castell DO, Harris LD. Hormonal control of gastroesophageal-sphincter strength. N Engl J Med. 1970;282:886–889. doi: 10.1056/NEJM197004162821602. [DOI] [PubMed] [Google Scholar]
- 19.Mungan Z, Pınarbaşı Şimşek B. Which drugs are risk factors for the development of gastroesophageal reflux disease? Turk J Gastroenterol. 2017;28:S38–S43. doi: 10.5152/tjg.2017.11. [DOI] [PubMed] [Google Scholar]
- 20.Nebel OT, Castell DO. Inhibition of the lower oesophageal sphincter by fat--a mechanism for fatty food intolerance. Gut. 1973;14:270–274. doi: 10.1136/gut.14.4.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Z, Nordenstedt H, Pedersen NL, Lagergren J, Ye W. Lifestyle factors and risk for symptomatic gastroesophageal reflux in monozygotic twins. Gastroenterology. 2007;132:87–95. doi: 10.1053/j.gastro.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinucci I, de Bortoli N, Giacchino M, Bodini G, Marabotto E, Marchi S, Savarino V, Savarino E. Esophageal motility abnormalities in gastroesophageal reflux disease. World J Gastrointest Pharmacol Ther. 2014;5:86–96. doi: 10.4292/wjgpt.v5.i2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dent J, Becher A, Sung J, Zou D, Agréus L, Bazzoli F. Systematic review: patterns of reflux-induced symptoms and esophageal endoscopic findings in large-scale surveys. Clin Gastroenterol Hepatol. 2012;10:863–873.e3. doi: 10.1016/j.cgh.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 24.Kahrilas PJ, Lin S, Chen J, Manka M. The effect of hiatus hernia on gastro-oesophageal junction pressure. Gut. 1999;44:476–482. doi: 10.1136/gut.44.4.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahrilas PJ, Shi G, Manka M, Joehl RJ. Increased frequency of transient lower esophageal sphincter relaxation induced by gastric distention in reflux patients with hiatal hernia. Gastroenterology. 2000;118:688–695. doi: 10.1016/s0016-5085(00)70138-7. [DOI] [PubMed] [Google Scholar]
- 26.Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999-2010. JAMA. 2012;307:491–497. doi: 10.1001/jama.2012.39. [DOI] [PubMed] [Google Scholar]
- 27.Doulami G, Triantafyllou S, Natoudi M, Albanopoulos K, Leandros E, Zografos G, Theodorou D. GERD-Related Questionnaires and Obese Population: Can They Really Reflect the Severity of the Disease and the Impact of GERD on Quality of Patients’ Life? Obes Surg. 2015;25:1882–1885. doi: 10.1007/s11695-015-1614-x. [DOI] [PubMed] [Google Scholar]
- 28.Stenard F, Iannelli A. Laparoscopic sleeve gastrectomy and gastroesophageal reflux. World J Gastroenterol. 2015;21:10348–10357. doi: 10.3748/wjg.v21.i36.10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hampel H, Abraham NS, El-Serag HB. Meta-analysis: obesity and the risk for gastroesophageal reflux disease and its complications. Ann Intern Med. 2005;143:199–211. doi: 10.7326/0003-4819-143-3-200508020-00006. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson BC, Somers SC, Fuchs CS, Kelly CP, Camargo CA Jr. Body-mass index and symptoms of gastroesophageal reflux in women. N Engl J Med. 2006;354:2340–2348. doi: 10.1056/NEJMoa054391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rey E, Moreno-Elola-Olaso C, Artalejo FR, Locke GR 3rd, Diaz-Rubio M. Association between weight gain and symptoms of gastroesophageal reflux in the general population. Am J Gastroenterol. 2006;101:229–233. doi: 10.1111/j.1572-0241.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 32.Feldman M, Richardson CT. Total 24-hour gastric acid secretion in patients with duodenal ulcer. Comparison with normal subjects and effects of cimetidine and parietal cell vagotomy. Gastroenterology. 1986;90:540–544. doi: 10.1016/0016-5085(86)91106-6. [DOI] [PubMed] [Google Scholar]
- 33.Caboclo JL, Cury Fde A, Borin AA, Caboclo LO, Ribeiro MF, de Freitas PJ, Andersson S. Gastric secretion elicited by conditioning in rats. Scand J Gastroenterol. 2009;44:672–679. doi: 10.1080/00365520802588083. [DOI] [PubMed] [Google Scholar]
- 34.Li CH, Hsieh TC, Hsiao TH, Wang PC, Tseng TC, Lin HH, Wang CC. Different risk factors between reflux symptoms and mucosal injury in gastroesophageal reflux disease. Kaohsiung J Med Sci. 2015;31:320–327. doi: 10.1016/j.kjms.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Bonfiglio F, Hysi PG, Ek W, Karhunen V, Rivera NV, Männikkö M, Nordenstedt H, Zucchelli M, Bresso F, Williams F, et al. A meta-analysis of reflux genome-wide association studies in 6750 Northern Europeans from the general population. Neurogastroenterol Motil. 2017:29. doi: 10.1111/nmo.12923. [DOI] [PubMed] [Google Scholar]
- 36.Gharahkhani P, Fitzgerald RC, Vaughan TL, Palles C, Gockel I, Tomlinson I, Buas MF, May A, Gerges C, Anders M, Becker J, Kreuser N, Noder T, Venerito M, Veits L, Schmidt T, Manner H, Schmidt C, Hess T, Böhmer AC, Izbicki JR, Hölscher AH, Lang H, Lorenz D, Schumacher B, Hackelsberger A, Mayershofer R, Pech O, Vashist Y, Ott K, Vieth M, Weismüller J, Nöthen MM; Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON); Esophageal Adenocarcinoma GenEtics Consortium (EAGLE); Wellcome Trust Case Control Consortium 2 (WTCCC2), Attwood S, Barr H, Chegwidden L, de Caestecker J, Harrison R, Love SB, MacDonald D, Moayyedi P, Prenen H, Watson RGP, Iyer PG, Anderson LA, Bernstein L, Chow WH, Hardie LJ, Lagergren J, Liu G, Risch HA, Wu AH, Ye W, Bird NC, Shaheen NJ, Gammon MD, Corley DA, Caldas C, Moebus S, Knapp M, Peters WHM, Neuhaus H, Rösch T, Ell C, MacGregor S, Pharoah P, Whiteman DC, Jankowski J, Schumacher J. Genome-wide association studies in oesophageal adenocarcinoma and Barrett’s oesophagus: a large-scale meta-analysis. Lancet Oncol. 2016;17:1363–1373. doi: 10.1016/S1470-2045(16)30240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron AJ, Lagergren J, Henriksson C, Nyren O, Locke GR 3rd, Pedersen NL. Gastroesophageal reflux disease in monozygotic and dizygotic twins. Gastroenterology. 2002;122:55–59. doi: 10.1053/gast.2002.30301. [DOI] [PubMed] [Google Scholar]
- 38.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in gastro-oesophageal reflux disease: a twin study. Gut. 2003;52:1085–1089. doi: 10.1136/gut.52.8.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reding-Bernal A, Sánchez-Pedraza V, Moreno-Macías H, Sobrino-Cossio S, Tejero-Barrera ME, Burguete-García AI, León-Hernández M, Serratos-Canales MF, Duggirala R, López-Alvarenga JC. Heritability and genetic correlation between GERD symptoms severity, metabolic syndrome, and inflammation markers in families living in Mexico City. PLoS One. 2017;12:e0178815. doi: 10.1371/journal.pone.0178815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu WF, Lam C, Del Bel R, Chan K, Miller L, Brown C, Chen Z, Cheng D, Patel D, Xu W, et al. Association between polymorphisms of the FOXF1 and MHC locus genes and gastroesophageal reflux disease (GERD) J Clin Oncol. 2014;32:Abstract 15. [Google Scholar]
- 41.Gharahkhani P, Tung J, Hinds D, Mishra A; Barrett’s and Esophageal Adenocarcinoma Consortium (BEACON), Vaughan TL, Whiteman DC, MacGregor S; BEACON study investigators. Chronic gastroesophageal reflux disease shares genetic background with esophageal adenocarcinoma and Barrett’s esophagus. Hum Mol Genet. 2016;25:828–835. doi: 10.1093/hmg/ddv512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tischoff I, Tannapfel A. Barrett’s esophagus: can biomarkers predict progression to malignancy? Expert Rev Gastroenterol Hepatol. 2008;2:653–663. doi: 10.1586/17474124.2.5.653. [DOI] [PubMed] [Google Scholar]
- 43.Ramzan Z, Nassri AB, Huerta S. The use of imaging and biomarkers in diagnosing Barrett’s esophagus and predicting the risk of neoplastic progression. Expert Rev Mol Diagn. 2014;14:575–591. doi: 10.1586/14737159.2014.919856. [DOI] [PubMed] [Google Scholar]
- 44.Zakko L, Wang KK. Genetically linking chronic gastroesophageal reflux disease: Barrett’s esophagus and esophageal adenocarcinoma. Ann Transl Med. 2016;4:290. doi: 10.21037/atm.2016.05.63. [DOI] [PMC free article] [PubMed] [Google Scholar]


