Abstract
Cyclin-dependent kinases (CDKs) are the central regulators of the eukaryotic cell cycle, and are conserved across eukaryotes. Their main and well-studied function lies in the regulation and the time-keeping of cell cycle entry and progression. Additionally, more and more non canonical functions of CDKs are being uncovered. One fairly recently discovered role of CDKs is the coordination of carbon and energy metabolism with proliferation. Evidence from different model organisms is accumulating that CDKs can directly and indirectly control fluxes through metabolism, for example by phosphorylating metabolic enzymes. In this mini-review, we summarize the emerging role of CDKs in regulating carbon and energy metabolism and discuss examples in different models from yeast to cancer cells.
Keywords: cell division cycle, proliferation, cyclin-dependent kinase, metabolism, energy homeostasis, metabolic fluxes, signaling
Introduction
The eukaryotic cell division cycle is a series of tightly coordinated processes that lead to duplication of DNA and accurate distribution of the genetic material into two daughter cells. The most important regulators driving and coordinating the cell cycle are the cyclin dependent kinases (CDKs) (Morgan, 2007, 2008). CDKs are well conserved between eukaryotes. Though the number of different isoforms varies between species, their structure and function is very similar (Malumbres, 2014): The cell cycle kinases themselves are usually present in constant concentrations, while their different regulatory subunits, the cyclins, are expressed only in specific phases of the cell cycle and thereby drive the cell cycle clock forward.
To drive cell cycle progression, CDKs phosphorylate many other cell cycle executers and regulators such as the cell cycle inhibitors Whi5 (de Bruin et al., 2004) and pRb (Lees et al., 1991), the origin recognition complex (Dahmann et al., 1995), or the anaphase-promoting complex (Rudner and Murray, 2000). However, when researchers started systematic and unbiased CDK targets screens (Ubersax et al., 2003; Chi et al., 2008; Dephoure et al., 2008; Holt et al., 2009; Errico et al., 2010), more and more targets not directly associated with the cell cycle were revealed. Many of these targets are involved in processes generally associated with cellular “house-keeping” such as translation, trafficking, and metabolism. The targets in metabolism include metabolic enzymes, metabolic regulators and molecules involved in organismal energy homeostasis (Figure 1, Table 1, Supplementary Table 1).
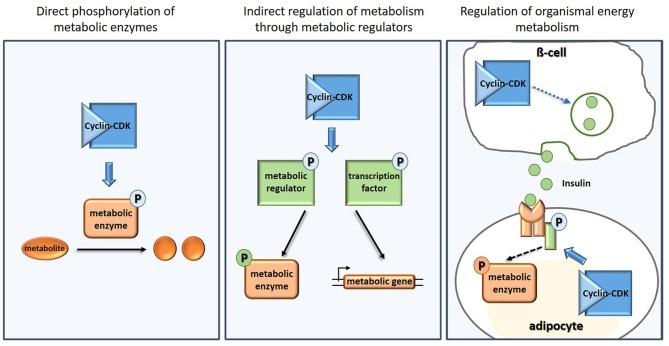
Figure 1.
Examples of how CDKs regulate carbon and energy metabolism directly (Left), indirectly (Middle), and on organismal level (Right).
Table 1.
Examples of cell cycle CDKs regulating carbon and energy metabolism.
| Publication | Organism | Cyclin-CDK | Metabolic Target | Cell cycle dependent? | Metabolic Effect |
|---|---|---|---|---|---|
| DIRECT PHOSPHORYLATION OF METABOLIC ENZYMES | |||||
| Wang et al., 2014 | Homo sapiens/MCF-10A cells | cyclin B1-CDK1 | CI subunits of the respiratory chain | Yes | Increase of mitochondrial respiration |
| Ewald et al., 2016 | Saccharomyces cerevisiae | CDK1 | Nth1 | Yes | Liquidation of trehalose/increase of internal glucose supply |
| Zhao et al., 2016 | Saccharomyces cerevisiae | CDK1 | Nth1, Gph1 | Yes | Liquidation of storage carbohydrates/increase of internal glucose supply |
| Harashima et al., 2016 | Arabidopsis thaliana | CDKA;1 | mMDH1, ALDH7B4, pfkB-like kinase, IMD1 | Unclear | Not determined |
| Wang et al., 2017 | Homo sapiens/T-ALL cell lines | cyclin D3-CDK6 | PFK1, PKM2 | Unclear | Reprogramming metabolism toward PPP and serine pathway/detox ROS |
| INDIRECT REGULATION OF METABOLISM | |||||
| Wang et al., 2006 | Mus musculus | cyclin D-CDK | NRF1 | Unclear | Inhibition of mitobiogenesis |
| Icreverzi et al., 2012 | Drosophila melanogaster | cyclin D-CDK4 | NRF-1 targets | Unclear | Regulation of mitobiogenesis |
| Harbauer et al., 2014 | Saccharomyces cerevisiae | Clb3- CDK1 | Tom6 | Yes | Increase in mitochondrial respiration |
| Lopez-Mejia et al., 2017 | Mus musculus/MEFs Mus musculus/C57BL6 | CDK4 | AMPKα2 | Unclear | Upregulation of glycolysis, inhibition of fatty acid oxidation |
| REGULATION OF ORGANISMAL ENERGY METABOLISM | |||||
| Annicotte et al., 2009 | Mus musculus /Min6 cells | CDK4 (-pRb-E2F) | Kir6.2 | Unclear | Insulin secretion |
| Lee et al., 2014 | Homo sapiens/U-2OS cells Mus musculus /C57BL6 | cyclin D1-CDK4 | GCN5 | No | Suppression of hepatic gluconeogenesis |
| Lagarrigue et al., 2016 | Mus musculus/ C57BL6 | cyclin D3-CDK4 | IRS2 | Unclear | Maintenance of insulin signaling in adipocytes |
| Kim et al., 2017 | Mus musculus | CDK2 | FOXO1 | Unclear | Regulation of β-cell mass and β-cell function |
The identification of metabolic targets of CDK highlights the tight links between metabolism, growth and the cell cycle, which are being increasingly appreciated in basic and clinical research (Vander Heiden et al., 2009; Galluzzi et al., 2013; Salazar-Roa and Malumbres, 2017; Ewald, 2018). The cell cycle is a series of macromolecular events, each having specific demands for anabolic precursors and energy. In this mini-review, we summarize the role of CDKs in meeting these metabolic demands by regulating carbon and energy metabolism in different organisms (Table 1).
CDKs directly regulating metabolic enzymes
As cells progress through the cell cycle, CDKs phosphorylate downstream regulators and proteins directly involved in executing DNA synthesis and cell division (Morgan, 2008). Since the discovery of CDKs, their target list is continually growing. The first mid-size screen searching for CDK substrates in the yeast S. cerevisiae was presented by Ubersax et al. (2003). This work defined a list of 181 proteins that were phosphorylated by CDK in extracts; many of these targets were not previously associated with the cell cycle, including several enzymes in carbohydrate and lipid metabolism. Two enzymes from lipid metabolism found in this list were later verified as CDK targets and were shown to be regulated in their activity by CDK (Santos-Rosa et al., 2005; Kurat et al., 2009).
The first large-scale untargeted phosphoproteomics experiments about a decade ago investigated the cell cycle and massively expanded the list of likely CDK targets. These studies revealed that neither the change of phosphorylation during the cell cycle, nor the list of direct CDK targets was limited to proteins generally considered to participate in the core cell cycle (Chi et al., 2008; Dephoure et al., 2008; Holt et al., 2009; Carpy et al., 2014). In fact, the list of CDK targets from Holt et al. (2009) was later re-analyzed with respect to metabolic targets (Zhao et al., 2016): Over a third of the 309 CDK targets fall into the broad category “metabolism”; at least a dozen of these are enzymes catalyzing reactions in central carbon, energy, or lipid metabolism.
Based on this initial evidence, two labs recently independently showed that yeast carbohydrate metabolism is regulated by CDK1 (the only cell cycle CDK in yeast) in a cell cycle dependent manner (Ewald et al., 2016; Zhao et al., 2016; Figure 1, Table 1). The enzymes Nth1 and Gph1 are activated by CDK to liquidate the carbohydrate storage molecules trehalose and glycogen, thereby generating glucose. This regulation is especially important in nutrient-limited environments, when cells are faced with sudden nutrient depletion (Ewald et al., 2016) or are approaching stationary phase (Zhao et al., 2016). Thus, CDK directly controls the increase of glycolytic flux at the G1/S transition to ensure sufficient carbon and energy supply during the yeast cell cycle.
A recent study in human cells also shows how CDK can control glycolytic flux (Wang et al., 2017), albeit in a different context. Wang et al. showed an interaction of CDK6-Cyclin D3 (G1/Interphase CDK in mammals) with nine out of eleven glycolytic enzymes in human cancer cells (Wang et al., 2017). The authors functionally characterized two of these enzymes, PFKP and PKM2. These enzymes catalyze the reaction of glucose-6-phosphate to fructose-bis-phosphate and the reaction from phospho-enol-pyruvate to pyruvate, respectively. PFKP and PKM are not only rate-controlling enzymes of glycolysis, but are also well known in the context of cancer (Al Hasawi et al., 2014; Lunt et al., 2015; Webb et al., 2016; Hsu and Hung, 2018). Both phosphorylations described in this study inhibit catalytic activity of the respective enzymes, presumably to re-direct flux from glycolysis into the pentose-phosphate-pathway and serine pathways to promote anabolism and antioxidant metabolism (Wang et al., 2017). Preventing flux into the pentose-phosphate-pathway in these cells led to depletion of antioxidants and a reactive oxygen induced apoptosis.
It remains to be shown whether this regulation is also physiologically relevant in healthy proliferating cells, and whether the CDK6 dependent catalytic activity of PFKP and PKM oscillates with the cell cycle. This is especially interesting since both the fructose-bis-phosphate and the pyruvate node are known to be regulated by multiple mechanisms, including other cell cycle regulators. For example, the ubiquitin ligases APC and SCF, both important cell cycle regulators, control the concentration of PFKFB3, which in turn produces fructose-2,6-bisphosphate, an activator of PFKP (Almeida et al., 2010; Tudzarova et al., 2011).
While the regulation of glycolysis and pentose-phosphate fluxes seems to be especially important for passing the restriction point and progressing through S-phase, mitochondrial metabolism takes center stage during mitosis (Bao et al., 2013; Harbauer et al., 2014). Consistent with the idea that mitochondrial ATP production is important for mitosis, CDK regulates mitochondrial metabolism both indirectly (see below) and directly in late cell cycle stages. Wang et al. showed in human cell lines that CDK1-Cyclin B1, which is important for entering and executing mitosis, partially localizes to the mitochondrial matrix (Wang et al., 2014). There, it phosphorylates several proteins of complex I of the respiratory chain. This leads to an activation of respiration and increased mitochondrial ATP production, which seems to be required for timely completing mitosis (Wang et al., 2014).
The three abovementioned mechanisms—regulation of glycolytic flux by the APC, diversion of intermediates into the pentose-phosphate-pathway through CDK regulation, and activation of mitochondrial respiration by CDK—may lead to cycling of carbon metabolism during the cell cycle to balance anabolic and energy fluxes: glycolytic flux is increasing throughout G1, nucleotide precursors for RNA and DNA metabolism are provided during late G1 and S phase, and mitochondrial ATP production is enhanced during mitosis. However, other studies have reported maximal respiration during late G1 phase (Schieke et al., 2008; Mitra et al., 2009), indicating there may be cell type or tissue specific regulatory mechanisms.
In plants, CDK also seems to target central carbon metabolism. A study in Arabidopsis thaliana recently identified several enzymes of glycolysis and mitochondrial metabolism as direct CDK-A (the CDK that drives G1/S/G2 in plants) substrates (Harashima et al., 2016). The function of these sites and their role in the cell cycle has yet to be determined, but this finding suggests that the direct regulation of central carbon metabolism by CDKs is conserved from yeast to mammals to plants.
CDKs targeting metabolic regulators
In addition to the direct phosphorylation of metabolic enzymes, CDK also targets metabolic pathways indirectly (Figure 1), by phosphorylating regulators of central carbon metabolism, regulators of mitochondria, and transporters.
Both human cells and yeast cells have been observed to increase respiration and mitochondrial ATP production during mitosis (Bao et al., 2013; Harbauer et al., 2014; Wang et al., 2014). However, in budding yeast a direct phosphorylation of respiratory chain components has not been shown as in human cells (see above). In this organism, CDK targets mitochondrial import. Specifically, the phosphorylation of the mitochondrial outer membrane transporter Tom6 enhances its import into the mitochondria, which further supports the assembly of other transporter components (Harbauer et al., 2014). This leads to more import of membrane proteins, including the fusion protein Mgm1. This in turn, like in human cells, leads to an increase in respiration and mitochondrial ATP production during the late phases of the cell cycle (Harbauer et al., 2014).
Not only protein import, but also other aspects of mitochondrial biogenesis (Sakamaki et al., 2006; Wang et al., 2006; Baltzer et al., 2009; Icreverzi et al., 2012) and mitochondrial morphology (Taguchi et al., 2007) have been shown to be regulated by CDK1 (active in G2/M) and CDK4 (active in G1) in flies and mammals. For example, the nuclear respiration factor (NRF1/2), responsible for the transcription of many nuclear-encoded mitochondrial genes, is regulated by CDK4-Cyclin D in both flies (Icreverzi et al., 2012) and mice (Wang et al., 2006). However, the effect of this regulation and the impact on mitochondrial metabolism remains unclear and may be different in each organism. The many additional layers of cross-talk between mitochondrial biogenesis and the cell cycle are extensively discussed in two recent reviews (Lopez-Mejia and Fajas, 2015; Horbay and Bilyy, 2016).
To globally target metabolic fluxes besides the mitochondria, cell cycle regulators might regulate metabolic master regulators such as TOR (target of rapamycin), PKA (protein-kinase A), or AMPK (adenylate-monophosphate activated kinase). Consistent with this idea, the PKA regulatory subunit Bcy1 was shown to be a likely CDK target in two screens in yeast (Ubersax et al., 2003; Holt et al., 2009). However, functional evidence that PKA or other metabolic master regulators are directly controlled by CDK in yeast is (to the best of our knowledge) still lacking. Such evidence was recently found in mouse embryonic fibroblasts: CDK4 directly phosphorylates and thereby inhibits the metabolic master regulator AMPK (specifically only the alpha2 subunit) (Lopez-Mejia et al., 2017). AMPK is the cell's energy barometer. Upon activation this kinase inhibits energy consuming processes and activates catabolism (Lin and Hardie, 2018). Lopez-Meija et al. showed that CDK inhibits AMPK to promote glycolysis and inhibit fatty acid oxidation. This seems to be yet another interesting mechanism by which rapidly proliferating cells can establish a “Warburg-metabolism” (Warburg, 1956) involving aerobic glycolysis, active anabolism and repressed catabolic fluxes (Vander Heiden et al., 2009). It will be interesting to investigate whether this specific inhibition of AMPK is also relevant in CDK4 hyper-activated cancer cells (Qie and Diehl, 2016).
In the same study, the inhibition of AMPK by CDK was further investigated in vivo at the organismal level. CDK4 knockout mice increase their oxidative metabolism and exercise capacity in an AMPK dependent manner (Lopez-Mejia et al., 2017), though it remains unclear if and how this effect is linked to growth or the cell cycle. The regulation of AMPK by CDK4 is thus one of the many mechanisms by which CDKs contribute to organismal energy homeostasis, as further detailed in the next section.
Organismal energy homeostasis—CDKs and insulin signaling
The role of CDKs—both canonical cell cycle, and non-cell cycle CDKs—in regulating organismal energy homeostasis was recognized even before their role in cellular metabolism was uncovered. CDKs act both as targets and as modulators of insulin and growth factor signaling (Figure 1), in proliferating and also in non-proliferating cells. This rather complex topic deserves its own review (see for example Lopez-Mejia et al., 2018); we here thus only provide a brief overview of some examples and emerging patterns.
The G1 kinase CDK4 plays a dual role in insulin signaling, functioning both in insulin-generating cells and in target cells. In mice, CDK4 was observed to regulate insulin secretion in insulin-producing pancreatic β-cells through the CDK4-pRb-E2F1 pathway (Annicotte et al., 2009). In response to high glucose, CDK4 kinase activity is increased through the insulin pathway resulting in pRb phosphorylation. Phosphorylated pRb releases the E2F1-DP complex which, in turn, activates the transcription of the Kir6.2 gene. Kir6.2 codes for a key component of the KATP channel involved in insulin secretion. CDK4, pRb and E2F1 expression was observed in nearly all insulin-producing β-cells, even though they were predominantly not proliferating (Annicotte et al., 2009). However, it remains unclear whether this pathway functions fully independently of the cell cycle in regulating insulin secretion.
Besides its role in insulin-generating β-cells, CDK4 was also described to act in insulin target cells such as hepatocytes and adipocytes (Lee et al., 2014; Lagarrigue et al., 2016). In these cells CDK is both a target and modulator of insulin signaling. For example, insulin activates hepatic cyclin D1-CDK4 which, in turn, phosphorylates the acetyltransferase GCN5. GCN5 subsequently acetylates PGC-1α thereby inhibiting the expression of gluconeogenic genes. This pathway involving cyclin D1-CDK4 is responsible for controlling glucose metabolism by suppressing hepatic glucose production in mice. Despite the activation of cyclin D1-CDK4 by insulin, the hepatocytes were not proliferating, indicating that the regulation of hepatic gluconeogenesis by cyclin D1-CDK4 is not cell cycle dependent (Lee et al., 2014). Other target cells of insulin, in which CDK4 was shown to play an important role, are adipocytes. In this cell type, insulin activates the cyclin D3-CDK4 complex which subsequently phosphorylates IRS2. This generates a positive feedback loop promoting insulin signaling (Lagarrigue et al., 2016). In the same study, Lagarrigue et al. showed that CDK4-deficient mice display decreased lipogenesis and increased lipolysis in white adipose tissue, indicating that CDK4 is essential for promoting anabolic metabolism in adipocytes (Lagarrigue et al., 2016).
Another cell cycle kinase shown to play an important role in insulin-generating pancreatic β-cells is CDK2 (the CDK responsible for S-phase entry and progression in mammals). In mice, pancreas-specific loss of CDK2 is associated with impaired glucose tolerance and defects in glucose-stimulated insulin secretion (Kim et al., 2017). Moreover, CDK2-deficient mice exhibit a reduced β-cell proliferation when faced with overnutrition or advancing age, indicating that CDK2 both regulates β-cell mass and β-cell function (Kim et al., 2017).
In addition to the cell cycle kinases CDK2 and CDK4, also non-cell cycle CDKs have been described to regulate organismal energy homeostasis in both insulin secreting cells and target cells. For example, CDK5 controls insulin secretion in pancreatic β-cells by phosphorylating and decreasing the activity of the L-type voltage-dependent Ca2+ channel (L-VDCC). Knockdown of CDK5 resulted in an enhanced insulin secretion under high glucose conditions suggesting that CDK5 exhibits a negative effect on insulin secretion in β-cells (Wei et al., 2005). In adipocytes, CDK5 regulates glucose uptake by the glucose transporter GLUT4 by several mechanisms (Okada et al., 2008; Lalioti et al., 2009). Silencing of CDK5 decreased glucose uptake by adipocytes indicating that CDK5 positively influences glucose uptake in this cell type (Lalioti et al., 2009). Another non-cell cycle CDK shown to be involved in insulin signaling of adipocytes is CDK8. In the absence of insulin, CDK8-CycC inhibits lipogenesis by phosphorylating SREBP-1c, promoting its degradation. This pathway is conserved from flies to mammals and seems to function completely independently of cell proliferation (Zhao et al., 2012).
Summary and outlook
The tight coordination between metabolism, growth and proliferation is becoming increasingly clear not only in cancer cells, but across different proliferating cell types in eukaryotic organisms. CDKs play an important role in coordinating metabolism with the cell cycle and ensuring that the carbon and energy demands of proliferating cells are met. Additionally, the G1 CDKs and non-cell cycle CDKs are important for the control of metabolism and energy homeostasis on organismal level, even in non-proliferating cells.
While it is clear that CDKs are involved in the regulation of carbon and energy metabolism across different cell types and species, data is not sufficiently dense yet to judge whether there are general patterns of metabolic regulation that are conserved. Likewise, it is still very much unclear which metabolic fluxes are actually needed to drive each cell cycle phase, and in which phase energy consumption is the highest. This may also be species and tissue dependent, i.e., different cell types may have different strategies to supply the appropriate amount of energy and anabolic precursors in a timely manner.
To address these challenging questions and to solve the roles of different CDKs in regulating metabolism in health and disease in vivo, carefully designed studies linking biochemical analysis to cellular and whole animal studies are needed. For obvious practical reasons, many CDK targets screens and investigations have been completed in vitro in nutrient environments that do not resemble the in vivo situation very well. However, as shown for carbohydrate storage in yeast, the coordination of carbon and energy metabolism with proliferation can be most important when resources are scarce or subject to strong fluctuations (Ewald et al., 2016; Zhao et al., 2016). Nutrient limitation is a constant threat to unicellular organisms (Smets et al., 2010), but can also occur in poorly vascularized solid tumors (Vander Heiden and DeBerardinis, 2017), or in tissues with strongly fluctuating energy requirement such as muscles.
In summary, among the many roles of the cyclin-dependent kinases, regulating carbon and energy metabolism is becoming more and more prominent. Further uncovering this interesting role of CDKs is not only essential for basic cell cycle biology, but may open new avenues for treatment of cancers with overactive CDKs.
Author contributions
JE designed the topic of the review. MS and JE wrote the manuscript. Both authors read and approved the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer DS and MS and handling Editor declared their shared affiliation.
Acknowledgments
We thank the members of the Ewald Lab for their support and proof-reading of the manuscript.
Footnotes
Funding. JE gratefully acknowledges funding from the Deutsche Forschungsgemeinschaft (Project 391105827), and the Daimler und Benz Foundation (Project 32-02/16). We further acknowledge Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of the University of Tübingen for the support of publication costs.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2018.00093/full#supplementary-material
References
- Al Hasawi N., Alkandari M. F., Luqmani Y. A. (2014). Phosphofructokinase: a mediator of glycolytic flux in cancer progression. Crit. Rev. Oncol. Hematol. 92, 312–321. 10.1016/j.critrevonc.2014.05.007 [DOI] [PubMed] [Google Scholar]
- Almeida A., Bolanos J. P., Moncada S. (2010). E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 107, 738–741. 10.1073/pnas.0913668107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annicotte J. S., Blanchet E., Chavey C., Iankova I., Costes S., Assou S., et al. (2009). The CDK4-pRB-E2F1 pathway controls insulin secretion. Nat. Cell Biol. 11, 1017–1023. 10.1038/ncb1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltzer C., Tiefenbock S. K., Marti M., Frei C. (2009). Nutrition controls mitochondrial biogenesis in the drosophila adipose tissue through delg and cyclin D/Cdk4. PLoS ONE 4:e6935. 10.1371/journal.pone.0006935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Mukai K., Hishiki T., Kubo A., Ohmura M., Sugiura Y., et al. (2013). Energy management by enhanced glycolysis in G1-phase in human colon cancer cells in vitro and in vivo. Mol. Cancer Res. 11, 973–985. 10.1158/1541-7786.MCR-12-0669-T [DOI] [PubMed] [Google Scholar]
- Carpy A., Krug K., Graf S., Koch A., Popic S., Hauf S., et al. (2014). Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast). Mol. Cell. Proteomics 13, 1925–1936. 10.1074/mcp.M113.035824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Welcker M., Hizli A. A., Posakony J. J., Aebersold R., Clurman B. E. (2008). Identification of CDK2 substrates in human cell lysates. Genome Biol. 9:R149. 10.1186/gb-2008-9-10-r149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C., Diffley J. F., Nasmyth K. A. (1995). S-Phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 5, 1257–1269. 10.1016/S0960-9822(95)00252-1 [DOI] [PubMed] [Google Scholar]
- de Bruin R. A., McDonald W. H., Kalashnikova T. I., Yates J., Wittenberg C. (2004). Cln3 activates G1-specific transcription via phosphorylation of the SBF transcription bound repressor Whi5. Cell 117, 887–898. 10.1016/j.cell.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villen J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., et al. (2008). A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767. 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A., Deshmukh K., Tanaka Y., Pozniakovsky A., Hunt T. (2010). Identification of substrates for cyclin dependent kinases. Adv. Enzyme Regul. 50, 375–399. 10.1016/j.advenzreg.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Ewald J. C. (2018). How yeast coordinates metabolism, growth and division. Curr. Opin. Microbiol. 45, 1–7. 10.1016/j.mib.2017.12.012 [DOI] [PubMed] [Google Scholar]
- Ewald J. C., Kuehne A., Zamboni N., Skotheim J. M. (2016). The yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression. Mol. Cell 62, 532–545. 10.1016/j.molcel.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Kepp O., Vander Heiden M. G., Kroemer G. (2013). Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 12, 829–846. 10.1038/nrd4145 [DOI] [PubMed] [Google Scholar]
- Harashima H., Dissmeyer N., Hammann P., Nomura Y., Kramer K., Nakagami H., et al. (2016). Modulation of plant growth in vivo and identification of kinase substrates using an analog-sensitive variant of CYCLIN-DEPENDENT KINASE A;1. BMC Plant Biol. 16:209. 10.1186/s12870-016-0900-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer A. B., Opalinska M., Gerbeth C., Herman J. S., Rao S., Schonfisch B., et al. (2014). Mitochondria. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science 346, 1109–1113. 10.1126/science.1261253 [DOI] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., Morgan D. O. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686. 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbay R., Bilyy R. (2016). Mitochondrial dynamics during cell cycling. Apoptosis 21, 1327–1335. 10.1007/s10495-016-1295-5 [DOI] [PubMed] [Google Scholar]
- Hsu M. C., Hung W. C. (2018). Pyruvate kinase M2 fuels multiple aspects of cancer cells: from cellular metabolism, transcriptional regulation to extracellular signaling. Mol. Cancer 17:35. 10.1186/s12943-018-0791-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icreverzi A., de la Cruz A. F., Van Voorhies W. A., Edgar B. A. (2012). Drosophila cyclin D/Cdk4 regulates mitochondrial biogenesis and aging and sensitizes animals to hypoxic stress. Cell Cycle 11, 554–568. 10.4161/cc.11.3.19062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Lee J. H., Merrins M. J., Gavrilova O., Bisteau X., Kaldis P., et al. (2017). Loss of cyclin-dependent Kinase 2 in the pancreas links primary beta-cell dysfunction to progressive depletion of beta-cell mass and diabetes. J. Biol. Chem. 292, 3841–3853. 10.1074/jbc.M116.754077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., et al. (2009). Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell 33, 53–63. 10.1016/j.molcel.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Lagarrigue S., Lopez-Mejia I. C., Denechaud P. D., Escote X., Castillo-Armengol J., Jimenez V., et al. (2016). CDK4 is an essential insulin effector in adipocytes. J. Clin. Invest. 126, 335–348. 10.1172/JCI81480 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lalioti V., Muruais G., Dinarina A., van Damme J., Vandekerckhove J., Sandoval I. V. (2009). The atypical kinase Cdk5 is activated by insulin, regulates the association between GLUT4 and E-Syt1, and modulates glucose transport in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 106, 4249–4253. 10.1073/pnas.0900218106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Dominy J. E., Choi Y. J., Jurczak M., Tolliday N., Camporez J. P., et al. (2014). Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 510, 547–551. 10.1038/nature13267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J. A., Buchkovich K. J., Marshak D. R., Anderson C. W., Harlow E. (1991). The retinoblastoma protein is phosphorylated on multiple sites by human Cdc2. Embo. J. 10, 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. C., Hardie D. G. (2018). AMPK: sensing glucose as well as cellular energy status. Cell Metabol. 27, 299–313. 10.1016/j.cmet.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Lopez-Mejia I. C., Castillo-Armengol J., Lagarrigue S., Fajas L. (2018). Role of cell cycle regulators in adipose tissue and whole body energy homeostasis. Cell. Mol. Life Sci. 75, 975–987. 10.1007/s00018-017-2668-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mejia I. C., Fajas L. (2015). Cell cycle regulation of mitochondrial function. Curr. Opin. Cell Biol. 33, 19–25. 10.1016/j.ceb.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Lopez-Mejia I. C., Lagarrigue S., Giralt A., Martinez-Carreres L., Zanou N., Denechaud P. D., et al. (2017). CDK4 phosphorylates AMPKalpha2 to inhibit its activity and repress fatty acid oxidation. Mol. Cell 68, 336.e6–349.e6. 10.1016/j.molcel.2017.09.034 [DOI] [PubMed] [Google Scholar]
- Lunt S. Y., Muralidhar V., Hosios A. M., Israelsen W. J., Gui D. Y., Newhouse L., et al. (2015). Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell 57, 95–107. 10.1016/j.molcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. (2014). Cyclin-dependent kinases. Genome Biol. 15:122. 10.1186/gb4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J. (2009). A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U.S.A. 106, 11960–11965. 10.1073/pnas.0904875106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. (ed.). (2007). The Cell Cycle: Principles of Control. London: New Science Press. [Google Scholar]
- Morgan D. O. (2008). SnapShot: cell-cycle regulators I. Cell 135, 764.e1– 764.e1. 10.1016/j.cell.2008.10.039 [DOI] [PubMed] [Google Scholar]
- Okada S., Yamada E., Saito T., Ohshima K., Hashimoto K., Yamada M., et al. (2008). CDK5-dependent phosphorylation of the Rho family GTPase TC10(alpha) regulates insulin-stimulated GLUT4 translocation. J. Biol. Chem. 283, 35455–35463. 10.1074/jbc.M806531200 [DOI] [PubMed] [Google Scholar]
- Qie S., Diehl J. A. (2016). Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. 94, 1313–1326. 10.1007/s00109-016-1475-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A. D., Murray A. W. (2000). Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 149, 1377–1390. 10.1083/jcb.149.7.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki T., Casimiro M. C., Ju X., Quong A. A., Katiyar S., Liu M., et al. (2006). Cyclin D1 determines mitochondrial function in vivo. Mol. Cell Biol. 26, 5449–5469. 10.1128/MCB.02074-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Roa M., Malumbres M. (2017). Fueling the cell division cycle. Trends Cell Biol. 27, 69–81. 10.1016/j.tcb.2016.08.009 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005). The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941. 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke S. M., McCoy J. P., Jr., Finkel T. (2008). Coordination of mitochondrial bioenergetics with G1 phase cell cycle progression. Cell Cycle 7, 1782–1787. 10.4161/cc.7.12.6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets B., Ghillebert R., De Snijder P., Binda M., Swinnen E., De Virgilio C., et al. (2010). Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56, 1–32. 10.1007/s00294-009-0287-1 [DOI] [PubMed] [Google Scholar]
- Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. (2007). Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529. 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- Tudzarova S., Colombo S. L., Stoeber K., Carcamo S., Williams G. H., Moncada S. (2011). Two ubiquitin ligases, APC/C-Cdh1 and SKP1-CUL1-F (SCF)-beta-TrCP, sequentially regulate glycolysis during the cell cycle. Proc. Natl. Acad. Sci. U.S.A. 108, 5278–5283. 10.1073/pnas.1102247108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., et al. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859–864. 10.1038/nature02062 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009). Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G., DeBerardinis R. J. (2017). Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669. 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li Z., Lu Y., Du R., Katiyar S., Yang J., et al. (2006). Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 103, 11567–11572. 10.1073/pnas.0603363103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Nicolay B. N., Chick J. M., Gao X., Geng Y., Ren H., et al. (2017). The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546, 426–430. 10.1038/nature22797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Fan M., Candas D., Zhang T. Q., Qin L., Eldridge A., et al. (2014). Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev. Cell 29, 217–232. 10.1016/j.devcel.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On respiratory impairment in cancer cells. Science 124, 269–270. [PubMed] [Google Scholar]
- Webb B., Forouhar F., Szu F. E., Seetharaman J., Tong L., Barber D. (2016). Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Biophys. J. 110, 206a−207a. 10.1016/j.bpj.2015.11.1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F. Y., Nagashima K., Ohshima T., Saheki Y., Lu Y. F., Matsushita M., et al. (2005). Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat. Med. 11, 1104–1108. 10.1038/nm1299 [DOI] [PubMed] [Google Scholar]
- Zhao G., Chen Y., Carey L., Futcher B. (2016). Cyclin-dependent kinase co-ordinates carbohydrate metabolism and cell cycle in S. cerevisiae. Mol. Cell 62, 546–557. 10.1016/j.molcel.2016.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Feng D., Wang Q., Abdulla A., Xie X. J., Zhou J., et al. (2012). Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 122, 2417–2427. 10.1172/JCI61462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.