Abstract
Research into proximate and ultimate mechanisms of individual cognitive variation in animal populations is a rapidly growing field that incorporates physiological, behavioural and evolutionary investigations. Recent studies in humans and laboratory animals have shown that the enteric microbial community plays a central role in brain function and development. The ‘gut–brain axis’ represents a multi-directional signalling system that encompasses neurological, immunological and hormonal pathways. In particular it is tightly linked with the hypothalamic–pituitary–adrenal axis (HPA), a system that regulates stress hormone release and influences brain development and function. Experimental examination of the microbiome through manipulation of diet, infection, stress and exercise, suggests direct effects on cognition, including learning and memory. However, our understanding of these processes in natural populations is extremely limited. Here, we outline how recent advances in predominantly laboratory-based microbiome research can be applied to understanding individual differences in cognition. Experimental manipulation of the microbiome across natal and adult environments will help to unravel the interplay between cognitive variation and the gut microbial community. Focus on individual variation in the gut microbiome and cognition in natural populations will reveal new insight into the environmental and evolutionary constraints that drive individual cognitive variation.
This article is part of the theme issue ‘Causes and consequences of individual differences in cognitive abilities’.
Keywords: microbiome, cognition, behaviour, stress, animal personality, diet
1. Background
The field of comparative cognition has long focused on describing cognitive mechanisms underlying behaviour across species, and has revealed insight into the convergent evolution of cognitive abilities among different taxa, such as corvids and apes [1]. Among other factors, comparative cognition has improved our understanding of the selection pressures that favour specific cognitive mechanisms, such as spatial memory in food-hoarding birds [2]. After many decades of intense study on differences between species, many researchers are increasingly exploring the causes and consequences of variation among individuals (e.g. [3]). Proximate causes associated with cognition, such as brain morphology (e.g. [2]) and hormonal responses (reviewed in [4]), have previously been identified, and along with behavioural measures, provide additional dimensions for quantifying cognition. Moreover, proximate mechanisms that are intrinsically linked with cognition provide insight into properties of cognition, such as phenotypic plasticity and the relative contribution of genetic, environmental and developmental effects. We argue that the microbiome is an important trait that should be explored as a mechanism explaining individual differences in cognition that have not been explored at all in natural populations outside of human studies.
The microbiome refers to microbial communities and their collective genetic material found in and on the body, and is most commonly studied within the gut. Studies have demonstrated a major role for the gastrointestinal system's microbiome in the development and maintenance of brain function [5], mediated by direct links with immune and endocrine systems (reviewed in [6]). What makes the relationship between the microbiome and brain especially significant from an evolutionary and ecological perspective is that research on humans and other animals including insects, birds and mammals suggests that the microbiome differs among individuals as a result of environmental factors, such as diet [7–9], infection [10], stress (reviewed in [11]), sociality [12,13] and host genotype [14,15], and that the microbiome can have direct effects on cognition, as demonstrated in laboratory mice (Mus musculus) and human infants (table 1 and references therein). In wild animal populations the roles of food and infectious diseases are important factors that have the potential to alter the microbiome of individuals. Furthermore, social interactions both influence stress and can facilitate the transmission of commensal and pathogenic bacteria across individuals [25,26], leading to differences in gut microbiota composition among social groups [13,27]. Here we describe how the study of microbial communities among individuals has almost entirely unexplored potential to explain individual differences in cognition and associated functional behaviours.
Table 1.
Associations between natural/experimentally-altered microbiomes and cognitive tasks. All studies were performed on mice, except for one on human infants [16] and one on human adults [17]. Conventionally colonized (CC) refers to mice born with natural microbiome, germ-free (GF) refers to mice born and kept free of any microbes and housed in sterile isolators, and ex-germ free (ex-GF) refers to mice born free of microbes and housed with CC mice post-weaning (three weeks old) to facilitate recolonization of microbes. Cognitive/behavioural results indicate if differences are significantly greater (>), significantly less (<) or non-significant (=). Neurological results are a brief overview and readers are referred to original papers for further details. BDNF, brain derived neurotrophic factor; OTUs, operational taxonomic units.
| microbiome treatment | cognition/behaviour assay method | cognitive/behaviour assay result | neurological/physiology assays results | reference |
|---|---|---|---|---|
| germ free (GF), recolonized (ex-GF) and conventionally colonized (CC) | choice of empty versus social chamber interactions with novel versus familiar mice food-preference test following demonstrator observation |
chamber sociability: GF < CC = ex-GF social recognition: GF and ex-GF < CC social observation of demonstrator: GF < CC = ex-GF social information transfer: GF = ex-GF = CC |
— | [18] |
| GF, ex-GF and CC | cued fear conditioning (tone + shock in experimental context (i.e. box)) | acquisition: GF = ex-GF = CC cue recall: GF < ex-GF = CC context recall: GF = CC = ex-GF extinction: GF < CC; ex-GF < CC |
genome-wide RNA sequencing of amygdala shows altered gene expressions | [19] |
| GF, CC, CC-pathogen infected (CC-in), CC-in with probiotics (CC-in-pro) | light–dark box novel object recognition T-maze tests performed following exposure to acute stress (AS) or no stress (NS) |
anxiety: CC-in = CC; GF = CC recognition and working memory (NS): CC-in = CC; GF < CC recognition and working memory (AS): CC-in < CC; GF < CC object and working memory (NS): CC-in-pro = CC-in object and working memory (AS): CC-in-pro > CC-in |
differences in corticosterone and hippocampal BDNF and c-Fos | [10] |
| anxious mouse strains fed different Bifidobacteria probiotics: B. longum and B. breve | object recognition task Barnes maze cued fear conditioning |
recognition memory: B. longum and B. breve > control mice spatial learning: B. longum = control = B. breve spatial memory: B. longum > control; B. breve = control fear conditioning learning and memory to context: B. longum and B. breve = control fear conditioning memory to cue and extinction: B. longum = control = B. breve |
no difference in basal corticosterone across treatment groups | [20] |
| microbiome manipulation through restricted diets: high fat (hf), high sugar (hs), normal chow (nc) | step-down latency and open field test novel object recognition/location Morris water maze |
anxiety and recognition memory: hf = hs = nc long-term spatial memory: hs < hf = nc reverse learning: hs = hf < nc |
— | [21] |
| beef (b) versus normal chow (nc) diet | novel apparatus neophobia hole-board box |
anxiety: b < nc working memory: b > nc spatial memory: b > nc |
— | [9] |
| high fat transplanted microbiome (hft), normal chow transplanted microbiome (nct) | elevated plus and open field assays cued fear conditioning |
anxiety: hft > nct cue acquisition, memory and extinction: hft < nct context memory: hft = nct |
differences in protein expression in brain (e.g. BDNF) | [7] |
| CC, antibiotic-treated (Abx), + exercise (exc), + probiotics (pro), + CC faecal transplant (ft) | novel object recognition | short-term memory: no difference across groups long-term memory: Abx and Abx-ft < CC Abx-exc and Abx-pro = CC CC-exc and CC-pro = CC |
differences in neurogenesis across treatment groups | [22] |
| CC, antibiotic-treated (Abx) | object recognition task Barnes maze |
recognition memory: Abx < CC spatial learning and memory: Abx = CC |
differences in protein and receptor expression in hippocampus, amygdala and hypothalamus | [23] |
| OTUs from gut microbiome | open field assay and light/dark box cued fear conditioning |
bacterial OTUs identified as either positively or negatively associated with anxiety and context memory | — | [24] |
| natural variation in microbiome age 1 (quantified as clusters of microbial taxa and diversity indexes) | Mullen Scales of Early Learning at age 1 and age 2 | higher scores associated with high levels of Bacteroides rather than high levels of faecalibacterium. High microbiome diversity associated with poor scores on visual reception, expressive and receptive language skills at age 2 | higher microbiome diversity associated with larger left precentral gyrus, left amygdala and right angular gyrus at age 2 | [16] |
| biopsied colon microbiome from patients with cirrhosis of the liver | psychometric test batteries for psychomotor speed, vasomotor coordination, attention, set shifting and inhibitory control | bacterial OTUs identified as either positively or negatively associated with cognitive performance across cognitive tests | bacterial OTUs identified as either positively or negatively associated with liver inflammation | [17] |
2. Communication between the microbiome and the brain
The gut–brain axis (GBA) describes bidirectional communication between the gastrointestinal tract and the brain through neural, endocrine and immune pathways. The physiological mechanisms through which this communication occurs are complex, not yet fully understood and have been reviewed elsewhere [6,28–31]. In brief, communication networks include the central nervous system (CNS), the enteric nervous system (ENS), autonomic nervous system (ANS) and the hypothalamic–pituitary–adrenal (HPA) axis. The microbiome can modulate the ENS through the excitability of the nervous system [6,32], the production of neurotransmitters such as gamma-aminobutyric acid (GABA) and serotonin [33], and metabolites such as short chain fatty acids that enable signals to reach the CNS through the vagus nerve [34], the longest cranial nerve linking the brainstem to the intestine. Messages from the brain to bacteria in the gut are received by receptors with binding specificity for brain-controlled signalling molecules (reviewed in [28]). Gut–brain communication is also tightly linked to the HPA axis, an endocrine system that coordinates the production and release of stress hormones, including corticosterone, and is known to influence cognition (reviewed in [4]). The development and regulation of the HPA axis is, at least in part, controlled by the microbiome [5,35,36], while the microbiome also responds to signals sent by the HPA axis. Signalling from the brain and the release of stress hormones can influence the permeability of the intestinal barrier maintained by the composition of the microbiome community. Disruption to gut permeability has knock-on effects on the uptake of nutrients and the blocking of pathogens and toxins, thus stimulating immune responses (reviewed in [28,31]). Experimental studies have shown that alterations within the GBA have a direct impact on behaviour, cognitive abilities and neuronal and protein expression in the brain, which we describe throughout this review.
The relationship between the brain and the microbiome is apparent from several studies that have demonstrated marked changes in host cognitive performance between normal, germ free (i.e. mice born without any microorganisms housed in sterile isolators) and experimentally altered microbial communities (i.e. restricted diets, antibiotics, microbiota transplantation and infectious vectors) (table 1 and references therein). Cognitive tasks include tests of learning, behavioural flexibility, fear recall, working and spatial memory and social learning (table 1), with significant differences in cognitive performance found even with small sample sizes (typically ranging from 9 to 15 individuals). These studies are frequently complemented with histology and gene expression assays that look at markers for brain cell activity and neuronal number in, for example, the hippocampus and amygdala (e.g. [10,19,23]). Neurotransmission in the hippocampal serotonergic system is disrupted in a sex-dependent manner in GF mice [35], a system associated with inhibitory control [37,38]. GF mice also have impaired fear-conditioned memory recall associated with differential gene expression in the amygdala compared to control mice, suggesting that an altered microbiome disrupts cellular signalling and causes neuronal hyperactivity [19]. These studies provide causal and functional explanations for links between cognitive and biological phenotypes that can be investigated further by looking at a range of variation in microbial communities across individuals to better understand the biological relevance of microbiome variation and its impact on cognition in natural populations.
3. Individual differences in the microbiome
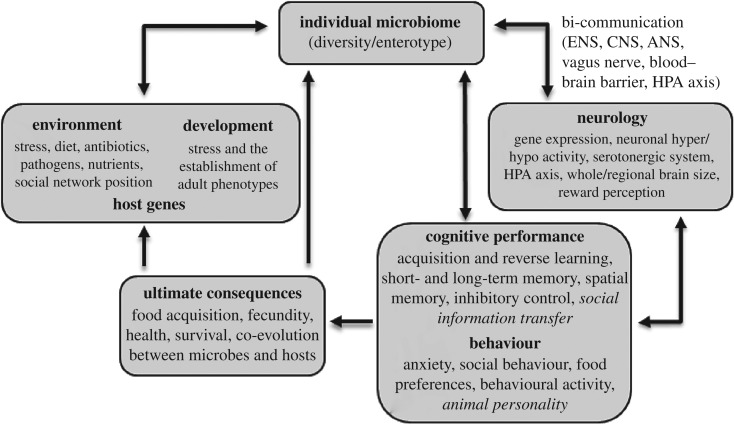
Much of what we know about individual differences in the microbiome stems from human studies that categorize the bacterial composition into enterotypes, that is, distinct profiles defined by the relative abundance of microbes from different taxonomic groups (e.g. [39]). Humans host an incredibly variable microbiome comprising between 1000 and 1150 different bacterial species [40–43] and a plethora of studies have addressed gastrointestinal diversity in an effort to define the core microbial communities associated with internal (e.g. age, heredity) and external (e.g. pharmaceutical, dietary) factors (e.g. [44]). Given the complexity of individual microbiome variation, comprehensive, metagenomic investigations have defined a core functional microbiome comprised of 100-fold more unique genes than in the human genome [45]. Studies are now documenting the microbiome within and between species in a range of animal taxa, including insects (e.g. [46]), birds (e.g. [47]) and other mammals (e.g. [48]). Species diversity within the gut microbiota can be quantified by sequencing the organisms found in faecal material, and in some cases through cloacal swabs (e.g. [49]) and directly from enteric tissue (e.g. [50]). Sequencing typically targets the small subunit ribosomal RNA gene (16S rRNA), a highly conserved gene that can be used as an evolutionary marker to identify taxa and their relative abundance. Alternatively, a shotgun sequencing approach captures all genes present and enables identification of the functional properties of genes within bacterial taxa. Clusters of gene sequences from the microbiota that have more than 97% shared sequence similarity in the 16S rRNA gene are known as operational taxonomic units (OTUs), and the proportion of OTUs that belong to distinct phyla varies across host taxa. Bacteroidetes and Firmicutes are the most common phyla in humans and other mammals, while Proteobacteria and Firmicutes are typically the most common in passerine birds (e.g. [47]). Variation in the microbiome may be determined by host genes [14,15], developmental conditions (e.g. [51]), natal environment (e.g. birth canal [52], nest environment [53]) and current environmental effects (e.g. diet, infection, acute stress (e.g. [9,10])), all of which can determine how resilient the microbiome is to perturbations that may alter its community composition. What causes this variation is at the heart of understanding the ecological and evolutionary significance of links between the microbiome and cognition (figure 1).
Figure 1.
Causes and consequences of individual variation in the microbiome in relation to environmental and developmental effects, and the subsequent impact on neurological, cognitive and behavioural traits. Arrows indicate potential causal directions of relationships and are not exhaustive (for example, development may directly affect cognition independent of microbiome). Italics refer to selected phenotypes that have yet to be investigated empirically (i.e. animal personality) and where null results have been found (i.e. social information transfer [18]).
4. Early life effects on the microbiome and cognition
Early life is a critical period for gut colonization [51], the establishment of the HPA axis, as well as neurodevelopment ([4] and references therein), and growing evidence suggests these systems are interrelated. Disruption to the HPA axis following developmental stressors can impair brain development and function (reviewed in [4,54]). This includes, among other effects, a reduction in neurogenesis following maternal separation [55] and social isolation [56] in rodents, and disrupted song learning through elevated stress hormones and nutritional restriction in birds (reviewed in [54]). Developmental stressors also reduce bacterial communities in primates [57] and rats [58], and GF mice develop exaggerated corticosterone responses [59] and disrupted neurogenesis [60]. Recolonization in mice can normalize adrenal responses, but only within an early developmental window [59,60]. Moreover, variation in microbiome diversity in one-year-old human infants can predict their cognitive abilities at two years of age [16]. Clearly, there are interactive effects of the microbiome and the HPA axis on the brain, yet studies that consider both systems as mechanisms influencing adult phenotypes are limited to rodents and primates, despite the well categorized work on developmental stress in other taxa such as birds [54]. Understanding the causal relationships between these pathways across taxa would provide a broader understanding of how early life experiences, such as dietary changes and maternal hormones, cause individual differences in brain function. Moreover, further work is needed to identify in what stages developmental effects on cognition can or cannot be reversed by, for example, newly colonized microbiota through natural transmission from the environment, or by targeted experimental approaches such as faecal transplantation. Indeed many animals engage in coprophagy by eating their own or conspecifics' faeces (reviewed in [61]). This behaviour is typically observed during periods when microbiome enhancement may be most critical, such as during early development (by eating mother or sibling excretions (e.g. [62]) and in response to environmental stressors (e.g. [63]).
Independent studies that have manipulated developmental history by either altering the microbiome in mice or altering corticosterone levels in birds have shown comparable effects on social behaviour. Unlike control mice, GF mice had no preference for novel as opposed to familiar mice, and had reduced attention times towards a demonstrator mouse during a social learning food-preference test. However, food choices following the social food-preference tasks were similar to CC mice, suggesting that disruption to social behaviour did not impair information use [18]. In separate studies, increased levels of corticosterone administered postnatally in food also caused changes in social learning strategies and information use in birds [64,65]. Moreover, corticosterone-exposed birds avoided associating with their parents and yet, like GF mice, performed as well as control individuals on a foraging task [65]. These studies demonstrate that although performance appears equivalent in foraging tasks, individuals may be achieving these outcomes using different strategies mediated by at least two systems that modulate one another, namely the microbiome and the adrenal response. Developmental stress caused either by a disrupted microbiome or by administering glucocorticoids alters with whom individuals associate and from whom they learn (see also [66], this issue). Although recent studies have shown that social networks predict microbial composition in wild primates [12,26], we predict that positions in social networks may also be mediated through microbiome, which in turn influences social transmission of microbes, both commensal and infectious, that may have beneficial [13,67] or detrimental effects [68–70] on health and cognition.
5. Adult variability in the microbiome and cognition
Cognitive plasticity enables individuals to acquire, retain and update information over time [71]. For example, seasonal changes in neuron proliferation can increase hippocampal volume and improve memory performance in food-caching animals (e.g. [2,72]). The microbiome is also an intrinsic trait that mediates plasticity of cognitive traits by altering phenotypes such as protein expression in the brain [10], adult hippocampal neurogenesis [22] and performance on cognitive tasks (table 1). Changes in the microbiome can occur through pathogen infection [10], dietary changes (e.g. [21]) and exercise [24], all of which are expected to vary across individual lifetimes, particularly in animals that are prone to contracting infections, for example, through social interactions, in animals that experience temporal and geographical changes in food availability and in animals that vary their energy expenditure during migration or breeding seasons. The captive environment can also alter microbiome profiles and therefore has implications for testing cognition in laboratory settings (e.g. [73,74]).
(a). Stress and infection
It is well established that stress can either enhance or block learning and memory, depending on the strength of the stressor and the individual's adrenal response (reviewed in [4]). Moreover, the effect of stress on cognition may be dependent on the individual's microbiome, and individual differences in cognition may not be observed or properly quantified unless tested under different environmental gradients. Infection with the enteric pathogen, Citrobacter rodentium, caused shifts in microbial communities in CC mice, impaired memory and resulted in an associated reduction in hippocampal expression of brain-derived neurotrophic factor (BDNF), but only if mice were exposed to acute stress prior to cognitive testing. By contrast, GF mice without infection showed impaired memory regardless of stress exposure [10]. Therefore, very different conclusions could be drawn regarding individual differences in cognition depending on gut microbiome and environmental conditions during cognitive testing. Individual differences in memory might only be observed if infected individuals were tested under stress.
The role of the microbiome as a proximate mechanism mediating differences in cognition due to stress and infection has yet to be investigated in wild populations, though correlated changes between faecal glucocorticoid metabolites and the microbiome of wild red squirrels (Tamiasciurus hudsonicus) were shown to be associated with an increase in Pasteurellaceae, a group of potential pathogens [75]. The causal direction of these relationships, whether individuals vary in their sensitivity to perturbation within a system and the degree of knock-on effects across systems are all unknown. Manipulative studies are required both in the natal environment and in adulthood, whereby one of these systems is altered (e.g. the microbiome) and the other system is measured (e.g. corticosterone concentrations) to identify the causal effects, individual differences in response to these alterations and the circumstances under which disruption affects individual cognition.
(b). Behavioural activity
A direct parallel can be drawn between exercise in humans and ‘behavioural activity’ in animals. Exercise in humans and rodents can increase adult neurogenesis (e.g. [24,76]) and alter microbiome profiles (e.g. [77]), and recent evidence points to the microbiome's role in mediating the effects of exercise on the generation of neural stem cells [22]. Exercise is important in reversing negative effects on neurogenesis as well as spatial and object recognition memory associated with microbial imbalance caused by antibacterial treatment in adult mice [22]. These processes may be relevant to animals during natal dispersal, or migrating animals such as birds that experience shifts in their microbiota as a result of long periods without feeding, or due to dietary changes across breeding grounds, stopover locations during migration and nutrient availability on non-breeding grounds [78]. Migratory staging could include increased use of farmlands [79] that can be contaminated with antibiotics present in animal feed [80]. Whether the effect of bacterial community modulation and activity levels on neurogenesis in mammals are also observed in birds has yet to be explored. Given that the hippocampus and spatial memory may be important in some species for effective migration [81], fluctuations in neurogenesis and memory associated with alterations in the gut microbiota could have the potential to influence the efficiency of individual migratory events. However, activity associated with long-distance flights may buffer effects on neurogenesis due to negative shifts in avian microbial populations. Differences in behavioural activity levels among individuals may also be associated with animal personalities, defined as consistent individual differences in behaviour over time [82]. Indeed, one notable behavioural measure that is frequently associated with the microbiome is anxiety, sometimes measured in terms of activity or exploratory behaviour (reviewed in [11]). Although not yet tested explicitly in the context of animal personality, we predict that variation in traits such as activity, anxiety or boldness may be associated with individual differences in microbiome profiles.
(c). Diet and foraging behaviour
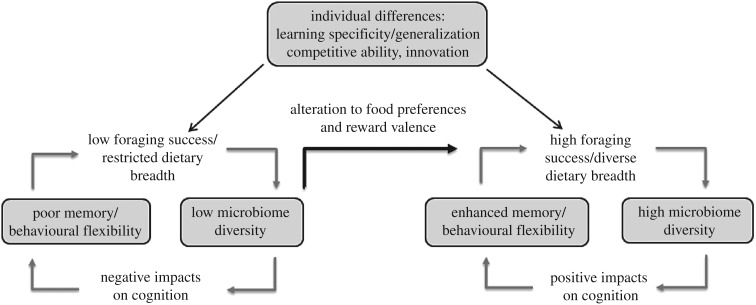
Diet as a consequence of foraging behaviour is a fundamental aspect of animal ecology and evolution, and access to food types can be influenced by a range of factors including cognition, competitive ability and innovativeness. Diet has been reported to alter microbiome composition, sometimes within 24 h [83]. Dietary manipulation has also led to impaired or enhanced cognition in mice models [7,9,21], and in one case has been shown to be independent of nutritional value [7]. Diet varies between and within species along spatial and temporal scales, which can cause shifts in gut microbiota as demonstrated in animals that engage in coprophagy [61], migratory birds during stopovers [84], wild mice across seasons [85] and animals brought into captivity [73], and therefore could impact cognition as a result. Microbiome shifts may also arise in the opposite causal direction when cognition directly affects an individual's dietary preferences or capacity to gain access to food, resulting in feedback loops between cognition and the microbiome via diet (figure 2). For instance, individual differences in learning specificity versus generalization of aposematic warning signals will affect the types of food individuals ingest when encountering novel prey [86]. If aversion to known aposematic signals is generalized to novel prey species that share similar cues, individuals may reduce the chances of ingesting toxic substances that could have adverse effects on gut microbiota, but generalized aversion may also restrict their dietary breadth of palatable prey, resulting in a less varied microbiome than individuals that sample prey that do not share the specific features of learned warning signals. These learning strategies and associated effects on the microbiome would be reliant on the range of variable prey types present in the environment (e.g. [86]).
Figure 2.
Positive feedback loops between microbiome diversity and cognition. A highly diverse microbiome improves memory and behavioural flexibility and associated foraging success, and as a result, dietary breadth maintains a diverse microbiome. By contrast, low microbial diversity impairs cognitive abilities, resulting in poor foraging success. However, the gut microbiome can influence host food preferences and reward valence as a means to increase microbial diversity, and to improve learning and memory for food sources, breaking out of the positive feedback loop (black arrow). Finally, individual differences in cognition (specificity versus generalization), competitive ability and innovativeness influence diet. Arrows represent directional influences on, or in response to, phenotypic traits (represented within grey boxes).
Individual microbiome profiles mediated by food access may be further determined by behaviours such as competitive ability, which has been shown to be negatively correlated with novel foraging behaviour [87] and learning [88] through the ‘necessity drives innovativeness' hypothesis. Consequently, competitive individuals may have greater access to easily accessible foods (e.g. bird feeders), while less competitive individuals are more likely to use alternative, more diverse food sources through innovative foraging behaviours. Multiple related phenotypic traits including innovation, cognitive traits such as learning and generalization and animal personality traits such as competitive ability, all have the potential to influence the microbiome composition mediated through diet, which in turn may feedback on cognition (figure 2).
(d). Nutrition
The effects of food nutritional value and foraging success on individual cognition are almost entirely unknown in natural populations (but see [89]), and studies on how these effects are mediated by the microbiome are restricted to the laboratory. High sugar and high fat diets in mice caused alterations in microbiota down to the genus level and impaired spatial memory and reversal learning, with effects most prominent in high-sugar groups [21]. The consequences of diet may be particularly relevant for species that consume a high proportion of sugar-rich foods such as nectar (e.g. hummingbirds) and fat-filled foods such as seeds and nuts (e.g. Clark's nutcracker, Nucifraga columbiana, and squirrels), while depending on spatial memory to remember which flowers have been depleted (e.g. [90]) and where food caches have been stored [2,72]. Alternatively, it may be found that, for example, species with sugar-rich diets have uniquely adapted physiological systems that benefit from having high proportions of sucrose-utilising bacteria. Other food types, such as protein, can increase microbial diversity and improve performance on tests of working and spatial memory [9]. Individuals within and across populations that vary in their propensity to forage on meat through innovative and opportunistic hunting behaviours [91,92] would facilitate individual variation in their microbiome. Positive feedback loops between cognition and foraging may be present whereby, for example, individuals with low microbial diversity have impaired behavioural flexibility and/or memory, and as a result are restricted to a more limited dietary niche, which in turn maintains low microbiome diversity (figure 2).
Food preferences can influence learning acquisition and memory retention depending on the nutritional content of the food types given (e.g. [93–95]), whether they can be metabolized [96] and if reward receptors for the metabolites are present in the brain [97], independent of taste perception [96]. The microbiome can alter reward signal pathways, release toxins that influence host mood, alter taste receptors and produce exact mammalian neurochemical analogues that are transmitted to the brain via the vagus nerve, all of which may contribute to changes in eating behaviour (reviewed in [98]). The absence of distinct commensal gut bacteria can modulate food preferences when Drosophila melanogaster (the common house-fly) is deficient in essential amino acids (eAA) [99]. By preferentially choosing foods containing Acetobacter pomorum and Lactobacilli species, individuals reversed the negative effects associated with eAA deficiency. This behaviour suggests that Drosophila can directly modulate their own microbiome, and this change in food preference could act as a mechanism to break free of positive feedback loops between poor foraging success and low microbial diversity (figure 2). Moreover, the metabolic profiles of the microbiome community could influence the strength of reward perception of different food types, thereby increasing reinforcement. As predicted by conditioning theory [100], we would expect faster learning and more robust memory associated with foods that contain specific microbes or nutrients. Given the bi-directional communication of the GBA (see §2, reviewed in [6,28–31]) and evidence indicating that the vagus nerve can stimulate gut inflammation and intestinal permeability, perhaps resulting in microbiome modulation (reviewed in [101]), it is tempting to speculate that neurotransmitters released by the brain as a result of obtaining these food types could be communicated to the microbiome via the vagus nerve. This proposition that the process of memory formation is signalled to the microbiome is an area of research that has yet to be explored.
When assessing links between the gut microbiota and cognition, it may be appropriate to disentangle the effects of nutrition versus microbiome on plasticity of cognitive performance. The transplantation of gut microbiota from experimental mice into control mice showed that cognitive changes associated with high fat diets were due to alterations in the microbiota, independent of food/nutrient ingestion [7]. By contrast, beef-fed mice with higher working memory than chow-fed mice ingested higher concentrations of taurine [9], an amino acid that can directly improve cognitive function [102], potentially independently of the microbiome. In a passerine bird, parents preferentially fed their nestlings taurine-rich spiders when chicks were four days old, and chicks supplemented with extra taurine had improved spatial memory [89]. Taurine has been shown to influence microbiome composition in mice [104], and since the microbiome also plays a role in the absorption and synthesis of micronutrients, the metabolism of compounds such as polyphenols [7,104] and taurine [103] may provide yet another pathway by which the microbiome influences cognition.
(e). Captivity
Individual differences in cognition are often quantified in environments where subjects are either reared in captivity or brought temporarily into captivity from the wild. Several aspects of the captive environment may influence individual cognitive performance, including neophobia, habituation and stress, but rarely is the effect of captivity on the microbiome considered. Captive-bred animals exhibit lower gut microbial diversity than their wild counterparts across a wide range of host taxa including seals [105], primates [74], birds [106], rodents [73] and fish [107], with some bacterial families being completely absent in captive animals [105]. In primates, the degree of impact reflects the intensity of captivity. Wild populations have the most natural microbiome, semi-captive sanctuary environments have an intermediate microbiome, and isolated captive populations have the least diverse microbiome [74]. Humans also follow this trend, whereby elderly populations confined to long-stay communities demonstrated decreased microbial diversity, lost taxa associated with positive health parameters and became frailer than those in short-stay communities or those who did not attend assisted living facilities at all [108]. The effects of reduced microbial diversity due to captivity on cognition are unknown, though we expect them to differ depending on the level of captivity. Subjects brought in from the wild are exposed to a restricted environment with an altered diet, and individuals held for longer durations may experience a greater decline in bacterial community diversity than those held for shorter durations, therefore the timing of cognitive tests may be crucial if associated changes in the microbiome do indeed influence performance on cognitive tasks. By comparison, studies on hand-reared captive populations with reduced microbial diversity may generate conservative estimates of species variation in cognition compared to wild populations. Moreover, we may expect repeatability in cognitive tasks to be more stable in captive populations as opposed to wild-caught populations if environmental and diet conditions in captivity are relatively stable compared to natural habitats. Administration of probiotics could be considered as a supplement for animals in captivity as they improved memory and emotional wellbeing in humans [109], normalized cognitive profiles, stress responses and gene expression in mice [10,20,22] and reduced gut leakiness and HPA response to acute stressors in rats [110].
6. Evolutionary consequences of the microbiome on cognition
So far we have focused largely on environmental effects that determine and alter individuals' microbiomes, and consequently cognition through developmental and phenotypic plasticity. There is also potential for selection to act on the microbiome and host genes that determine cognitive abilities. The role of the microbiome as a driver of the evolution of cognition has been explored theoretically within the context of the social brain hypothesis [111], suggesting that selection favours social complexity as a means to increase transmission of beneficial microbes causing host–symbiont coevolution on RNA regulation in the brain [112]. Furthermore, there is growing realization that the microbiome plays an important role in epigenetic control of gene expression associated with cognition and anxiety [112]. It is difficult to identify to what extent the microbiome is heritable as any apparent stability may be caused by environment effects shared by parent and offspring (e.g. [113,114]). Nevertheless, even if microorganisms are transferred from the environment, the bacterial community that establishes itself is filtered by and adapted to host traits including gut physiology, enzymatic phenotypes, immune function and dietary profiles, which is suggestive of co-diversification [46]. Indeed, individual variation in the microbiome has been shown to be associated specifically with host genetic variation [14,15], providing evidence that host genomes are adapting to their microbiome and vice versa. Further support for co-diversification comes from phylogenetic comparative studies in the mammalian lineage where diet and microbiome profiles have evolved convergently [115,116]. Moreover, because microbiome diversity is linked to fitness in mammals [117], life-history traits including clutch size, lay date and adult survival in birds [118] and fecundity in Drosophila [99], there is likely an adaptive component to microbiome variation that may function to optimize food preferences, social behaviour, stress responses, immune functions and cognitive abilities to mutually benefit both host and microbes.
7. Conclusion
We have proposed several potential causal links between the gut microbiome, ecology, physiology, behaviour and cognition in natural animal populations, derived from the rapidly increasing plethora of studies from humans and laboratory animals. Because the microbiome is ubiquitous across species and variable among individuals, it is a promising avenue of research for investigating cognition in the context of individual differences across a broad range of taxa from evolutionary, developmental and phenotypic plasticity perspectives. We encourage further research into unexplored questions as to how natural variation in the microbiome is determined by early life effects and throughout adulthood due to socio-ecological factors as well as interactions with adrenal and immunological systems. Furthermore, studying multiple proximate mechanisms in tandem is necessary to uncover the interacting and causal relationships between cognition, behaviour, diet and the microbiome, and will help to elucidate the environmental and genetic effects determining individual differences in cognition and coevolutionary processes between microbes and hosts.
Acknowledgements
We would like to thank Paul Ross and Catherine Stanton for insightful discussions and two anonymous reviewers for their valuable comments. We also thank Joah Madden for constructive comments on the manuscript and for organizing the Causes and Consequences of Individual Differences in Cognition Workshop from which this special issue arose.
Data accessibility
This article has no additional data.
Authors' contributions
G.L.D. wrote the manuscript with written and intellectual contributions from A.C.C., C.N.J. and J.L.Q. The idea for the paper was conceived by G.L.D. and J.L.Q. All authors gave final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
G.L.D., A.C.C. and J.L.Q. have received funding from the European Research Council under the European Union's Horizon 2020 Programme (FP7/2007-2013)/ERC Consolidator Grant ‘Evoecocog’ Project no. 617509, awarded to J.L.Q., and Marie Curie Career Integration grant no. PCIG12-GA-2012-334383 awarded to J.L.Q. C.N.J. is funded by the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie IF grant agreement no. 708986. However, the funders had no role in content, data collection, or the decision to submit for publication.
References
- 1.Emery NJ, Clayton NS. 2004. The mentality of crows: convergent evolution of intelligence in corvids and apes. Science 306, 1903–1907. ( 10.1126/science.1098410) [DOI] [PubMed] [Google Scholar]
- 2.Sherry DF, Vaccarino AL, Buckenham K, Herz RS. 1989. The hippocampal complex of food-storing birds. Brain Behav. Evol. 34, 308–317. ( 10.1159/000116516) [DOI] [PubMed] [Google Scholar]
- 3.Croston R, Branch CL, Kozlovsky DY, Dukas R, Pravosudov VV. 2015. Heritability and the evolution of cognitive traits. Behav. Ecol. 26, 1447–1459. ( 10.1093/beheco/arv088) [DOI] [Google Scholar]
- 4.Lupien SJ, McEwen BS, Gunnar MR, Heim C. 2009. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat. Rev. Neurosci. 10, 434–445. ( 10.1038/nrn2639) [DOI] [PubMed] [Google Scholar]
- 5.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl Acad. Sci. USA 108, 3047–3052. ( 10.1073/pnas.1010529108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer EA. 2011. Gut feelings: the emerging biology of gut–brain communication. Nat. Rev. Neurosci. 12, 453–466. ( 10.1038/nrn3071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard E IV, Taylor CM, Welsh DA, Berthoud HR. 2015. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol. Psychiatry 77, 607–615. ( 10.1016/j.biopsych.2014.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan D, Yu Z. 2014. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 5, 108–119. ( 10.4161/gmic.26945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Dowd SE, Scurlock B, Acosta-Martinez V, Lyte M. 2009. Memory and learning behavior in mice is temporally associated with diet-induced alterations in gut bacteria. Physiol. Behav. 96, 557–567. ( 10.1016/j.physbeh.2008.12.004) [DOI] [PubMed] [Google Scholar]
- 10.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, MacQueen G, Sherman PM. 2011. Bacterial infection causes stress-induced memory dysfunction in mice. Gut 60, 307–317. ( 10.1136/gut.2009.202515) [DOI] [PubMed] [Google Scholar]
- 11.Foster JA, McVey Neufeld KA. 2013. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312. ( 10.1016/j.tins.2013.01.005) [DOI] [PubMed] [Google Scholar]
- 12.Perofsky AC, Lewis RJ, Abondano LA, Di Fiore A, Meyers LA. 2017. Hierarchical social networks shape gut microbial composition in wild Verreaux's sifaka. Proc. R. Soc. B 284, 20172274 ( 10.1098/rspb.2017.2274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl Acad. Sci. USA 108, 19 288–19 292. ( 10.1073/pnas.1110474108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson AK, et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18 933–18 938. ( 10.1073/pnas.1007028107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKnite AM, et al. 2012. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS ONE 7, e39191 ( 10.1371/journal.pone.0039191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson AL, et al. 2018. Infant gut microbiome associated with cognitive development. Biol. Psychiatry 83, 148–159. ( 10.1016/j.biopsych.2017.06.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajaj JS, et al. 2012. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G675–G685. ( 10.1152/ajpgi.00152.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. 2014. Microbiota is essential for social development in the mouse. Mol. Psychiatry 19, 146–148. ( 10.1038/mp.2013.65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoban AE, Stilling RM, Moloney G, Shanahan F, Dinan TG, Clarke G, Cryan JF. 2017. The microbiome regulates amygdala-dependent fear recall. Mol. Psychiatry. 23, 1134–1144. ( 10.1038/mp.2017.100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savignac HM, Tramullas M, Kiely B, Dinan TG, Cryan JF. 2015. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behav. Brain Res. 287, 59–72. ( 10.1016/j.bbr.2015.02.044) [DOI] [PubMed] [Google Scholar]
- 21.Magnusson KR, Hauck L, Jeffrey BM, Elias V, Humphrey A, Nath R, Perrone A, Bermudez LE. 2015. Relationships between diet-related changes in the gut microbiome and cognitive flexibility. Neuroscience 300, 128–140. ( 10.1016/j.neuroscience.2015.05.016) [DOI] [PubMed] [Google Scholar]
- 22.Mohle L, et al. 2016. Ly6Chi monocytes provide a link between antibiotic-induced changes in gut microbiota and adult hippocampal neurogenesis. Cell Rep. 15, 1945–1956. ( 10.1016/j.celrep.2016.04.074) [DOI] [PubMed] [Google Scholar]
- 23.Frohlich EE, et al. 2016. Cognitive impairment by antibiotic-induced gut dysbiosis: analysis of gut microbiota–brain communication. Brain Behav. Immun. 56, 140–155. ( 10.1016/j.bbi.2016.02.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang SS, et al. 2014. Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol. Neurodegener. 9, 36 ( 10.1186/1750-1326-9-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archie EA, Tung J. 2015. Social behavior and the microbiome. Curr. Opin. Behav. Sci. 6, 28–34. ( 10.1016/j.cobeha.2015.07.008) [DOI] [Google Scholar]
- 26.Tung J, et al. 2015. Social networks predict gut microbiome composition in wild baboons. eLife 4, e05224 ( 10.7554/eLife.05224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grieneisen LE, Livermore J, Alberts S, Tung J, Archie EA. 2017. Group living and male dispersal predict the core gut microbiome in wild baboons. Integr. Comp. Biol. 57, 770–785. ( 10.1093/icb/icx046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carabotti M, Scirocco A, Maselli MA, Severi C. 2015. The gut–brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 28, 203–209. [PMC free article] [PubMed] [Google Scholar]
- 29.Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712. ( 10.1038/nrn3346) [DOI] [PubMed] [Google Scholar]
- 30.Forsythe P, Bienenstock J, Kunze WA. 2014. Vagal pathways for microbiome–brain–gut axis communication. In Microbial endocrinology: the microbiota–gut–brain axis in health and disease (eds Lyte M, Cryan JF), pp. 115–133. New York, NY: Springer New York. [DOI] [PubMed] [Google Scholar]
- 31.Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP. 2015. Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell Neurosci. 9, 392 ( 10.3389/fncel.2015.00392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunze WA, Mao YK, Wang B, Huizinga JD, Ma X, Forsythe P, Bienenstock J. 2009. Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium-dependent potassium channel opening. J. Cell. Mol. Med. 13, 2261–2270. ( 10.1111/j.1582-4934.2009.00686.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer LM, Aravind L, Coon SL, Klein DC, Koonin EV. 2004. Evolution of cell−cell signaling in animals: did late horizontal gene transfer from bacteria have a role? Trends Genet. 20, 292–299. ( 10.1016/j.tig.2004.05.007) [DOI] [PubMed] [Google Scholar]
- 34.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl Acad. Sci. USA 108, 16050–16055. ( 10.1073/pnas.1102999108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. 2012. The microbiome–gut–brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 18, 666–673. ( 10.1038/mp.2012.77) [DOI] [PubMed] [Google Scholar]
- 36.Neufeld KM, Kang N, Bienenstock J, Foster JA. 2011. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 23, 255–e119 ( 10.1111/j.1365-2982.2010.01620.x) [DOI] [PubMed] [Google Scholar]
- 37.Brunner D, Hen R. 1997. Insights into the neurobiology of impulsive behavior from serotonin receptor knockout mice. Ann. N. Y. Acad. Sci. 836, 81–105. ( 10.1111/j.1749-6632.1997.tb52356.x) [DOI] [PubMed] [Google Scholar]
- 38.Homberg JR, Pattij T, Janssen MCW, Ronken E, de Boer SF, Schoffelmeer ANM, Cuppen E. 2007. Serotonin transporter deficiency in rats improves inhibitory control but not behavioural flexibility. Eur. J. Neurosci. 26, 2066–2073. ( 10.1111/j.1460-9568.2007.05839.x) [DOI] [PubMed] [Google Scholar]
- 39.Arumugam M, et al. 2011. Enterotypes of the human gut microbiome. Nature 473, 174–180. ( 10.1038/nature09944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697. ( 10.1126/science.1177486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lloyd-Price J, Abu-Ali G, Huttenhower C. 2016. The healthy human microbiome. Genome Med. 8, 51 ( 10.1186/s13073-016-0307-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308, 1635–1638. ( 10.1126/science.1110591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gill SR, et al. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. ( 10.1126/science.1124234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. ( 10.1038/nature11234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. ( 10.1038/nature08821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE. 2014. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol. Ecol. 23, 1268–1283. ( 10.1111/mec.12611) [DOI] [PubMed] [Google Scholar]
- 47.Hird SM, Sanchez C, Carstens BC, Brumfield RT. 2015. Comparative gut microbiota of 59 neotropical bird species. Front. Microbiol. 6, 1403 ( 10.3389/fmicb.2015.01403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ. 2017. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 14319 ( 10.1038/ncomms14319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Videvall E, Strandh M, Engelbrecht A, Cloete S, Cornwallis CK. 2017. Measuring the gut microbiome in birds: comparison of faecal and cloacal sampling. Mol. Ecol. 18, 424–434. ( 10.1111/1755-0998) [DOI] [PubMed] [Google Scholar]
- 50.Roggenbuck M, et al. 2014. The microbiome of new world vultures. Nat. Commun. 5, 5498 ( 10.1038/ncomms6498) [DOI] [PubMed] [Google Scholar]
- 51.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA 108, 4578–4585. ( 10.1073/pnas.1000081107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bokulich NA, et al. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82 ( 10.1126/scitranslmed.aad7121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacob S, Parthuisot N, Vallat A, Ramon-Portugal F, Helfenstein F, Heeb P, Martin L. 2015. Microbiome affects egg carotenoid investment, nestling development and adult oxidative costs of reproduction in Great tits. Funct. Ecol. 29, 1048–1058. ( 10.1111/1365-2435.12404) [DOI] [Google Scholar]
- 54.MacDougall-Shackleton SA, Spencer KA. 2012. Developmental stress and birdsong: current evidence and future directions. J. Ornithol. 153, 105–117. ( 10.1007/s10336-011-0807-x) [DOI] [Google Scholar]
- 55.Mirescu C, Peters JD, Gould E. 2004. Early life experience alters response of adult neurogenesis to stress. Nat. Neurosci. 7, 841–846. ( 10.1038/nn1290) [DOI] [PubMed] [Google Scholar]
- 56.Lu L, Bao G, Chen H, Xia P, Fan X, Zhang J, Pei G, Ma L. 2003. Modification of hippocampal neurogenesis and neuroplasticity by social environments. Exp. Neurol. 183, 600–609. ( 10.1016/S0014-4886(03)00248-6) [DOI] [PubMed] [Google Scholar]
- 57.Bailey MT, Coe CL. 1999. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev. Psychobiol. 35, 146–155. ( 10.1002/(SICI)1098-2302(199909)35:2%3C146::AID-DEV7%3E3.0.CO;2-G) [DOI] [PubMed] [Google Scholar]
- 58.O'Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EMM, Cryan JF, Dinan TG. 2009. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol. Psychiatry 65, 263–267. ( 10.1016/j.biopsych.2008.06.026) [DOI] [PubMed] [Google Scholar]
- 59.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. 2004. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J. Physiol. 558, 263–275. ( 10.1113/jphysiol.2004.063388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, O'Leary OF. 2015. Adult hippocampal neurogenesis is regulated by the microbiome. Biol. Psychiatry 78, e7–e9. ( 10.1016/j.biopsych.2014.12.023) [DOI] [PubMed] [Google Scholar]
- 61.Soave O, Brand CD. 1991. Coprophagy in animals: a review. Cornell Vet. 81, 357–364. [PubMed] [Google Scholar]
- 62.Korner M, Diehl JMC, Meunier J. 2016. Growing up with feces: benefits of allo-coprophagy in families of the European earwig. Behav. Ecol. 27, 1775–1781. ( 10.1093/beheco/arw113) [DOI] [Google Scholar]
- 63.Beerda B, Schilder MBH, Van Hooff JARA, De Vries HW, Mol JA. 1999. Chronic stress in dogs subjected to social and spatial restriction. I. Behavioral responses. Physiol. Behav. 66, 233–242. ( 10.1016/S0031-9384(98)00289-3) [DOI] [PubMed] [Google Scholar]
- 64.Boogert NJ, Zimmer C, Spencer KA. 2013. Pre- and post-natal stress have opposing effects on social information use. Biol. Lett. 9, 20121088 ( 10.1098/rsbl.2012.1088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farine D, Spencer K, Boogert N. 2015. Early-life stress triggers juvenile zebra finches to switch social learning strategies. Curr. Biol. 25, 2184–2188. ( 10.1016/j.cub.2015.06.071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boogert NJ, Lachlan RF, Spencer KA, Templeton CN, Farine DR. 2018. Stress hormones, social associations and song learning in zebra finches. Phil. Trans. R. Soc. B 373, 20170290 ( 10.1098/rstb.2017.0290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lombardo MP. 2008. Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav. Ecol. Sociobiol. 62, 479–497. ( 10.1007/s00265-007-0428-9) [DOI] [Google Scholar]
- 68.Gegear RJ, Otterstatter MC, Thomson JD. 2006. Bumble-bee foragers infected by a gut parasite have an impaired ability to utilize floral information. Proc. R. Soc. B 273, 1073–1078. ( 10.1098/rspb.2005.3423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kavaliers M, Colwell DD, Galea LAM. 1995. Parasitic infection impairs spatial learning in mice. Anim. Behav. 50, 223–229. ( 10.1006/anbe.1995.0234) [DOI] [Google Scholar]
- 70.Soler JJ, Peralta-Sanchez JM, Martin-Vivaldi M, Martin-Platero AM, Flensted-Jensen E, Moller AP. 2012. Cognitive skills and bacterial load: comparative evidence of costs of cognitive proficiency in birds. Naturwissenschaften 99, 111–122. ( 10.1007/s00114-011-0875-z) [DOI] [PubMed] [Google Scholar]
- 71.Shettleworth SJ. 2010. Cognition, evolution and behaviour. New York, NY: Oxford University Press. [Google Scholar]
- 72.Sherry DF, Hoshooley JS. 2010. Seasonal hippocampal plasticity in food-storing birds. Phil. Trans. R. Soc. B 365, 933–943. ( 10.1098/rstb.2009.0220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kohl KD, Skopec MM, Dearing MD. 2014. Captivity results in disparate loss of gut microbial diversity in closely related hosts. Conserv. Physiol. 2, cou009 ( 10.1093/conphys/cou009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clayton JB, et al. 2016. Captivity humanizes the primate microbiome. Proc. Natl Acad. Sci. USA 113, 10 376–10 381. ( 10.1073/pnas.1521835113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stothart MR, Bobbie CB, Schulte-Hostedde AI, Boonstra R, Palme R, Mykytczuk NCS, Newman AEM. 2016. Stress and the microbiome: linking glucocorticoids to bacterial community dynamics in wild red squirrels. Biol. Lett. 12, 20150875 ( 10.1098/rsbl.2015.0875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.So JH, Huang C, Ge M, Cai G, Zhang L, Lu Y, Mu Y. 2017. Intense exercise promotes adult hippocampal neurogenesis but not spatial discrimination. Front. Cell. Neurosci. 11, 13 ( 10.3389/fncel.2017.00013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monda V, et al. 2017. Exercise modifies the gut microbiota with positive health effects. Oxid. Med. Cell Longev. 2017, 3831972 ( 10.1155/2017/3831972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waite DW, Ujvari B, Hoye BJ, Klaassen M. 2017. Active migration is associated with specific and consistent changes to gut microbiota in Calidris shorebirds. J. Anim. Ecol. 87, 428-437. ( 10.1111/1365-2656.12784) [DOI] [PubMed] [Google Scholar]
- 79.Prop J, Black JM. 1998. Food intake, body reserves and reproductive success of barnacle geese Branta leucopsis staging in different habitats. In Research on Arctic geese (eds Mehlum F, Black JM, Madsen J), pp. 175–193. Oslo, Norway: Norsk Polarinstitutt. [Google Scholar]
- 80.Burkholder J, Libra B, Weyer P, Heathcote S, Kolpin D, Thorne PS, Wichman M. 2007. Impacts of waste from concentrated animal feeding operations on water quality. Environ. Health Perspect. 115, 308–312. ( 10.1289/ehp.8839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pravosudov VV, Kitaysky AS, Omanska A. 2006. The relationship between migratory behaviour, memory and the hippocampus: an intraspecific comparison. Proc. R. Soc. B 273, 2641–2649. ( 10.1098/rspb.2006.3624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reale D, Reader SM, Sol D, McDougall PT, Dingemanse NJ. 2007. Integrating animal temperament within ecology and evolution. Biol. Rev. 82, 291–318. ( 10.1111/j.1469-185X.2007.00010.x) [DOI] [PubMed] [Google Scholar]
- 83.David LA, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. ( 10.1038/nature12820) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lewis WB, Moore FR, Wang S. 2016. Changes in gut microbiota of migratory passerines during stopover after crossing an ecological barrier. Auk 134, 137–145. ( 10.1642/AUK-16-120.1) [DOI] [Google Scholar]
- 85.Maurice CF, Knowles SC, Ladau J, Pollard KS, Fenton A, Pedersen AB, Turnbaugh PJ. 2015. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 9, 2423–2434. ( 10.1038/ismej.2015.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ihalainen E, Rowland HM, Speed MP, Ruxton GD, Mappes J. 2012. Prey community structure affects how predators select for Mullerian mimicry. Proc. R. Soc. B 279, 2099–2105. ( 10.1098/rspb.2011.2360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cole EF, Quinn JL. 2012. Personality and problem-solving performance explain competitive ability in the wild. Proc. R. Soc. B 279, 1168–1175. ( 10.1098/rspb.2011.1539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mery F, Kawecki TJ. 2003. A fitness cost of learning ability in Drosophila melanogaster. Proc. R. Soc. Lond. B 270, 2465–2469. ( 10.1098/rspb.2003.2548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold KE, Ramsay SL, Donaldson C, Adam A. 2007. Parental prey selection affects risk-taking behaviour and spatial learning in avian offspring. Proc. R. Soc. B 274, 2563–2569. ( 10.1098/rspb.2007.0687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henderson J, Hurly TA, Healy SD. 2001. Rufous hummingbirds' memory for flower location. Anim. Behav. 61, 981–986. ( 10.1006/anbe.2000.1670) [DOI] [Google Scholar]
- 91.Estok P, Zsebok S, Siemers BM. 2010. Great tits search for, capture, kill and eat hibernating bats. Biol. Lett. 6, 59 ( 10.1098/rsbl.2009.0611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mitani JC, Watts DP. 2001. Why do chimpanzees hunt and share meat? Anim. Behav. 61, 915–924. ( 10.1006/anbe.2000.1681) [DOI] [Google Scholar]
- 93.Burke CJ, Waddell S. 2011. Remembering nutrient quality of sugar in Drosophila. Curr. Biol. 21, 746–750. ( 10.1016/j.cub.2011.03.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sclafani A, Ackroff K. 2016. Operant licking for intragastric sugar infusions: differential reinforcing actions of glucose, sucrose and fructose in mice. Physiol. Behav. 153, 115–124. ( 10.1016/j.physbeh.2015.10.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simcock NK, Gray H, Bouchebti S, Wright GA. 2017. Appetitive olfactory learning and memory in the honeybee depend on sugar reward identity. J. Insect. Physiol. 106, 71–77. ( 10.1016/j.jinsphys.2017.08.009) [DOI] [PubMed] [Google Scholar]
- 96.Dus M, Min S, Keene AC, Lee GY, Suh GSB. 2011. Taste-independent detection of the caloric content of sugar in Drosophila. Proc. Natl Acad. Sci. USA 108, 11 644–11 649. ( 10.1073/pnas.1017096108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyamoto T, Slone J, Song X, Amrein H. 2012. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 151, 1113–1125. ( 10.1016/j.cell.2012.10.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alcock J, Maley CC, Aktipis CA. 2014. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 36, 940–949. ( 10.1002/bies.201400071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leitao-Goncalves R, et al. 2017. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 15, e2000862 ( 10.1371/journal.pbio.2000862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rescorla RA, Wagner A. 1972. A theory of Pavlovian conditioning: variations in the effectiveness of reinforcement and nonreinforcement. In Classical conditioning II: current research (eds Black AH, Prokasy WF), pp. 64–99. New York, NY: Appleton-Century-Crofts. [Google Scholar]
- 101.Bonaz B, Bazin T, Pellissier S. 2018. The vagus nerve at the interface of the microbiota–gut–brain axis. Front. Neurosci. 12, 49 ( 10.3389/fnins.2018.00049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim HY, et al. 2014. Taurine in drinking water recovers learning and memory in the adult APP/PS1 mouse model of Alzheimer's disease. Sci. Rep. 4, 7467 ( 10.1038/srep07467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu H, Guo Z, Shen S, Shan W. 2016. Effects of taurine on gut microbiota and metabolism in mice. Amino Acids 48, 1601–1617. ( 10.1007/s00726-016-2219-y) [DOI] [PubMed] [Google Scholar]
- 104.Vauzour D. 2012. Dietary polyphenols as modulators of brain functions: biological actions and molecular mechanisms underpinning their beneficial effects. Oxid. Med. Cell Longev. 2012, 914273 ( 10.1155/2012/914273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nelson TM, Rogers TL, Carlini AR, Brown MV. 2013. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145. ( 10.1111/1462-2920.12022) [DOI] [PubMed] [Google Scholar]
- 106.Wienemann T, Schmitt-Wagner D, Meuser K, Segelbacher G, Schink B, Brune A, Berthold P. 2011. The bacterial microbiota in the ceca of Capercaillie (Tetrao urogallus) differs between wild and captive birds. Syst. Appl. Microbiol. 34, 542–551. ( 10.1016/j.syapm.2011.06.003) [DOI] [PubMed] [Google Scholar]
- 107.Dhanasiri AKS, Brunvold L, Brinchmann MF, Korsnes K, Bergh O, Kiron V. 2011. Changes in the intestinal microbiota of wild Atlantic cod Gadus morhua L. upon captive rearing. Microb. Ecol. 61, 20–30. ( 10.1007/s00248-010-9673-y) [DOI] [PubMed] [Google Scholar]
- 108.Jeffery IB, Lynch DB, O'Toole PW. 2015. Composition and temporal stability of the gut microbiota in older persons. ISME J. 10, 170–182. ( 10.1038/ismej.2015.88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lawrence K, Hyde J. 2017. Microbiome restoration diet improves digestion, cognition and physical and emotional wellbeing. PLoS ONE 12, e0179017 ( 10.1371/journal.pone.0179017) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Ait-Belgnaoui A, et al. 2012. Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 37, 1885–1895. ( 10.1016/j.psyneuen.2012.03.024) [DOI] [PubMed] [Google Scholar]
- 111.Dunbar RIM. 1998. The social brain hypothesis. Evol. Anthropol. 6, 178–190. ( 10.1002/(SICI)1520-6505(1998)6:5%3C178::AID-EVAN5%3E3.0.CO;2-8) [DOI] [Google Scholar]
- 112.Stilling RM, Bordenstein SR, Dinan TG, Cryan JF. 2014. Friends with social benefits: host–microbe interactions as a driver of brain evolution and development? Front. Cell. Infect. Microbiol. 4, 147 ( 10.3389/fcimb.2014.00147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lucas FS, Heeb P. 2005. Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caeruleus nestlings. J. Avian Biol. 36, 510–516. ( 10.1111/j.0908-8857.2005.03479.x) [DOI] [Google Scholar]
- 114.Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H. 2012. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc. Natl Acad. Sci. USA 109, 13 034–13 039. ( 10.1073/pnas.1110994109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320, 1647–1651. ( 10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974. ( 10.1126/science.1198719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suzuki TA. 2017. Links between natural variation in the microbiome and host fitness in wild mammals. Integr. Comp. Biol. 57, 756–769. ( 10.1093/icb/icx104) [DOI] [PubMed] [Google Scholar]
- 118.Benskin CM, Rhodes G, Pickup RW, Mainwaring MC, Wilson K, Hartley IR. 2015. Life history correlates of fecal bacterial species richness in a wild population of the blue tit Cyanistes caeruleus. Ecol. Evol. 5, 821–835. ( 10.1002/ece3.1384) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.