Abstract
Background
Human respiratory syncytial virus (RSV) is the leading cause of severe acute respiratory infection in infants and young children, which is characterized by repeated infections. However, the role of amino acid substitutions in repeated infections remains unclear. Hence, this study aimed to elucidate the genetic characteristics of RSV in children with repeated infections using molecular analyses of F and G genes.
Methods
We conducted a cohort study of children younger than 5 years in the Philippines. We collected nasopharyngeal swabs from children with acute respiratory symptoms and compared F and G sequences between initial and subsequent RSV infections.
Results
We examined 1802 children from May 2014 to January 2016 and collected 3471 samples. Repeated infections were observed in 25 children, including 4 with homologous RSV-B reinfections. Viruses from the 4 pairs of homologous reinfections had amino acid substitutions in the G protein mostly at O-glycosylation sites, whereas changes in the F protein were identified at antigenic sites V (L173S) and θ (Q209K), considered essential epitopes for the prefusion conformation of the F protein.
Conclusions
Amino acid substitutions in G and F proteins of RSV-B might have led to antigenic changes, potentially contributing to homologous reinfections observed in this study.
Keywords: respiratory syncytial virus, RSV, repeated infection
The Q209K substitution in antigenic site θ and L173S substitution in antigenic site V of F protein and changes in O-linked glycosylation pattern in G protein of RSV-B may contribute to the homologous reinfections in children.
Human respiratory syncytial virus (RSV), a member of genus Orthopneumovirus of the Pneumoviridae family, is a principal etiologic agent of acute lower respiratory infections (ALRIs), including bronchiolitis and pneumonia, among infants and young children. Shi et al estimated that 33.1 million episodes of RSV-ALRI, with 3.2 million hospital admissions and 59600 in-hospital deaths, occurred in children younger than 5 years in 2015 [1]. RSV is classified into 2 subgroups, RSV-A and RSV-B, based on their antigenic and sequence differences. Both subgroups are further classified into genotypes based on the sequence variability of the mucin-like second variable region of the G gene. While RSV-A can be categorized into more than 10 genotypes [2, 3], including the ON1 genotype with a 72-nucleotide duplication in the second variable region of the G gene, RSV-B can be divided into 9 genotypes, including the BA genotype with a 60-nucleotide duplication in the second variable region of the G gene, which has more than 12 minor groups [2, 4–7]. In recent years, the commonly circulating strains worldwide are the ON1 genotype for RSV-A and the BA genotype for RSV-B [7–11].
While the majority of children are considered to have been exposed to RSV before the age of 2 years, natural infection does not induce life-long immunity, permitting repeated infections [12, 13]. RSV F and G proteins are primary targets for protective immune responses [14, 15]. Antibodies against the RSV F protein can prevent severe disease in humans as demonstrated using a monoclonal antibody, palivizumab, as a passive prophylaxis [16]. Despite a long history of efforts to develop vaccines for RSV, no licensed vaccines are currently available [17]. One of the promising vaccines is the vaccine targeting the prefusion conformation of the F protein. The prefusion F protein drives an irreversible conformational change required for membrane fusion by transitions to the postfusion conformation [18]. The neutralizing activity against RSV in human sera is primarily derived from prefusion F protein-specific antibodies. Although antigenic sites θ, III, and V are the major neutralization-sensitive epitopes on the prefusion F protein [19, 20], the effect of amino acid substitutions in antigenic sites of the prefusion conformation of the F protein on repeated infection is largely unclear [21]. Hall et al reported that repeated infections frequently occur throughout life, which was detected by challenge experiments using the same subgroup, indicating that there is only partial cross-immunity against different strains within the same subgroup [22]. A few studies on repeated infections with homologous and heterologous subgroups of RSV have detected repeated infections by measuring the serum antibody or analyzing only partial G gene sequences of viruses from repeated infections with the same genotype [23–29]. Although the F protein is the primary target of neutralizing antibodies, analysis of the F gene of RSV in repeated infections was not conducted in those studies. The study reported here aimed to elucidate the genetic characteristics of RSV detected in children with repeated infections by molecular analysis of full-length F and G genes.
MATERIALS AND METHODS
Study Population and Samples
The study was conducted in 2 municipalities of Caibiran and Kawayan in Biliran Province, Eastern Visayas Region of the Philippines, as part of a prospective cohort study on acute respiratory infections in children younger than 5 years. Although 4012 children were enrolled in this cohort study, we examined only 1802 children who remained in the cohort from May 2014 to January 2016 for the analysis. We collected nasopharyngeal swabs (NPSs) from children with acute respiratory symptoms when they visited health care facilities or when they presented with acute respiratory symptoms, within 7 days after onset, during biweekly household visit by trained study nurses. The sampling rates among children with acute respiratory symptoms in health care facilities and household were approximately 90% and 10%, respectively. After the sample collection, NPSs in viral transport medium were stored at 4°C and transported with ice packs to the Research Institute for Tropical Medicine (RITM) in Manila for further analysis. We obtained informed consent from guardians of all participants. This study was approved by the Institutional Review Board of RITM (2008-05-10, 2013-2) and the Ethics Committee of Tohoku University Graduate School of Medicine (2012-1-5, 2012-1-63).
Viral RNA Extraction and cDNA Synthesis
We extracted viral RNA using the QIAamp MinElute Virus Spin kit (Qiagen, Hilden, Germany). The viral RNA was reverse transcribed to complementary DNA (cDNA) using Moloney Murine Leukemia Virus reverse transcriptase and random primers (Invitrogen, Carlsbad, CA).
Polymerase Chain Reaction and Gene Sequencing
RSV screening was performed by real-time polymerase chain reaction (PCR), and positive samples were further classified into RSV-A or RSV-B by PCR and sequencing of the second variable region of C-terminus of the G gene, as previously described [9]. The full-length open reading frame of the G and F genes of RSV-B was amplified using previously published primers for the G and F genes [30, 31]. Moreover, PCR products were purified using illustra ExoProStar (GE Healthcare, Amersham, UK), followed by nucleotide sequencing using the BigDye Terminator version 1.1 Cycle Sequencing kit and Genetic Analyzer 3730 (Applied Biosystems, Foster City, CA). The nucleotide sequences of the F gene (LC311524–LC311573, LC377910, and LC377911) and the G gene (LC311360–LC311409) in this study were submitted to GenBank. We also included sequences from our previous studies in the Philippines in the analysis [9, 32, 33].
Phylogenetic Analysis
The phylogenic trees of the F and G genes were generated by the maximum likelihood method using MEGA software version 6.06 [34]. We evaluated the statistical support using the bootstrap method with 1000 replicates, and bootstrap values ≥70% are shown on the branches of the consensus tree.
Analysis of Deduced Amino Acid Sequence and Prediction of N- and O-Glycosylation Sites
In this study, the alignment and position numbering of amino acid sequences of the G and F proteins of RSV-B were based on the reference sequences BA/100/04 (accession number: DQ227395) and CH-18357 (accession number: JX198143), respectively. We used NetOGlyc 4.0 and NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetOGlyc/ and http://www.cbs.dtu.dk/services/NetNGlyc/) to predict potential O-linked and N-linked glycosylation sites. While the O-linked glycosylation is based on amino acids Ser and Thr, the N-linked glycosylation is based on the amino acid configuration of Asn–Xaa–Ser/Thr, where Xaa can be any amino acids, except for Pro.
Mapping of Amino Acid Substitutions in Antigenic Sites of the F and G Proteins
We identified amino acid substitutions by comparing the deduced amino acid sequences of the F and G proteins from initial and subsequent RSV repeated infections. All identified substitutions were checked for match with the reported antigenic sites of the F protein [20]. The 3-dimensional structure of the F protein (PDB ID: 4JHW) was downloaded from the Protein Data Bank (http://www.pdb.org) [35]. Furthermore, the 3-dimensional model was visualized using MacPyMOL Molecular Graphics System, Version 1.7.4.5 Schrödinger, LLC (http://www.pymol.org). The homology modeling of the 3-dimensional structure of the G protein was performed using SWISS-MODEL (http://swissmodel.expasy.org).
RESULTS
Detection and Genotyping of RSV
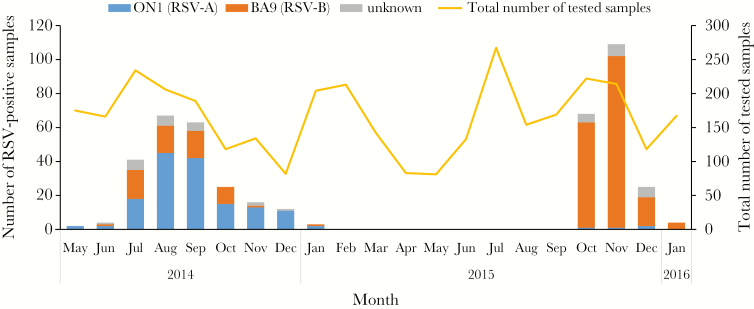
In this study, 439 (12.6%) samples tested positive for RSV among 3471 NPSs. Of these RSV-positive samples, 154 (35.1%) were RSV-A and 246 (56.0%) were RSV-B. From May 2014 to January 2016, 2 RSV epidemics were observed (Figure 1); the first epidemic occurred between May 2014 and January 2015 and the second between October 2015 and January 2016. The majority of RSV detected in the first epidemic was RSV-A (150/233; 64.4%), whereas RSV-B (184/206; 89.3%) was predominant in the second epidemic. The sequencing results of the second variable region of the G gene revealed that all RSV-A and RSV-B strains were classified as ON1 and BA9 genotypes, respectively (Figure 1).
Figure 1.
The monthly distribution of respiratory syncytial virus (RSV) in Caibiran and Kawayan in Biliran Province in the Philippines. Blue, ON1 (RSV-A) viruses; orange, BA9 (RSV-B) viruses; gray, viruses with undetermined subgroups.
Detection of Repeated Infection
During the study period, we detected repeated RSV infections in 25 children (Supplementary Table 1), including 21 children with heterologous subgroup infections by RSV-A and RSV-B and 4 children with homologous subgroup infections by RSV-B. We detected no repeated infections with RSV-A. None of the children with repeated infections had congenital abnormalities or underlying diseases, such as chronic lung, heart, blood, kidney, and brain diseases. No children died during the study period. The median interval period for repeated infections was 13 (range: 3–17) months. The shortest interval between repeated infections was 3 months and was due to heterologous subgroup infections by RSV-B and RSV-A; both episodes occurred in the same epidemic period in 2014 (child 25). None of the children with RSV-B homologous reinfections were hospitalized and none were exposed to tobacco smoking in their household.
Phylogenetic Analysis of RSV-B Including Homologous Subgroup Reinfection
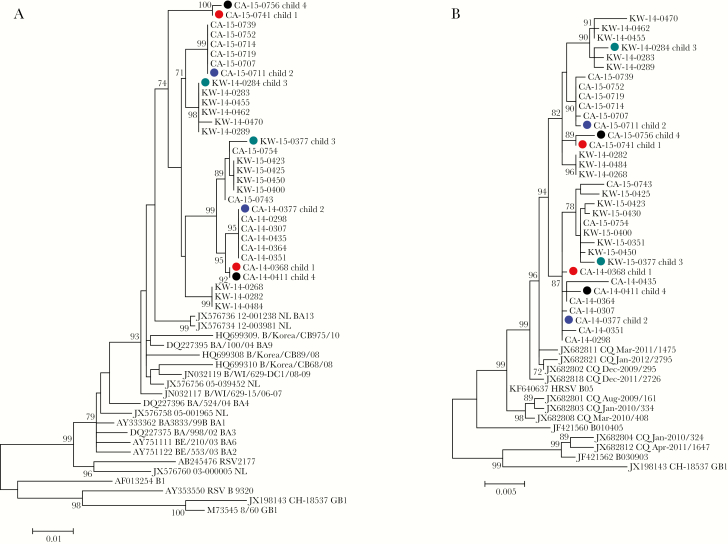
We assessed whole G and F genes sequences and constructed phylogenetic trees, including sequences of representative strains, which were detected in the same month (July and August 2014; October 2015) with repeated infections (Figure 2), to compare genetic sequences of RSV-B detected from homologous reinfections. Each pair of the viruses detected from 4 children with homologous reinfections with RSV-B in 2014 and 2015 was distributed in distinct clusters in both phylogenetic trees of the G and F genes (Figure 2).
Figure 2.
Phylogenetic trees of complete sequences of the G gene (A) and F gene (B) of respiratory syncytial virus-B (RSV-B) from children with homologous reinfections and representative strains detected in the Philippines in 2014 and 2015. RSV-B sequences from children with repeated infections are indicated by a solid circle in red (child 1), blue (child 2), green (child 3), and black (child 4).
Amino Acid Analysis of Homologous Reinfections With RSV-B
Comparison of G and F protein sequences of RSV-B between initial and subsequent homologous infections in 4 children revealed common amino acid substitutions at positions 107, 136, and 254 in the G protein and positions 173 and 209 in the F protein. Based on these amino acid substitutions, viruses were divided into 2 clusters, namely ATI and TRT (Table 1). Viruses in the ATI cluster showed a characteristic pattern of amino acid substitutions of A107, T136, and I254 in the G protein and L173 and Q209 in the F protein, whereas TRT viruses harbored T107, R136, and T254 in the G protein and S173 and K209 in the F protein. In Caibiran, 3 children (child 1, 2, and 4) developed repeated infections with RSV-B. Viruses detected from these children in 2014 were ATI viruses, whereas those identified in 2015 were TRT viruses. In contrast, initial and subsequent viruses from a child (child 3) in Kawayan revealed the opposite amino acid substitution pattern to that of viruses from 3 children in Caibiran (Table 1).
Table 1.
Amino Acid Substitutions in the G and F Proteins of Respiratory Syncytial Virus-B from Children With Homologous Reinfections
| Child ID | Sex | Age, mo | Location-Year | Amino Acid Position | |||||
|---|---|---|---|---|---|---|---|---|---|
| G Protein | F Protein | ||||||||
| Clustera | 107 | 136 | 254 | 173 | 209 | ||||
| 1 | F | 13 | CA-2014 | ATI | A | T | I | L | Q |
| 28 | CA-2015 | TRT | T | R | T | S | K | ||
| 2 | F | 12 | CA-2014 | ATI | A | T | I | L | Q |
| 27 | CA-2015 | TRT | T | R | T | S | K | ||
| 3 | F | 13 | KW-2014 | TRT | T | R | T | S | K |
| 27 | KW-2015 | ATI | A | T | I | L | Q | ||
| 4 | M | 32 | CA-2014 | ATI | A | T | I | L | Q |
| 46 | CA-2015 | TRT | T | R | T | S | K | ||
Abbreviations: CA, Caibiran; F, female; KW, Kawayan; M, male .
aATI cluster with the characteristic pattern of amino acid substitutions of A107, T136, I254 in the G protein and L173, Q209 in the F protein; TRT cluster with T107, R136, T254 in the G protein and S173, K209 in the F protein.
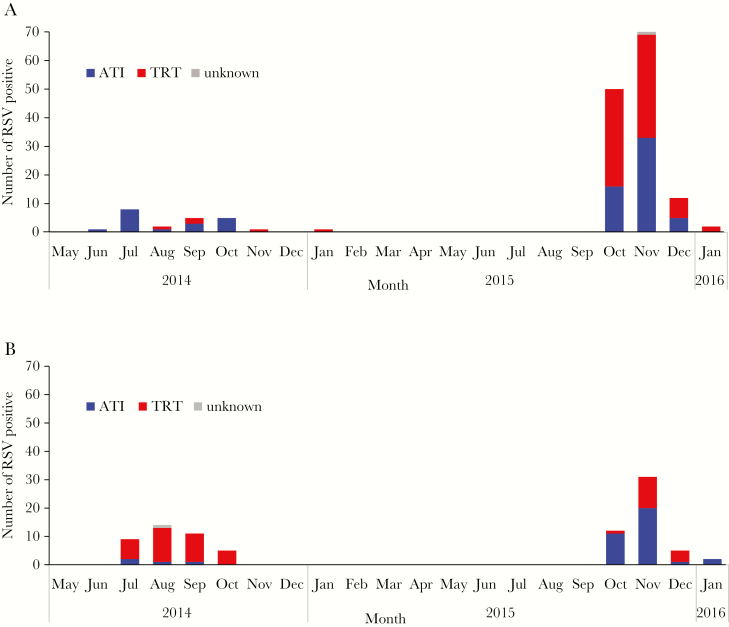
Because the characteristic combination of amino acid substitutions in both G and F proteins was consistent in each strain among samples collected in this study, the temporal distributions of ATI and TRT viruses were demonstrated based on amino acid I or T at position 254 located in the second variable region of the G gene (Figure 3). These specific combinations of amino acid substitutions reflect the dominant circulating strains in the 2 municipalities (Figure 3). In Caibiran, ATI virus (18/23; 78.3%) was the dominant circulating strain in 2014 but shifted to TRT virus (79/134; 59.0%), which was detected in a higher proportion in 2015 (Figure 3A). In Kawayan, TRT virus (34/39; 87.2%) was dominant in 2014 but changed to ATI virus (34/50, 68.0%) as the dominant circulating strain in the following year (Figure 3B).
Figure 3.
The monthly distribution of respiratory syncytial virus-B (RSV-B) with characteristic amino acid substitutions at position 254 in the G protein. Samples were collected in Caibiran (A) and Kawayan (B). Blue, ATI viruses with I254; red, TRT viruses with T254; gray, viruses with undetermined genotypes.
Temporal Analysis of Amino Acid Substitutions in RSV-B Strains in the Philippines
Supplementary Figure 1 shows the analysis of the deduced amino acid sequences of RSV-B G and F proteins from samples in this study and our previous studies in the Philippines from 2008 to 2015. The amino acid substitution patterns in the G protein revealed that TRT was the major combination pattern in the Philippines until 2013 but changed to ATI in 2014 (Supplementary Figure 1A). While amino acid substitution patterns in the F protein demonstrated that S173 and Q209 (S–Q) were the major combination until 2012, these changed to S173 and K209 (S–K) in 2013 (Supplementary Figure 1B). Subsequently, the new combination L173 and Q209 (L–Q) in the F protein was detected in ATI virus, which was dominant in Caibiran in 2014, whereas S–K was the major combination pattern in TRT virus, which was dominant in Kawayan in 2014 (Supplementary Figure 1B). As the predominant circulating strain changed in the second epidemic, L–Q became the major combination pattern in Kawayan, and a mixed combination of S–K and L–Q pattern was observed in Caibiran (Supplementary Figure 1B).
Changes in Predicted N- and O-glycosylation Sites in the Homologous Reinfection With RSV-B
Variations in predicted glycosylation sites in the G protein were compared between initial and subsequent RSV-B homologous reinfections (Table 2). Among these changes, substitutions at positions 107, 136, and 254 were identified as common sites between initial and subsequent RSV-B homologous reinfections, which might have led to either gain or loss of potential O-glycosylation sites. In this study, we observed no changes in potential N-linked and O-linked glycosylation sites of the F protein between viruses from initial and subsequent homologous reinfections.
Table 2.
Variations in Predicted O-glycosylation Sites in the G Protein of Respiratory Syncytial Virus-B (RSV-B) From Children With Repeated Infections
| Child ID | Location-Year | Amino Acid Position | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 87 | 107 | 136 | 137 | 227 | 239 | 254 | 265 | 280 | 281 | 290 | 303 | ||
| 1 | CA-2014 | – | – | O | O | – | O | – | O | O | O | O | – |
| CA-2015 | O | O | – | O | O | – | O | O | – | – | O | – | |
| 2 | CA-2014 | – | – | O | O | – | O | – | O | O | O | O | O |
| CA-2015 | – | O | – | – | O | O | O | – | O | O | – | – | |
| 3 | KW-2014 | – | O | – | O | O | O | O | O | O | O | – | – |
| KW-2015 | – | – | O | O | O | O | – | O | O | O | O | – | |
| 4 | CA-2014 | – | – | O | O | – | O | – | O | O | O | O | – |
| CA-2015 | O | O | – | O | O | – | O | O | – | – | O | – | |
Variations in the predicted gain or loss of potential O-glycosylation sites in the G protein are represented by circles and dashes, respectively. The amino acid positions that characterized RSV-B from children with repeated infections in 2014 and 2015 are (107, 136, 254) shown in bold. The numbering of amino acid positions in the G protein sequence was based on reference strain CH-18537 (JX198143).
Abbreviations: CA, Caibiran; KW, Kawayan.
Mapping of Amino Acid Substitutions in the F and G Proteins
The amino acids at positions 173 and 209 are indicated in the 3-dimensional structure of the prefusion conformation of the F protein of RSV shown in Supplementary Figure 2. The Q209K substitution is located on the surface of antigenic site θ, a metastable site specific to the prefusion state of the RSV F protein [19]. Furthermore, the L173S substitution is localized on antigenic site V, which is also identified as a prefusion conformation-specific neutralizing epitope [20]. Amino acid alignments of representative RSV F protein sequences from the subgroups A and B are presented in Supplementary Figure 3. Both S173 and K209 were consensus amino acids in RSV-A. None of the amino acid substitutions on antigenic sites I, II, III, and IV were detected in children with homologous RSV reinfections. Amino acid positions 107, 136, and 254 are indicated in the 3-dimensional structure of the G protein of RSV shown in Supplementary Figure 4. A107T and T136R are located in the mucin-like first variable region, while I254T is located in the mucin-like second variable region (Supplementary Figure 4) [36].
DISCUSSION
In this cohort study from 2014 to 2016, we observed 25 children with repeated infections with RSV, of whom 21 had heterologous subgroup infections, that is, RSV-A–RSV-B infections, and 4 had homologous subgroup infections with RSV-B. Muelenaer et al reported that antibody responses after infections with RSV-A were more cross-reactive than those after infection with RSV-B [37]. However, in this study, the initial infections in 20 children were caused by RSV-A. In addition, previous studies have demonstrated that homologous reinfections with RSV-A were more frequent than RSV-B [25–28], which could be potentially attributed to a higher degree of divergence among various genotypes of RSV-A and the induction of more complete or long-lasting subgroup-specific immune response against RSV-B than RSV-A [27, 28]. However, in this study, all homologous reinfections were due to RSV-B during the 2 consecutive epidemics, which probably occurred because a majority of viruses in the second epidemic were RSV-B with only a few RSV-A (Figure 1).
Although repeated infections with homologous subgroups of RSV have been reported [23, 25, 27–29, 38], no study has focused on the possible role of amino acid substitutions in both F and G proteins for reinfections. Each pair of viruses collected from children with repeated infections exhibited characteristic amino acid substitutions at positions 107, 136, and 254 in the G protein and 173 and 209 in the F protein. Reportedly, the F and G proteins are primary targets of human neutralizing antibody responses, and antibodies against the F protein are more vital than antibodies against the G protein as neutralizing antibodies [14, 39]. The majority of the F protein in formalin-inactivated or UV-inactivated virus, or the purified F protein is in the postfusion conformation [40]. However, most of the neutralizing antibodies elicited by natural infection predominantly target F protein in the prefusion conformation [20, 40]. Neutralizing antibodies, such as palivizumab and 101F, bind to antigenic sites II and IV, respectively, which are present both in the pre- and postfusion conformation of the F protein [41, 42]. Mutations at antigenic sites II and IV were not observed in this study (Supplementary Figure 3). Over 60% of the highly potent neutralizing antibodies from healthy adult donors target antigenic sites θ and V, which represent 2 of the 3 prefusion conformation specific sites (III, V, and θ) in the F protein [20]. In this study, the amino acid substitutions were observed at antigenic sites θ and V (Supplementary Figure 2). The site θ is reportedly an essential neutralizing epitope on the prefusion F protein [19]. The amino acid at position 209 is one of the antigenic sites recognized by the prefusion-specific neutralizing antibody, D25, which was isolated from humans [43]. Moreover, a monoclonal antibody, MEDI8897, was optimized from the antibody D25, targeting site θ, which interacts with the amino acid at position 209 in the F protein, and the Q209K substitution in the F protein was attributed to a 1.6-fold increase in the half maximal inhibitory concentration (IC50) [44]. The amino acid position 173, which is located on antigenic site VIII [45] and is currently consolidated in antigenic site V [20], is also a prefusion-specific neutralizing epitope that is recognized by the monoclonal antibody hRSV90 [45]. An S173R mutation in the F protein resulted in the loss of binding for hRSV90 antibody [45]. Overall, L173S and Q209K substitutions in the F protein might be attributed to repeated infections observed in this study. However, these studies were only performed in vitro to analyze the antibody-antigen interaction, and the biological role of these substitutions in repeated infections remains unknown. If L173S and Q209K substitutions are involved in repeated infections, these findings might have critical implications for the development of vaccines and monoclonal antibodies for prophylaxis.
Amino acid substitutions observed in this study included positions 107 and 136 in the G protein, which are located in the mucin-like first variable region, and 254, which is located in the mucin-like second variable region (Supplementary Figure 4) [36]. These 3 amino acid positions were not the same as the substitutions at positions at 223, 247, and 258, which were identified in RSV-B virus that caused homologous reinfections in a previous study by Zlateva et al [27, 46]. Furthermore, the loss or gain of glycosylation sites might potentially support the continued circulation of RSVs by escaping from host antibodies and cytotoxic T cells (CD8) [15]. In this study, changes in the O-glycosylation pattern at amino acid positions 107, 136, and 254 of the G protein of RSV-B might have altered its antigenicity and facilitated homologous reinfections.
In previous studies, BA9 had been the major genotype of RSV-B in the Philippines since 2008, which caused several epidemics in the country [9, 33]. The temporal amino acid variations in the F protein at positions 173 and 209 were determined by the sequence analysis of RSV-B detected in the Philippines from 2008 to 2015 (Supplementary Figure 1). Notably, RSV-B with the S–K amino acid combination was detected in China after 2010 [47], and the L–Q amino acid combination was detected in sequences from China and England after 2013, as deposited in GenBank. In the Philippines, RSV-B with the S–K amino acid combination in the F protein was detected in 2008 and RSV-B with the L–Q combination was detected after 2014 (Supplementary Figure 1). The amino acid substitutions at position 173 and 209 in the F protein of RSV-B encoded Ser (173S) and Lys (209K), which are consensus amino acid residues with F protein of RSV-A (Supplementary Figure 3). During the study period, 2 genetic variants (ATI and TRT) of RSV-B were cocirculating at different proportions in 2 locations, Caibiran and Kawayan in Biliran Island. In 2014, ATI virus with L–Q combination in F protein was dominant in Caibiran, but, in 2015, TRT virus with the S–K combination and ATI virus with L–Q combination were mixed. However, in Kawayan, TRT virus was dominant in the first epidemic but changed to ATI virus as the dominant circulating strain in the second epidemic. Regarding the homologous reinfection with RSV-B, the possibility of repeated infection by chance with the major epidemic strain regardless of antigenic change cannot be excluded. The waning of immunity in children may also play a role in repeated infections, but, due to the unavailability of serum samples, we could not evaluate this in the present study. Further analysis of other homologous infections for RSV would be necessary to define the role of relevant amino acid substitutions in homologous reinfections.
In conclusion, substitutions at L173S and Q209K in the F protein and changes in the O-linked glycosylation pattern in the G protein in RSV-B might contribute to the homologous reinfections observed in this study.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We would like to thank the staff of Biliran cohort project, Research Institute for Tropical Medicine-Tohoku Collaborating Research Center, and Virology Department of Tohoku University for sample collection and laboratory work.
Financial support. This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases from the Japan Agency for Medical and Research and Development (AMED) (grant number JP18fm0108013); the Science and Technology Research Partnership for Sustainable Development from AMED and Japan International Cooperation Agency (grant number JP16jm0110001); and Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (grant number JP16H02642).
Potential conflicts of interest. All authors report no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Shi T, McAllister DA, O’Brien KL, et al. . Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017; 390:946–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 1998; 79(Pt 9):2221–9. [DOI] [PubMed] [Google Scholar]
- 3. Eshaghi A, Duvvuri VR, Lai R, et al. . Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One 2012; 7:e32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trento A, Viegas M, Galiano M, et al. . Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol 2006; 80:975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dapat IC, Shobugawa Y, Sano Y, et al. . New genotypes within respiratory syncytial virus group B genotype BA in Niigata, Japan. J Clin Microbiol 2010; 48:3423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khor CS, Sam IC, Hooi PS, Chan YF. Displacement of predominant respiratory syncytial virus genotypes in Malaysia between 1989 and 2011. Infect Genet Evol 2013; 14:357–60. [DOI] [PubMed] [Google Scholar]
- 7. Bashir U, Nisar N, Mahmood N, Alam MM, Sadia H, Zaidi SS. Molecular detection and characterization of respiratory syncytial virus B genotypes circulating in Pakistani children. Infect Genet Evol 2017; 47:125–31. [DOI] [PubMed] [Google Scholar]
- 8. Hu P, Zheng T, Chen J, et al. . Alternate circulation and genetic variation of human respiratory syncytial virus genotypes in Chengdu, West China, 2009–2014. J Med Virol 2017; 89:32–40. [DOI] [PubMed] [Google Scholar]
- 9. Malasao R, Okamoto M, Chaimongkol N, et al. . Molecular characterization of human respiratory syncytial virus in the Philippines, 2012–2013. PLoS One 2015; 10:e0142192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moreira FB, Rosario CS, Santos JS, et al. . Molecular characterization and clinical epidemiology of human respiratory syncytial virus (HRSV) A and B in hospitalized children, Southern Brazil. J Med Virol 2017; 89:1489–93. [DOI] [PubMed] [Google Scholar]
- 11. Otieno JR, Kamau EM, Agoti CN, et al. . Spread and evolution of respiratory syncytial virus A genotype ON1, Coastal Kenya, 2010–2015. Emerg Infect Dis 2017; 23:264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henderson FW, Collier AM, Clyde WA Jr, Denny FW. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N Engl J Med 1979; 300:530–4. [DOI] [PubMed] [Google Scholar]
- 13. Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 1986; 140:543–6. [DOI] [PubMed] [Google Scholar]
- 14. Olmsted RA, Elango N, Prince GA, et al. . Expression of the F glycoprotein of respiratory syncytial virus by a recombinant vaccinia virus: comparison of the individual contributions of the F and G glycoproteins to host immunity. Proc Natl Acad Sci U S A 1986; 83:7462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collins P, Karron R. Respiratory syncytial virus and metapneumovirus. In: Knipe DM, Howley PM, eds. Fields virology. 6th ed Philadelphia: Lippincott Williams & Wilkins, 2013:1086–123. [Google Scholar]
- 16. The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 1998; 102:531–7. [PubMed] [Google Scholar]
- 17. Higgins D, Trujillo C, Keech C. Advances in RSV vaccine research and development—a global agenda. Vaccine 2016; 34:2870–5. [DOI] [PubMed] [Google Scholar]
- 18. Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 2012; 4:613–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ngwuta JO, Chen M, Modjarrad K, et al. . Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 2015; 7:309ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilman MS, Castellanos CA, Chen M, et al. . Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol 2016; 1:pii:eaaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kimura H, Nagasawa K, Kimura R, et al. . Molecular evolution of the fusion protein (F) gene in human respiratory syncytial virus subgroup B. Infect Genet Evol 2017; 52:1–9. [DOI] [PubMed] [Google Scholar]
- 22. Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis 1991; 163:693–8. [DOI] [PubMed] [Google Scholar]
- 23. Mufson MA, Belshe RB, Orvell C, Norrby E. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J Clin Microbiol 1987; 25:1535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sullender WM, Mufson MA, Prince GA, Anderson LJ, Wertz GW. Antigenic and genetic diversity among the attachment proteins of group A respiratory syncytial viruses that have caused repeat infections in children. J Infect Dis 1998; 178:925–32. [DOI] [PubMed] [Google Scholar]
- 25. Parveen S, Broor S, Kapoor SK, Fowler K, Sullender WM. Genetic diversity among respiratory syncytial viruses that have caused repeated infections in children from rural India. J Med Virol 2006; 78:659–65. [DOI] [PubMed] [Google Scholar]
- 26. Scott PD, Ochola R, Sande C, et al. . Comparison of strain-specific antibody responses during primary and secondary infections with respiratory syncytial virus. J Med Virol 2007; 79:1943–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zlateva KT, Vijgen L, Dekeersmaeker N, Naranjo C, Van Ranst M. Subgroup prevalence and genotype circulation patterns of human respiratory syncytial virus in Belgium during ten successive epidemic seasons. J Clin Microbiol 2007; 45:3022–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamaguchi M, Sano Y, Dapat IC, et al. . High frequency of repeated infections due to emerging genotypes of human respiratory syncytial viruses among children during eight successive epidemic seasons in Japan. J Clin Microbiol 2011; 49:1034–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kutsaya A, Teros-Jaakkola T, Kakkola L, et al. . Prospective clinical and serological follow-up in early childhood reveals a high rate of subclinical RSV infection and a relatively high reinfection rate within the first 3 years of life. Epidemiol Infect 2016; 144:1622–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Niekerk S, Venter M. Replacement of previously circulating respiratory syncytial virus subtype B strains with the BA genotype in South Africa. J Virol 2011; 85:8789–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agenbach E, Tiemessen CT, Venter M. Amino acid variation within the fusion protein of respiratory syncytial virus subtype A and B strains during annual epidemics in South Africa. Virus Genes 2005; 30:267–78. [DOI] [PubMed] [Google Scholar]
- 32. Kadji FM, Okamoto M, Furuse Y, et al. . Differences in viral load among human respiratory syncytial virus genotypes in hospitalized children with severe acute respiratory infections in the Philippines. Virol J 2016; 13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohno A, Suzuki A, Lupisan S, et al. . Genetic characterization of human respiratory syncytial virus detected in hospitalized children in the Philippines from 2008 to 2012. J Clin Virol 2013; 57:59–65. [DOI] [PubMed] [Google Scholar]
- 34. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berman HM, Westbrook J, Feng Z, et al. . The protein data bank. Nucleic Acids Res 2000; 28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi Y, Mason CS, Jones LP, Crabtree J, Jorquera PA, Tripp RA. Antibodies to the central conserved region of respiratory syncytial virus (RSV) G protein block RSV G protein CX3C-CX3CR1 binding and cross-neutralize RSV A and B strains. Viral Immunol 2012; 25:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muelenaer PM, Henderson FW, Hemming VG, et al. . Group-specific serum antibody responses in children with primary and recurrent respiratory syncytial virus infections. J Infect Dis 1991; 164:15–21. [DOI] [PubMed] [Google Scholar]
- 38. Scott PD, Ochola R, Ngama M, et al. . Molecular analysis of respiratory syncytial virus reinfections in infants from coastal Kenya. J Infect Dis 2006; 193:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Connors M, Collins PL, Firestone CY, Murphy BR. Respiratory syncytial virus (RSV) F, G, M2 (22K), and N proteins each induce resistance to RSV challenge, but resistance induced by M2 and N proteins is relatively short-lived. J Virol 1991; 65:1634–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Magro M, Mas V, Chappell K, et al. . Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A 2012; 109:3089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol 1989; 63:2941–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLellan JS, Chen M, Chang JS, et al. . Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol 2010; 84:12236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McLellan JS, Chen M, Leung S, et al. . Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 2013; 340:1113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu Q, McLellan JS, Kallewaard NL, et al. . A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci Transl Med 2017; 9:pii:eaaj1928. [DOI] [PubMed] [Google Scholar]
- 45. Mousa JJ, Kose N, Matta P, Gilchuk P, Crowe JE Jr. A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol 2017; 2:16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zlateva KT, Lemey P, Moës E, Vandamme AM, Van Ranst M. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J Virol 2005; 79:9157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xia Q, Zhou L, Peng C, et al. . Detection of respiratory syncytial virus fusion protein variants between 2009 and 2012 in China. Arch Virol 2014; 159:1089–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.