For years, drug developers have followed the “one drug, one target” paradigm: Identify and synthesize a molecule that acts on a single protein or other biological target to treat a specific disease. This approach is costly. On average, it takes at least a decade and $2.6 billion to bring a new drug from lab bench to pharmacy shelf, according to a report from the Pharmaceutical Research & Manufacturers of America. And fewer than 12% of drugs that enter clinical trials earn U.S. Food & Drug Administration approval.

Credit: Aleutie/Shutterstock.
The biology of disease isn’t quite so simple, however, and often involves more than just a single faulty protein. As researchers have accepted this notion, they have begun to shift their drug development strategy: Some developers are now characterizing diseases more broadly by measuring gene expression patterns—how and when genetic instructions are decoded—in cells and tissues and examining changes to those patterns in diseased cells and tissues. They hope to improve drug development success rates by screening vast libraries of compounds for those that reverse expression patterns so they look like those found in healthy tissue.
Recognizing that such gene expression data is available in public databases, a growing group of computational drug discovery start-ups and academic research laboratories has begun to sift through them for compounds that might have efficacy against diseases far different from their original targets—an approach that skips having to build compounds from the ground up. Even large pharmaceutical companies have begun embracing this type of “big data” analytics. They are hoping that this approach might uncover surprising new uses for old drugs—and simultaneously slash the cost of drug development by using computational methods to see what is hiding in plain sight.
Giving old drugs new life
Over the past decade or so, “people were starting to realize all these data were available, and computers were getting more powerful to do this kind of work,” says Atul Butte, director of the Bakar Computational Health Sciences Institute at the University of California, San Francisco. One demonstration emerged in 2013, while Butte was at the Stanford University School of Medicine. He and his colleagues used a data mining technology they developed to search hundreds of thousands of gene expression profiles from public data, such as the National Center for Biotechnology Information’s Gene Expression Omnibus. These profiles spanned a broad range of cells and tissues: healthy and diseased, as well as treated and untreated with drugs.
The researchers used their method to examine data from small-cell lung cancer (SCLC) cells and tissue models that had been treated with various drug compounds and looked for samples with gene expression patterns that more closely matched the patterns of healthy lung cells—a reversal from the diseased cells. They then compiled a list of the compounds that led to these reversals. The idea is that, for example, if certain genes consistently show high expression in SCLC cells, a drug that lowers their expression may be able to treat the condition despite being approved for another disease. At the top of the list of promising compounds were tricyclic antidepressants.
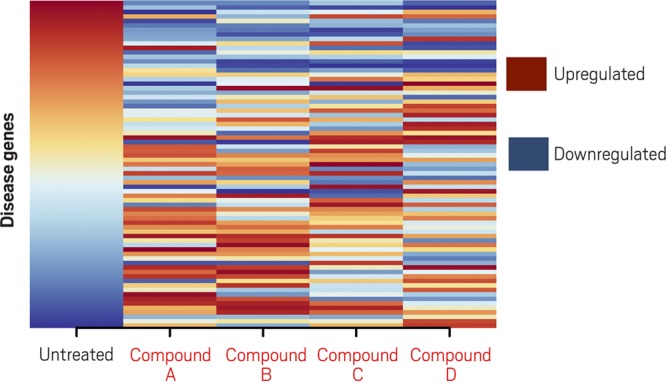
Candidate drug compounds (A, B, C, D) change the gene expression pattern in cancer cells, compared with that of untreated cells (first column). Those that reverse the expression pattern relative to untreated cells, seen as an inversion of the color pattern, are candidate treatments. Credit: Adapted from Chen et al., Nat. Commun. 2017, DOI: 10.1038/ncomms16022, under Creative Commons license.
The researchers homed in on one candidate, a tricyclic antidepressant called imipramine. Butte and his team saw that imipramine spurred human SCLC cells growing in laboratory dishes and in mice to undergo programmed cell suicide, or apoptosis. The drug also slowed or prevented metastases in mice carrying SCLC tumors. It blocked the growth of other neuroendocrine tumors, too, including a pediatric tumor known as a neuroblastoma. Encouraged by the team’s results, Butte’s Stanford colleague Joel Neal led researchers in launching clinical trials of desipramine, a molecule similar to imipramine, in patients with SCLC and other highly aggressive neuroendocrine tumors. But after Phase II trials revealed unacceptable side effects, including dizziness, drowsiness, and fatigue, the team had to stop developing the drug.
Still, the approach saves researchers the time and effort of developing a compound from scratch, says Gini Deshpande, founder and CEO of NuMedii, which licensed Butte’s team’s data mining technology, developed it further, and renamed it Artificial Intelligence for Drug Discovery (AIDD). “It enables us to find novel connections and identify novel biology that can form the basis for drug discovery,” she says. The company is continuing to look for new compounds to test.
And the desipramine trial is not necessarily a dead end, she adds: “You can imagine modifying imipramine to have fewer side effects.” Since FDA has already approved for human use many of the drugs in public databases, researchers can bring them to clinical trials much faster than they could a novel compound. The entire process of identifying a therapeutic target and candidate compounds that hit the target, plus carrying out the preclinical studies needed to ensure activity in relevant preclinical models and safe testing in humans, takes about six years on average, Deshpande says. The Stanford researchers launched their first clinical trial less than two years after the data-mining technology identified the lead compounds.
NuMedii is also looking beyond SCLC. In a collaboration with a pharmaceutical company then known as Aptalis, NuMedii filed a patent in 2014 for the use of β−blockers—frequently used to treat high blood pressure—to treat gastrointestinal disorders, including inflammatory bowel disease, ulcerative colitis, and Crohn’s disease.
NuMedii is far from alone in using data mining to give old drugs new life. Marina Sirota, who is also at the Bakar Institute and helped develop the algorithm with Butte, has continued to use the algorithm; she is looking for drug leads for liver cancer, basal cell carcinoma, and other conditions. Her team applied the method in a trial with a small number of patients to correctly predict that a class of drugs that narrows blood vessels may be able to treat an autoimmune disease called dermatomyositis.
On the other side of the country, Connecticut-based BioXcel uses similar data-mining approaches to repurpose existing drugs, a handful of which are currently in Phase I and Phase II trials. These include compounds that may be able to treat blood cancers, dementia, and bipolar disorder. In the U.K., Healx is mining big data with an eye toward rare diseases. The company recently identified an antidepressant called tianeptine as a possible treatment for CDKL5 syndrome, a genetic disorder that leads to impaired neurological development and uncontrollable seizures. Pharnext in France uses big data analytics to generate combinations of existing drugs that could treat diseases. One of its candidates is currently in Phase III clinical trials for the treatment of another rare neurological disorder, called Charcot-Marie-Tooth disease, with results expected at the end of this year.
Data mining for cell lines and more
Once researchers identify their candidate compounds, big data approaches could further streamline the drug development process by ensuring researchers test candidates on the right cell lines in preclinical studies. One of the main reasons drugs fail, says Bin Chen of Michigan State University, is that groups choose cell lines, tissue models, and animal models for their early testing that don’t accurately reflect the disease in humans. For instance, although researchers have used mice to study inflammatory diseases, a recent study found that genomic responses to inflammation in these mice don’t correlate well with genomic responses to inflammation in humans.
Chen leads a team that’s targeting metastatic cancer—cancer that has spread in the body beyond the original tumor. Although somestudies have evaluated how closely cell lines reflect what happens in the original tumor, he says, none have looked at how closely cell lines model metastatic cancer; until now, there has been a dearth of data on metastatic disease, making such studies difficult.
Using artificial intelligence to analyze public data, Chen’s team developed a statistical measure of how well the gene expression profiles of human breast cancer cell lines correlate with those of tumor samples from patients with metastatic breast cancer. In a preliminary study, they compared the profiles of 57 breast cancer cell lines with those of tumor samples from patients and found significant differences. In other words, researchers developing metastatic breast cancer drugs need to recognize the limitations of cell lines and may need to use or develop other preclinical models altogether.
NuMedii wants to streamline even earlier stages of drug development by using AIDD to identify different subtypes of a disease so that the company can tailor treatments to them from the outset. Typically, researchers identify subtypes after a clinical trial, comparing the biomarkers present in patients who responded to the drug with those in patients who did not and using those to define subtypes. “We try to flip that on its head,” Deshpande says. “What are the molecular mechanisms that are changing in different patient subsets, so you know from the get-go who you’re developing the drug for?” NuMedii has begun teasing apart the subtypes of idiopathic pulmonary fibrosis, a chronic disease that causes scarring in the lungs.
Mine or bust
Deshpande believes the pharmaceutical industry’s dismal success rates leave it little choice but to consider data mining as an alternative to the traditional one-drug, one-target approach. “You spend $1 billion to $4 billion to discover one successful drug.” When you consider the number of diseases, she says, “the math doesn’t add up.” Big data approaches could allow drug developers to weed out less-than-stellar candidates early on. “Even if we end up with some failures, getting to failure fast is imperative as an industry,” she says. “If you’re going to fail, fail quickly for way lower costs.”
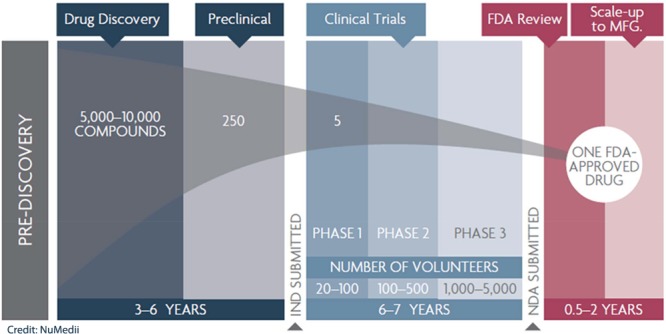
Very few drugs actually make it to clinical approval. Credit: NuMedii.
To be sure, big data approaches still face commercial hurdles, especially when it comes to repurposing drugs. To move forward in clinical trials, Chen and colleagues have spent the past year trying to license a patent-protected drug that their data mining system predicted could treat a disease other than the one for which it was originally intended. (Chen wouldn’t disclose the drug.) “It is not easy to convince a big company to license their compound to an academic lab or a start-up,” Chen says. For now, Chen’s team is trying artificial intelligence methods to work around the problem by redesigning the compound, making it different enough from the original to avoid patent infringement.
Hermann Mucke, founder and owner of H.M. Pharma Consultancy, a life sciences consulting firm in Vienna, attributes the hesitation that Chen and many of his clients face from big pharmaceutical companies partly to “corporate ego,” especially when it comes to repurposing compounds that have been shelved after failing in clinical trials. “No one wants to be reminded of this defeat,” he says. Those looking to repurpose drugs might find more luck licensing compounds from smaller start-ups, which need the revenue and have more capacity to take on another project.
Mucke also notes that it’s important not to overstate the direct cost savings of repurposing drugs. For an individual project, drug repurposing cuts out early stage development costs, “but that’s not very much in relation to the Phase II and Phase III studies.”
Researchers mining big data for drug candidates face not only commercial challenges but scientific ones too. Despite all the data on how compounds affect gene expression in cells and animal models, such data in clinical trial patients are lacking. “In a perfect world, we would have patients treated with drugs and their expression profile before and after treatment,” Aravind Subramanian of Broad Institute of MIT & Harvard says. He and his colleagues have developed the Connectivity Map (CMap), which he likens to Google for biologically active compounds. For instance, users could input a disease’s gene expression pattern, and CMap would generate a ranked list of compounds that reverse that pattern. So far, the tool has been used in about 30 studies to identify drug candidates, but the data have been based mostly on cancer cells rather than patients.
Also, although researchers are able to identify compounds that reverse gene expression patterns in disease, it’s more challenging to uncover the mechanisms. “We see that they work, but we don’t always know why,” UCSF’s Sirota says.
Drug candidates mined from big data have yet to receive FDA approval, and it remains to be seen whether such approaches will ultimately outperform the traditional approach. Still, Deshpande remains “hugely optimistic.” Since NuMedii launched in 2010, she has noticed a groundswell of other start-ups applying big data approaches to drug discovery. Even large pharmaceutical companies, like Novartis and GlaxoSmithKline, have followed suit. Once “a couple drugs get across the finish line” using big data, the pharmaceutical industry will adopt the approach even more broadly, she says. “It’s early days, but we’ve got promising hits.”
Melissa Pandika is a freelance contributor toChemical & Engineering News, the weekly newsmagazine of the American Chemical Society.
In collaboration with C&EN.


