Key Points
The prevalence of Ph-like ALL is lower in children with NCI SR ALL compared with HR ALL.
Ph-like ALL in SR patients harbors few targetable kinase fusions and has improved outcome compared with HR Ph-like ALL.
Abstract
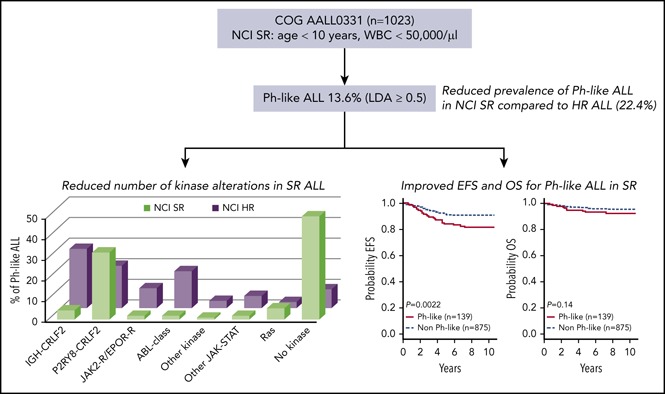
Philadelphia chromosome–like acute lymphoblastic leukemia (Ph-like ALL; BCR-ABL1–like ALL) in children with National Cancer Institute (NCI) intermediate- or high-risk (HR) ALL is associated with poor outcome. Ph-like ALL is characterized by genetic alterations that activate cytokine receptor and kinase signaling and may be amenable to treatment with tyrosine kinase inhibitors. The prevalence, outcome, and potential for targeted therapy of Ph-like ALL in standard-risk (SR) ALL is less clear. We retrospectively analyzed a cohort of 1023 SR childhood B-ALL consecutively enrolled in the Children's Oncology Group AALL0331 clinical trial. The Ph-like ALL gene expression profile was identified in 206 patients, and 67 patients with either BCR-ABL1 (n = 6) or ETV6-RUNX1 (n = 61) were excluded from downstream analysis, leaving 139 of 1023 (13.6%) as Ph-like. Targeted reverse transcription polymerase chain reaction assays and RNA-sequencing identified kinase-activating alterations in 38.8% of SR Ph-like cases, including CRLF2 rearrangements (29.5% of Ph-like), ABL-class fusions (1.4%), JAK2 fusions (1.4%), an NTRK3 fusion (0.7%), and other sequence mutations (IL7R, KRAS, NRAS; 5.6%). Patients with Ph-like ALL had inferior 7-year event-free survival compared with non–Ph-like ALL (82.4 ± 3.6% vs 90.7 ± 1.0%, P = .0022), with no difference in overall survival (93.2 ± 2.4% vs 95.8 ± 0.7%, P = .14). These findings illustrate the significant differences in the spectrum of kinase alterations and clinical outcome of Ph-like ALL based on presenting clinical features and establish that genomic alterations potentially targetable with approved kinase inhibitors are less frequent in SR than in HR ALL.
Visual Abstract
Introduction
Although long-term survival for childhood acute lymphoblastic leukemia (ALL) now exceeds 85%, several genetically defined subgroups continue to experience an increased risk of treatment failure and poor survival.1,2 One such high-risk (HR) subtype is Philadelphia chromosome–like (Ph-like ALL) or BCR-ABL1–like ALL. Ph-like ALL patients are defined as having a gene expression signature similar to BCR-ABL1-positive ALL, but lack the BCR-ABL1 fusion gene, and commonly harbor genetic alterations targeting B-lymphoid transcription factors, including IKZF1 (Ikaros).3,4 The prevalence of Ph-like ALL increases with age, ranging from 10% to 15% of children to 20% to 25% of young adults (21-39 years) and adults (>40 years) with ALL, and is associated with poor outcome in all ages.5-13 The genomic landscape of Ph-like ALL is defined by a diverse array of genetic alterations that dysregulate cytokine receptor and kinase signaling pathways and may be amenable to treatment with tyrosine kinase inhibitors (TKIs), analogous to the successful treatment of BCR-ABL1-positive ALL with ABL1 inhibitors.14,15 A main theme emerging from these studies is that despite the large number of individual kinase alterations identified in Ph-like ALL, the majority converge on a limited number of pathways that may be targeted effectively in vivo using chemotherapy combined with ABL-class or JAK/STAT-class TKIs.16
In published series of children and adults, ∼50% of children with Ph-like ALL harbor genomic rearrangements that result in the overexpression of CRLF2, with frequent concomitant sequence mutations in Janus kinases or other regulators of JAK-STAT signaling, particularly JAK2,17-20 that are potentially amenable to treatment with JAK inhibitors such as ruxolitinib.21,22 CRLF2 rearrangements (CRLF2-R) occur by either translocation of CRLF2 into the immunoglobulin heavy chain enhancer locus (IGH-CRLF2) or through focal deletion of a portion of the PAR1 pseudo-autosomal region of chromosome X/Y, resulting in P2RY8-CRLF2 fusion. Another major Ph-like ALL subgroup includes cases with alterations that activate JAK-STAT signaling and are candidates for JAK inhibitor therapy, including rearrangements of JAK2, EPOR, TYK2, and IL2RB, collectively present in ∼10% of Ph-like ALL in cohorts of children and adolescents with HR ALL as defined by age and white blood cell (WBC) count.9,11,23-25 One of the most therapeutically targetable subgroups of Ph-like ALL includes patients that harbor ABL-class gene fusions (ABL1, ABL2, CSF1R, LYN, PDGFRA, and PDGFRB), present in 15% to 20% of childhood Ph-like ALL patients and potentially amenable to treatment with the ABL1 inhibitor imatinib or the dual ABL1/SRC inhibitor dasatinib.11,12,26,27 The efficacy of dasatinib and ruxolitinib with combination chemotherapy is currently being evaluated in Children’s Oncology Group (COG) clinical trials AALL1131 (#NCT02883049) and AALL1521 (#NCT02723994), in St. Jude Children’s Research Hospital TXVII (#NCT03117751), and in adults with relapsed/refractory ALL at the MD Anderson Cancer Center (#NCT02420717).28
Comprehensive genomic profiling of Ph-like ALL in children enrolled in COG protocols has focused on patients with National Cancer Institute (NCI) HR ALL (age ≥10 years or WBC count ≥50 ×109/L) or those with standard-risk (SR) ALL (age 1-9.99 years and WBC count <50 ×109/L) and elevated minimal residual disease (MRD) at the end of induction.9,11 A single institution study of 344 patients from St. Jude Total Therapy XV demonstrated that conventional salvage treatment with MRD-based risk-directed therapies was able to overcome the poor prognostic influence of Ph-like ALL. Of note, the spectrum of kinase alterations in St. Jude Ph-like ALL patients is different from those reported from COG, with significantly less CRLF2-R identified (28% vs 50% for COG), likely because of differences in ethnicity.29 Other groups have characterized predominantly B-other ALL lacking common genomic rearrangements and have focused on intermediate risk ALL as defined by MRD or have not performed comprehensive unbiased sequencing to fully characterize the potential spectrum of genomic alterations.25,30-32 More recently, additional genomic studies have identified new oncogenic subtypes in ALL consisting of cases with DUX4/ERG deregulation, MEF2D and ZNF384 rearrangements, and ETV6-RUNX1-like expression.33-38 However, the prevalence, genomic landscape, and prognostic significance of Ph-like ALL in a large cohort of SR ALL patients lacking HR features is still unclear. This is important to assess, as two-thirds of patients with B-ALL have SR features and approximately equal relapse events occur in children initially diagnosed as SR and HR. To address this question, we analyzed an unselected cohort of 1023 consecutive cases of SR ALL enrolled in COG clinical trial AALL0331 using our recently published comprehensive algorithm for identifying Ph-like ALL and associated kinase alterations, including whole transcriptome sequencing for all Ph-like cases without an identified driver genomic alteration.9
Methods
Patients and samples
We retrospectively studied 1023 newly diagnosed patients with SR B-ALL consecutively enrolled in COG AALL0331 (#NCT00103285) between 2006 and 2008.39 Samples were selected based on availability of cryopreserved leukemia blasts from patients who met the eligibility criteria. Patients and/or their parent(s)/guardian(s) provided informed consent for clinical trial participation, banking, and future research. All analyses were approved by the Institutional Review Boards of Nationwide Children’s Hospital, the University of New Mexico, and St. Jude Children’s Research Hospital.
Identification of Ph-like ALL
All cases were analyzed using an 8-gene TaqMan low-density array (LDA) polymerase chain reaction (PCR) assay on a 384-well microfluidic card as previously described.9,10 An integrated score between 0 and 1 was generated from the 8-gene assay, and any sample with a predictive score of ≥0.5 was considered to be Ph-like and is referred to herein as LDApos (supplemental Table 1; available on the Blood Web site).
Identification of CRLF2 rearrangements
Details for the algorithm used to identify kinase alterations are provided in supplemental Figure 1. Any case determined to have elevated CRLF2 expression (CRLF2high; defined as ΔCt ≤6 by LDA) was assessed for P2RY8-CRLF2 by TaqMan PCR on the LDA card, with fluorescence in situ hybridization (FISH) for IGH-CRLF2 performed as described previously for CRLF2high cases without P2RY8-CRLF2.17 All CRLF2high cases were assessed for JAK1 or JAK2 mutations by Sanger sequencing as previously described.17 Identified coding variants were confirmed to be somatic by comparison with matched remission DNA.
RT-PCR for kinase fusions
The remaining Ph-like CRLF2low cases (defined as ΔCt >6 by LDA), and all CRLF2high cases without an identified CRLF2-R, were subject to multiplex and singleplex reverse transcription–PCR (RT-PCR) assays for 41 known kinase fusions involving ABL1, ABL2, CSF1R, JAK2, NTRK3, and PDGFRB as previously described.9 Primers are listed in supplemental Table 2.
Transcriptome sequencing analysis
CRLF2low cases without a kinase fusion identified by RT-PCR and CRLF2high cases without an identified CRLF2-R or kinase fusion underwent total-stranded whole transcriptome sequencing (RNA-sequencing) using the TruSeq library preparation on the Illumina HiSeq 4000 platform. Samples were analyzed for kinase fusions using the algorithms CICERO11 and FusionCatcher.40 Sequence mutations were analyzed using the Genome Analysis Toolkit pipeline.41 Sequencing metrics are provided in supplemental Table 3.
Statistical analysis
Associations between categorical values were examined using the χ2 test. Associations between Ph-like status and treatment outcomes from diagnosis (event-free [EFS] and overall survival [OS] and day 29 MRD) were estimated using the Kaplan-Meier method and log-rank test.42 An event was defined as a failure to achieve remission, a relapse after remission, death from any cause, or the development of a second malignancy. A multivariable analysis of EFS and OS was performed using the Cox proportional-hazards regression model.
Results
Prevalence of Ph-like ALL
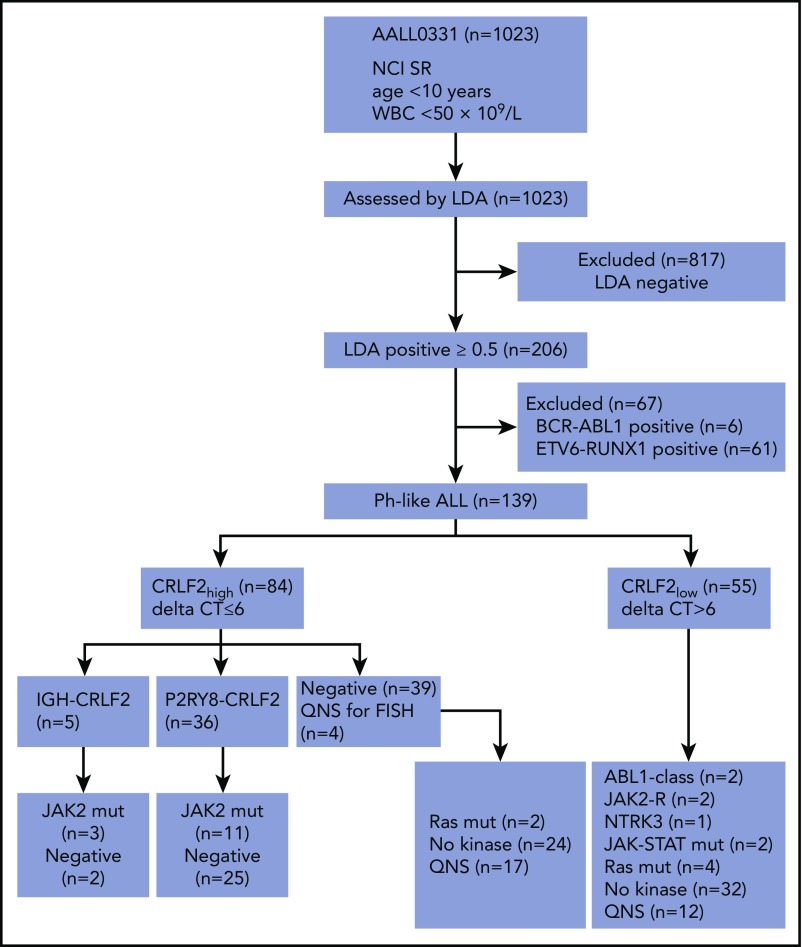
Of 1023 patients analyzed, 206 (20.1%) were identified as Ph-like by the LDA card. Cases with known genetic subgroups including BCR-ABL1 (n = 6) and ETV6-RUNX1 (n = 61 of 318 ETV6-RUNX1 cases) were excluded from downstream analysis because either (a) a targetable kinase lesion was already identified (BCR-ABL1) or (b) targetable kinase fusions have not been identified in comprehensive genome/RNA-sequencing analyses of ETV6-RUNX1 ALL.33,38,43 The remaining 139 patients with Ph-like ALL (13.6%) were tested for kinase alterations according to our previously published algorithm, with RNA-sequencing performed for cases without an identified genomic alteration (Figure 1; supplemental Figure 1).
Figure 1.
Screening algorithm. Testing pipeline developed for downstream characterization of Ph-like ALL cases for the identification of kinase alterations. QNS, quantity not sufficient.
Genomic landscape of CRLF2 rearrangements
Among the 139 Ph-like ALL cases, 84 (60.4%) were CRLF2high. P2RY8-CRLF2 was identified in 36 CRLF2high Ph-like ALL cases using the LDA card. Of the 48 remaining CRLF2high cases, 44 had material for FISH analysis, with IGH-CRLF2 rearrangement identified in 5 cases (Table 1; Figure 2). Notably, 36 of the 39 (92.3%) CRLF2high cases that were negative for a CRLF2-R showed gain of the CRLF2 locus by either duplication of an X or Y chromosome on karyotype or increased copy number of CRLF2 by FISH, potentially accounting for the high CRLF2 expression observed in the absence of a rearrangement. Only 20 of 46 CRLF2low Ph-like ALL cases with evaluable material harbored X or Y chromosomal duplication. Overall, CRLF2 rearrangement was present in 41 of 84 (48.8%) LDApos CRLF2high cases, with an overall frequency of 29.5% in Ph-like ALL (41/139). Of the 41 CRLF2-R cases, 14 harbored a concomitant activating mutation in JAK2 (34.1% of CRLF2-R). No mutations in JAK1 were identified in this cohort (Figure 2). We also identified 13 cases with P2RY8-CRLF2 that lacked the Ph-like gene expression profile signature. These cases had LDA values that ranged from 0.32 to 0.49 with no JAK1 or JAK2 mutations identified and were not considered Ph-like.
Table 1.
Frequency of kinase alterations in SR ALL
| Subgroup | NCI SR, n (%) (n = 1023) |
|---|---|
| Ph-like ALL | 139 (13.6) |
| CRLF2-R | 41 (4) |
| IGH-CRLF2 | 5 (0.5) |
| P2RY8-CRLF2 | 36 (3.5) |
| CRLF2-R with JAK-STAT mutation | 14 (1.7) |
| CRLF2-R without JAK-STAT mutation | 27 (2.6) |
| Non-CRLF2 | 98 (9.6) |
| ABL1-class fusions | 2 (0.2) |
| JAK2-R | 2 (0.2) |
| EPOR-R | 0 |
| NTRK3-R | 1 (0.1) |
| Other JAK-STAT | 2 (0.2) |
| Ras | 6 (0.6) |
| No kinase identified | 56 (5.5) |
| Not tested | 29 (2.8) |
ABL1-class: ZMIZ1-ABL1, EBF1-PDGFRB. JAK2-R: ETV6-JAK2, PAX5-JAK2. NTRK3: ETV6-NTRK3. Other JAK-STAT: IL7R. Ras: KRAS, NRAS.
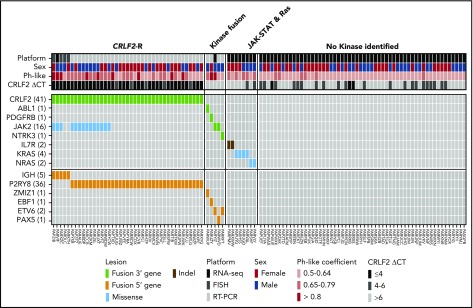
Figure 2.
Heat map showing kinase alterations identified in SR Ph-like ALL. Data are shown for 110 Ph-like ALL patients with material available for testing. The Ph-like coefficient was obtained from the LDA card using an algorithm including 8 genes that are highly expressed in Ph-like ALL.
Additional kinase alterations in Ph-like ALL
The 98 Ph-like cases without CRLF2-R, including 55 CRLF2low cases and 43 CRLF2high cases lacking a CRLF2-R, were assessed for kinase alterations by multiplex RT-PCR for 41 known fusions (n = 83) and RNA-sequencing (n = 65). Overall, 95/98 (96.9%) cases were studied by either RT-PCR only (n = 30), both RT-PCR and RNA-sequencing (n = 53), or by RNA-sequencing only (n = 12). Three cases lacked material for downstream analyses (supplemental Figure 1). Among the 83 cases subjected to RT-PCR, 4 fusions were identified: ZMIZ1-ABL1, EBF1-PDGFRB, PAX5-JAK2, and ETV6-NTRK3. Of the remaining 91 unclassified cases, 65 with available material were subject to RNA-sequencing to identify additional kinase alterations. From this analysis, we identified 1 case with an ETV6-JAK2 fusion, 2 cases with an IL7R sequencing mutation, 4 cases with a KRAS mutation, and 2 cases with an NRAS mutation (Figure 2).
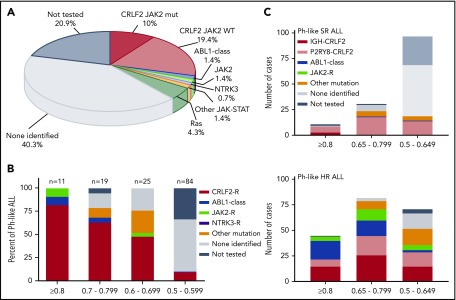
In summary, 13 Ph-like ALL cases without a CRLF2-R with evaluable material harbored a genomic alteration predicted to activate kinase or cytokine receptor signaling, including 2 cases with ABL-class fusions (1 ABL1 and 1 PDGFRB; 1.4% of Ph-like), 2 cases with a JAK2 fusion (1.4%), 1 case with an NTRK3 fusion (0.7%), alterations activating JAK-STAT signaling (IL7R; 1.4%) or Ras signaling (KRAS, NRAS; 4.3%). No cases were identified with truncating rearrangement of the erythropoietin receptor. Fifty-six of 139 (40.3%) Ph-like cases (including 24/84 [28.6%] CRLF2high and 32/55 [58.2%] CRLF2low) lacked a kinase alteration by RNA-sequencing analysis, 50 of which had an LDA value ≤0.65. Twenty-nine cases (21.3% of Ph-like) lacked material for complete evaluation by RNA-sequencing and remain classified as unknown (Table 1; Figure 3A).
Figure 3.
Targetable kinase alterations in Ph-like ALL. (A) Summary of genetic subgroups in Ph-like ALL. (B) Frequency of targetable alterations according to the LDA Ph-like coefficient. CRLF2-R, IGH-CRLF2, and P2RY8-CRLF2; ABL1-class fusion, ZMIZ1-ABL1 and EBF1-PDGFRB; JAK2 fusion, ETV6-JAK2 and PAX5-JAK2; other kinase fusion, ETV6-NTRK3; other sequence mutation, IL7R, KRAS, and NRAS. (C) Distribution of kinase alterations by LDA value in SR and HR Ph-like ALL.9
In total, 54 of 139 Ph-like ALL cases (38.8%) harbored a genetic alteration activating kinase or cytokine receptor signaling. Based on LDA coefficient, 11 of 139 Ph-like cases (7.9% of Ph-like) had an LDA ≥0.8, whereas the majority of cases had an LDA value between 0.5 and 0.649 (97 of 139, 69.8%). Cases with higher LDA values were more likely to harbor targetable kinase alterations, with all 11 cases with LDA values ≥0.8 harboring a CRLF2-R (n = 9) or other kinase fusion (n = 2). Only 10.7% (9/84) of cases with an LDA value between 0.5 and 0.59 harbored a targetable alteration, including CRLF2-R (n = 8) and NTRK3 fusion (n = 1; P < .0001) (Figure 3B). We also observed differences between the spectrum of kinase alterations in SR vs HR ALL9 (Figure 3C).
Among the 56 Ph-like ALL cases that lacked a kinase alteration by RNA-sequencing analysis, 6 cases harbored fusions in additional genes not involved in kinase and cytokine receptor signaling, including KMT2A-MLLT10, TCF3-PBX1, IGH-DUX4, CUX1-NUTM1, CREBBP-SLX4, and PAX5-ZCCHC7 (supplemental Table 1). Five of these cases harbored LDA values below 0.6, with the exception being PAX5-ZCCHC7 (LDA value of 0.758).
Treatment outcomes
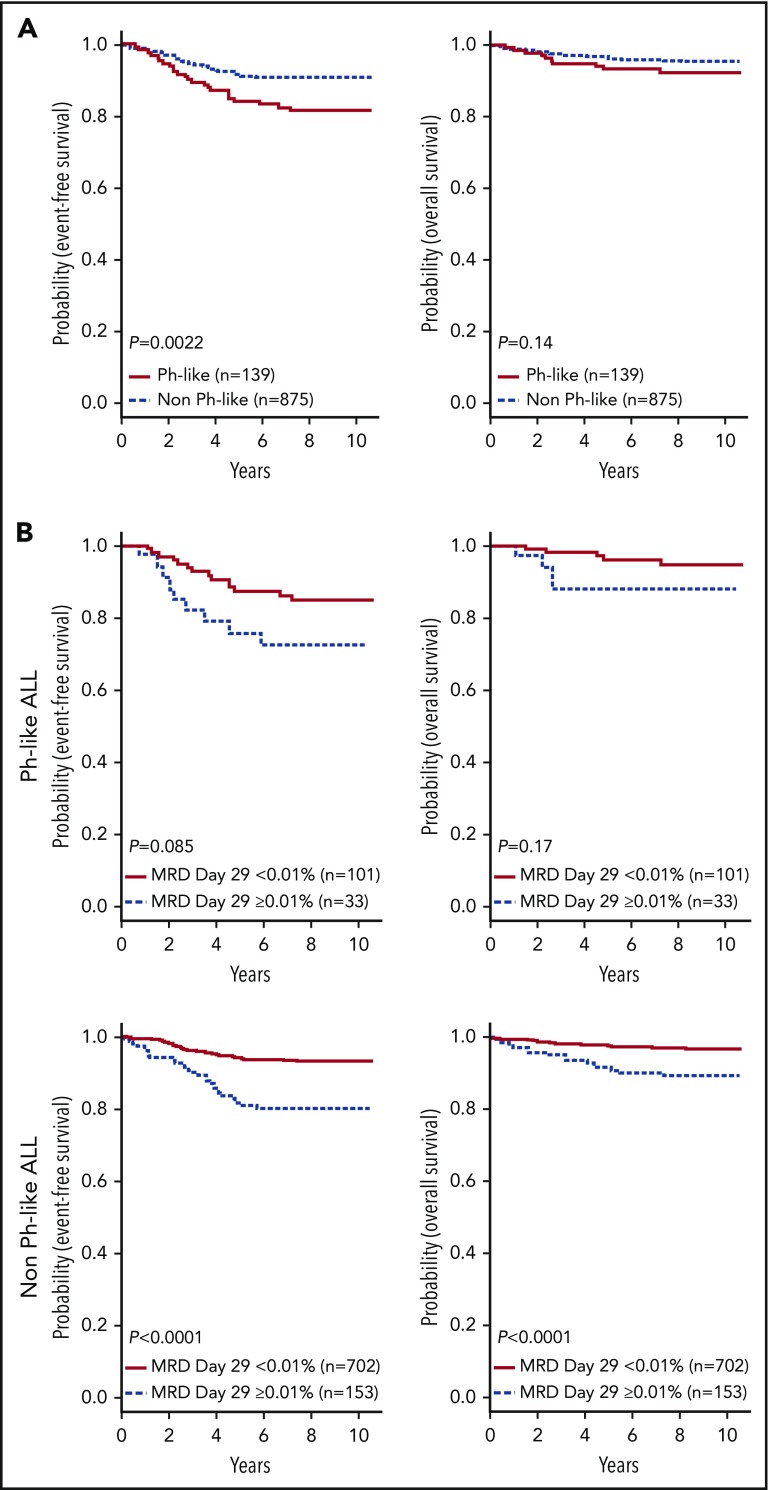
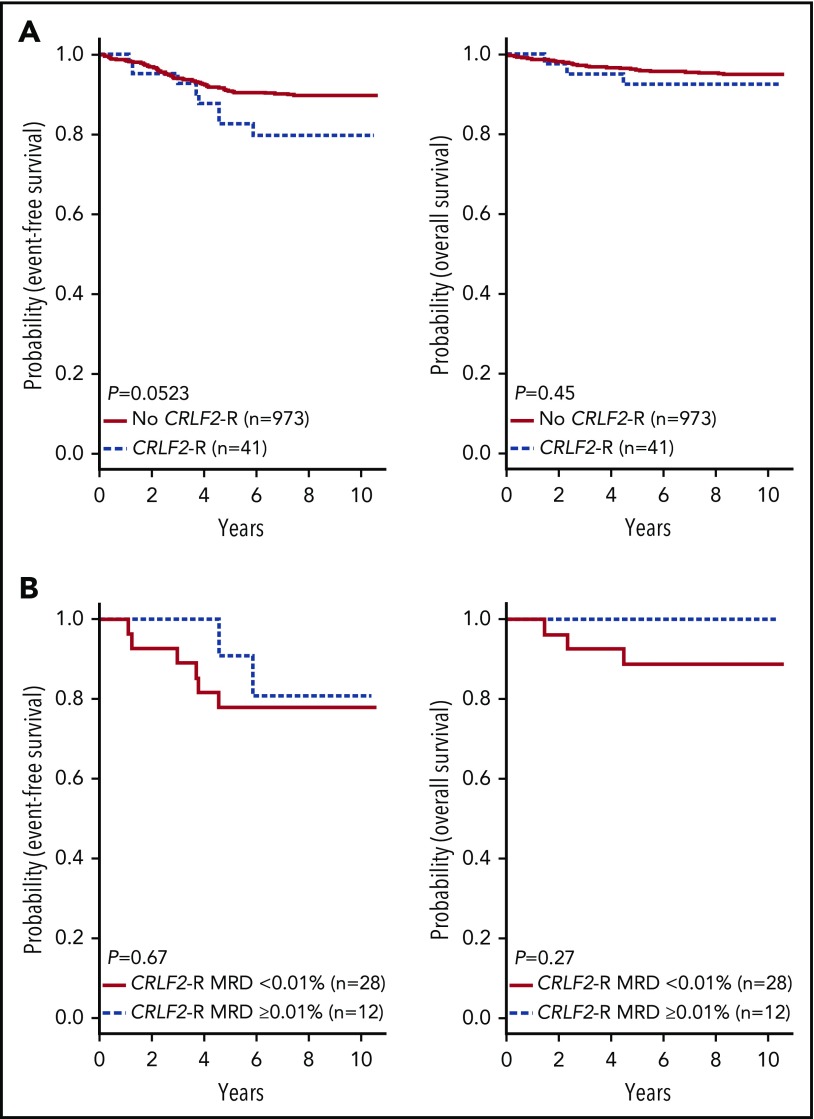
In contrast to HR ALL,11 no differences in sex or WBC count were observed between Ph-like and non-Ph-like ALL patients (supplemental Table 4). The SR ALL patients with Ph-like ALL had inferior 7-year EFS rates compared with non-Ph-like ALL (82.4 ± 3.6% vs 90.7 ± 1.0%, P = .0022), with no significant difference in OS (93.2 ± 2.4% vs 95.8 ± 0.7%, P = .14) (Figure 4A). Ph-like ALL cases were more likely to have end induction MRD ≥0.01% compared with non-Ph-like cases, but this did not reach statistical significance (24.6% vs 17.9%, P = .064). Among all patients, positive MRD ≥0.01% was associated with inferior outcome compared with patients with MRD <0.01% in both 7-year EFS (78.4 ± 0.7% vs 92.4 ± 0.7%, P < .0001) and OS rates (89.8 ± 2.5% vs 97.2 ± 0.6%, P < .0001). However, positive MRD was not predictive of EFS (72.4 ± 8.6% vs 86.0 ± 3.6%, P = .085) or OS in Ph-like ALL (87.9 ± 6.1% vs 95.7 ± 2.3%, P = .17) (Figure 4B), likely because of the small patient numbers. The presence of Ph-like ALL and a CRLF2-R was associated with inferior 7-year EFS (79.8 ± 6.7% vs 90.0 ± 1.1%, P = .0523) but not OS (92.4 ± 4.4% vs 95.6 ± 0.7%, P = .45) (Figure 5A). Notably, 70% (28/40) of these Ph-like ALL CRLF2-R cases that had an excellent response with MRD <0.01% at the end of induction therapy had a 7-year EFS of only 78.0 ± 8.2% (Figure 5B). In multivariable analyses, Ph-like ALL (P = .032) and positive MRD (P < .0001) were independently associated with poor EFS, whereas only positive MRD (P < .0001) was predictive of poor OS (supplemental Table 5).
Figure 4.
Outcome of children with SR B-ALL enrolled in COG AALL0331. (A) Kaplan-Meier estimates of EFS and OS of patients with Ph-like ALL compared with non Ph-like ALL (including ETV6-RUNX1, excluding BCR-ABL1). (B) Kaplan-Meier estimates of positive (≥0.01%) vs negative MRD (<0.01%) at end of induction for patients with Ph-like ALL (top) and non Ph-like ALL (bottom).
Figure 5.
Outcome of CRLF2-R patients enrolled in COG AALL0331. Kaplan-Meier estimates of EFS and OS of patients with Ph-like ALL and CRLF2-R compared with patients without a CRLF2-R (excluding BCR-ABL1) (A); positive (≥0.01%) vs negative MRD (<0.01%) at end of induction for patients with Ph-like ALL and a CRLF2-R (B).
Discussion
Multiple groups have reported the treatment failure and poor outcome associated with Ph-like ALL in children and adults.5-13 Studies in children with NCI HR ALL demonstrate a high prevalence of Ph-like ALL and associated targetable alterations, guiding the development of clinical trials testing the efficacy of appropriate TKI therapy in HR ALL. In this comprehensive retrospective study of 1023 patients with NCI SR B-ALL and mature outcome data enrolled in COG AALL0331, we report the spectrum of targetable alterations and prognostic significance of Ph-like ALL in SR patients.
We show that Ph-like ALL is less common in an unselected cohort of COG SR patients compared with both HR ALL and SR ALL patients with MRD >0.01% at end of induction (13.6% vs 22.4% vs 17.0%; Table 2),9 and comparable to 10.0% observed in SR patients treated in St. Jude Children’s Research Hospital protocols TXV and TXVI.11 Many published studies have estimated the frequency of Ph-like ALL by screening B-other patients with a limited panel of known fusions and commonly without performing FISH for CRLF2-R, thus likely underestimating the true incidence of kinase alterations.25,30-32 Here, we have performed comprehensive genomic analysis on every Ph-like ALL case with available material, including PCR assays that can identify P2RY8-CRLF2 and 41 kinase fusions including a total of 3 different splice variants, CRLF2 FISH, and/or unbiased RNA-sequencing to accurately determine the frequency of Ph-like ALL and associated kinase alterations. This is the most comprehensive report investigating the prevalence of Ph-like ALL in childhood SR ALL as defined by the NCI criteria and confirms that older age and presenting WBC count are important clinical features of this subtype.
Table 2.
Comparison of kinase alterations in HR and SR ALL
| Subgroup | NCI HR,9 n (%) (n = 884) | NCI SR9 MRD+, n (%) (n = 505) | NCI SR, n (%) (n = 1023) | P * |
|---|---|---|---|---|
| Ph-like ALL | 198 (22.4) | 86 (17.0) | 139 (13.6) | <.0001 |
| CRLF2-R | 96 (10.9) | 29 (5.7) | 41 (4) | <.0001 |
| IGH-CRLF2 | 56 (6.3) | 5 (1.0) | 5 (0.5) | <.0001 |
| P2RY8-CRLF2 | 40 (4.5) | 24 (4.8) | 36 (3.5) | .29 |
| CRLF2-R with JAK-STAT mutation | 46 (5.2) | 17 (3.4) | 14 (1.7) | <.0001 |
| CRLF2-R without JAK-STAT mutation | 50 (5.7) | 12 (2.4) | 27 (2.6) | .001 |
| Non-CRLF2 | 102 (11.5) | 57 (11.3) | 98 (9.6) | |
| ABL1-class fusions | 35 (4.0) | 5 (1.0) | 2 (0.2) | <.0001 |
| JAK2-R | 9 (1.0) | 5 (1.0) | 2 (0.2) | .029 |
| EPOR-R | 11 (1.2) | 0 | 0 | <.0001 |
| Other kinase | 7 (0.8) | 6 (1.2) | 1 (0.1) | .028 |
| Other JAK-STAT | 12 (1.4) | 6 (1.2) | 2 (0.2) | .0049 |
| Ras | 6 (0.7) | 11 (2.2) | 6 (0.6) | >.99 |
| No kinase identified | 18 (2.0) | 24 (4.8) | 56 (5.5) | .0001 |
| Not tested | 4 (0.5) | 0 | 29 (2.8) | <.0001 |
ABL1-class: ABL1, ABL2, CSF1R, PDGFRB. Other kinase: NTRK3, LYN, FLT3. Other JAK-STAT: IL7R, SH2B3, JAK1. Ras: KRAS, NRAS.
Comparing NCI HR patients treated on AALL1131 vs NCI SR patients treated on AALL0331.
We observed striking differences in the distribution of LDA values between SR and HR Ph-like ALL patients, with a much lower frequency of cases with LDA ≥0.8 (8% vs 23%), and a higher frequency of cases with LDA value 0.5 to 0.649 (70% vs 36%) (Figure 3C).9 We also observed a difference in the spectrum of kinase alterations in SR compared with HR Ph-like ALL patients. Rearrangement of CRLF2 was lower in SR Ph-like ALL (29.4% of Ph-like ALL, 4% overall) compared with HR ALL (45% to 50% of Ph-like, 10.9% overall; Table 2).9,11 Accordingly, only 49% of CRLF2high cases in this study harbored a confirmed CRLF2-R. The moderate increase in CRLF2 expression observed in the cases lacking a CRLF2-R was likely because of duplication of the CRLF2 locus with gain of an X or Y chromosome (present in 92% of cases), which has also been observed in prior studies.44 The distribution of CRLF2 fusion partners among Ph-like ALL patients also differed markedly, with a ratio of 7:1 P2RY8:IGH in the current study compared with 1:1.4 in HR ALL, resulting in a significantly higher prevalence of the poor prognosis IGH-CRLF2 rearrangement in HR ALL compared with SR ALL (6.3% vs 0.5%).9,17 This finding is similar to our previous analysis of CRLF2-R in SR-ALL that showed a ratio of 4.6:1 P2RY8:IGH and an overall frequency of 1.6% for IGH-CRLF2 among 561 SR-ALL cases.44 However, that study did not include SR-ALL cases with the favorable genetic subsets defined by trisomies of chromosome 4 and 10 or ETV6-RUNX1 fusion that collectively account for more than half of NCI SR ALL cases. The increased proportion of P2RY8-CRLF2 over IGH-CRLF2 in this study is also comparable to that observed in ALL patients treated on the ALL–Berlin-Frankfurt-Münster (BFM) 2000 and Italian Association of Pediatric Haematology and Oncology (AIEOP)–BFM ALL 2009 protocols and confirms previous reports that IGH-CRLF2 is associated with increased age at diagnosis compared with P2RY8-CRLF2 (median age 14 vs 4 years).17,44-47 We also observed a lower frequency of CRLF2-R with JAK mutations in SR ALL: 34.1% of CRLF2-R compared with ∼50% in HR ALL.9,11,17
Significant differences in the prevalence of additional targetable alterations were also identified, with a 10-fold decrease in SR compared with HR patients (3.5% vs ∼30% of Ph-like ALL). In this study, only 5 of 1023 patients (0.5% of cohort) harbored a kinase fusion that would be considered amenable to TKI therapy, including 2 cases with an ABL-class fusion amenable to treatment with an ABL-class inhibitor, 2 cases with a JAK2 fusion amenable to treatment with a JAK2 inhibitor such as ruxolitinib, and 1 case with an NTRK3 fusion that could be treated with a TRK inhibitor. Interestingly, no cases with a truncating rearrangement of EPOR were present in this cohort, which is a recurrent alteration in childhood HR and adult Ph-like ALL.9-11 A substantial number of Ph-like ALL cases in the SR cohort (40.3% of Ph-like) did not harbor a targetable fusion by RNA-sequencing analysis, a notable increase from 9% among HR ALL patients.9,11 A majority of these cases (50/56) had a low positive LDA value (0.5-0.65 on a scale of 0-1).
We identified 61 of 318 (19.2%) ETV6-RUNX1 cases to have a low but positive LDA value, the majority being 0.5 to 0.6 (55/61 cases). The LDA card was originally trained to identify all actionable lesions activating kinase signaling in HR ALL cases, a cohort that is underrepresented for alterations common in SR ALL, such as ETV6-RUNX1. As such, this training did not take into account the expression of the 8 genes included in the LDA in ETV6-RUNX1 ALL. Using the entire gene expression profile of Ph+ cases to predict Ph-like ALL, ETV6-RUNX1 cases are not predicted as Ph-like (data not shown) and do not harbor kinase-activating alterations and should not be considered Ph-like for risk stratification and therapeutic targeting purposes.
HR Ph-like ALL patients experience high rates of treatment failure and death compared with non Ph-like ALL patients enrolled in the same HR COG protocols.8,11 In this study of SR ALL, patients with Ph-like ALL had inferior EFS compared with non Ph-like ALL, but no difference in OS was observed. Furthermore, the outcome of SR patients with Ph-like ALL is significantly better than that of HR Ph-like ALL patients, with 5-year EFS rates of 84.1% compared with 62.6% (COG AALL0232).8 Despite the improved outcome, Ph-like ALL in SR patients was associated with elevated MRD levels at end of induction, as previously observed in COG and St. Jude protocols.11,29 The presence of Ph-like ALL was independently associated with poor EFS, but not OS. Thus, the overall outcomes for Ph-like ALL patients identified in SR ALL are better than patients identified in HR cohorts.
We previously performed more limited genomic analyses, including RT-PCR for P2RY8-CRLF2 and CRLF2 FISH, on 562 SR-ALL patients treated on COG P9905, which used less intensive chemotherapy than AALL0331.44 In that study, CRLF2-R was not associated with inferior EFS (hazard ratio 1.26, P = .484), perhaps because the favorable cases with trisomy of 4 and 10 or ETV6-RUNX1 were not included. In the current study, the 7-year EFS for SR Ph-like ALL cases with CRLF2-R was only 79.3 ± 6.8%. Furthermore, CRLF2-R cases had similar EFS rates regardless of MRD status at the end of induction therapy. This is in contrast to patients treated on AIEOP-BFM protocols, where the presence of P2RY8-CRLF2 was only prognostic in the MRD-positive population.48 Although the number of cases is small, these data suggest that it may not be appropriate to treat SR Ph-like ALL CRLF2-R cases with the relatively low-intensity COG SR ALL therapy.
In summary, we demonstrate reduced frequency of Ph-like ALL and targetable alterations and superior outcome in SR ALL patients compared with HR ALL. These findings underscore the significant differences in the prevalence, genomic landscape, and prognostic significance of Ph-like ALL based on age and presenting WBC count. Although the likeliness of identifying a kinase lesion is much lower in SR compared with HR ALL, screening of cases that lack subtype-defining chromosomal alterations should be considered, particularly in those cases that have suboptimal responses to remission induction therapy.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the staff of the Flow Cytometry and Cell Sorting Core Facility and the Genome Sequencing Facility of the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital. The authors also thank Joshua Stokes from Biomedical Communications at St. Jude Children’s Research Hospital for graphic design.
This work was supported by grants from the St. Baldrick's Foundation Consortium Award (S.P.H., J.M.G.-F., C.G.M., C.L.W., and M.L.L.); the Leukemia & Lymphoma Society Specialized Center of Research (W.L.C., S.P.H., J.M.G.-F., C.L.W., and C.G.M.); the National Cancer Institute National Institutes of Health (U10 CA180886 and U10 CA180899; R50 CA211542 [R.C.H.] and R35 CA197695 [C.G.M.]); the National Institute of General Medical Sciences, National Institutes of Health (P50 GM 115279); the American Society of Hematology; and the COG Hematopoietic Malignancies Program (K.G.R.). M.L.L. is a UCSF Benioff Chair of Children’s Health and the Deborah and Arthur Ablin Professor of Pediatric Molecular Oncology at Benioff Children’s Hospital. S.P.H. is the Jeffrey E. Perelman Distinguished Chair in Pediatrics at the Children's Hospital of Philadelphia. C.G.M. is the William E. Evans Endowed Chair at St. Jude Children’s Research Hospital.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.G.R. prepared the manuscript; K.G.R., I-M.C., K.P., E.S., H.J., M.V., Y.Z., and D.P.-T. performed experiments; K.G.R., S.C.R., R.C.H., Y.D., Z.G., J.Z., M.D., N.A.H., A.J.C., M.J.B, B.L.W., C.L.W., J.M.G.-F., C.G.M., and S.P.H. analyzed data; E.A.R., L.A.M., K.W.M., W.L.C., M.L.L., and S.P.H. provided patient samples; and all authors edited the manuscript.
Conflict-of-interest disclosure: R.C.H., I-M.C., and C.L.W. are coinventors on US Patent No. 8 568 974 B2 “Identification of novel subgroups of high risk pediatric precursor-B acute lymphoblastic leukemia, outcome correlations and diagnostic and therapeutic methods.” The remaining authors declare no competing financial interests.
Correspondence: Charles G. Mullighan, Department of Pathology, 262 Danny Thomas Pl, MS342, St. Jude Children’s Research Hospital, Memphis, TN 38105; e-mail: charles.mullighan@stjude.org; and Stephen P. Hunger, Children’s Hospital of Philadelphia, CTRB 3060, 3501 Civic Center Blvd, Philadelphia, PA 19104; e-mail: hungers@email.chop.edu.
References
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541-1552. [DOI] [PubMed] [Google Scholar]
- 2.Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35(9):975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullighan CG, Su X, Zhang J, et al. ; Children’s Oncology Group. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boer JM, Koenders JE, van der Holt B, et al. Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica. 2015;100(7):e261-e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herold T, Schneider S, Metzeler KH, et al. Adults with Philadelphia chromosome-like acute lymphoblastic leukemia frequently have IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2017;102(1):130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain N, Roberts KG, Jabbour E, et al. Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood. 2017;129(5):572-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loh ML, Zhang J, Harvey RC, et al. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children’s Oncology Group TARGET Project. Blood. 2013;121(3):485-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reshmi SC, Harvey RC, Roberts KG, et al. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children’s Oncology Group. Blood. 2017;129(25):3352-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts KG, Gu Z, Payne-Turner D, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol. 2017;35(4):394-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwab C, Ryan SL, Chilton L, et al. EBF1-PDGFRB fusion in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): genetic profile and clinical implications. Blood. 2016;127(18):2214-2218. [DOI] [PubMed] [Google Scholar]
- 13.Tasian SK, Hurtz C, Wertheim GB, et al. High incidence of Philadelphia chromosome-like acute lymphoblastic leukemia in older adults with B-ALL. Leukemia. 2017;31(4):981-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz KR, Carroll A, Heerema NA, et al. ; Children’s Oncology Group. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia. 2014;28(7):1467-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biondi A, Schrappe M, De Lorenzo P, et al. Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol. 2012;13(9):936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts KG, Yang YL, Payne-Turner D, et al. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv. 2017;1(20):1657-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41(11):1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell LJ, Capasso M, Vater I, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114(13):2688-2698. [DOI] [PubMed] [Google Scholar]
- 20.Yoda A, Yoda Y, Chiaretti S, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2010;107(1):252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maude SL, Tasian SK, Vincent T, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120(17):3510-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tasian SK, Doral MY, Borowitz MJ, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120(4):833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iacobucci I, Li Y, Roberts KG, et al. Truncating erythropoietin receptor rearrangements in acute lymphoblastic leukemia. Cancer Cell. 2016;29(2):186-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schinnerl D, Fortschegger K, Kauer M, et al. The role of the Janus-faced transcription factor PAX5-JAK2 in acute lymphoblastic leukemia. Blood. 2015;125(8):1282-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steeghs EMP, Jerchel IS, de Goffau-Nobel W, et al. JAK2 aberrations in childhood B-cell precursor acute lymphoblastic leukemia. Oncotarget. 2017;8(52):89923-89938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weston BW, Hayden MA, Roberts KG, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31(25):e413-e416. [DOI] [PubMed] [Google Scholar]
- 28.Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood. 2017;130(19):2064-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts KG, Pei D, Campana D, et al. Outcomes of children with BCR-ABL1–like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol. 2014;32(27):3012-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boer JM, Marchante JR, Evans WE, et al. BCR-ABL1-like cases in pediatric acute lymphoblastic leukemia: a comparison between DCOG/Erasmus MC and COG/St. Jude signatures. Haematologica. 2015;100(9):e354-e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boer JM, Steeghs EM, Marchante JR, et al. Tyrosine kinase fusion genes in pediatric BCR-ABL1-like acute lymphoblastic leukemia. Oncotarget. 2017;8(3):4618-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imamura T, Kiyokawa N, Kato M, et al. Characterization of pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia with kinase fusions in Japan. Blood Cancer J. 2016;6(5):e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Z, Churchman M, Roberts K, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirabayashi S, Ohki K, Nakabayashi K, et al. ; Tokyo Children’s Cancer Study Group (TCCSG). ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102(1):118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilljebjörn H, Fioretos T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood. 2017;130(12):1395-1401. [DOI] [PubMed] [Google Scholar]
- 36.Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YF, Wang BY, Zhang WN, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, McCastlain K, Yoshihara H, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48(12):1481-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattano LA, Devidas M, Friedmann AM. Outstanding outcome for children with standard risk-low (SR-Low) acute lymphoblastic leukemia (ALL) and no benefit to intensified Peg-asparaginase (PEG-ASNase) therapy: results of Children’s Oncology Group (COG) Study AALL0331 [abstract]. Blood. 2014;124(21). Abstract 793. [Google Scholar]
- 40.Nicorici D, Satalan M, Edgren H, et al. FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data [published online ahead of print 19 November 2014]. bioRxiv. doi:10.1101/011650. [Google Scholar]
- 41.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50(3):163-170. [PubMed] [Google Scholar]
- 43.Papaemmanuil E, Rapado I, Li Y, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen IM, Harvey RC, Mullighan CG, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119(15):3512-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell LJ, Jones L, Enshaei A, et al. Characterisation of the genomic landscape of CRLF2-rearranged acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2017;56(5):363-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115(26):5393-5397. [DOI] [PubMed] [Google Scholar]
- 47.Schmäh J, Fedders B, Panzer-Grümayer R, et al. Molecular characterization of acute lymphoblastic leukemia with high CRLF2 gene expression in childhood. Pediatr Blood Cancer. 2017;64(10):e26539. [DOI] [PubMed] [Google Scholar]
- 48.Palmi C, Vendramini E, Silvestri D, et al. Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26(10):2245-2253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.