Abstract
Background
Staphylococcus aureus is a contagious, opportunistic pathogen that causes clinical or subclinical mastitis in dairy cattle. The genetic background and antimicrobial resistance of isolates from Ethiopian dairy farms has not been studied. Therefore, the aim of this study was to characterize S. aureus from Ethiopian hand milked dairy cows, by spa, MLST and virulence factor typing, and by assessment of antimicrobial susceptibility. A total of 79 S. aureus isolates from intramammary infections was studied. A PCR was used to detect lukM-lukF’ and pvl genes encoding the bovine and human associated bi-component leukocidins, and the toxic shock syndrome toxin gene-1 (tst). Antimicrobial susceptibility was determined using the broth microdilution method.
Results
Twenty different spa types were identified, most isolates were t042 (58%), and the closely related t15786 (11%). The proportion of isolates positive for lukM-lukF’, tst and pvl was low at 0.04, 0.10 and 0.09 respectively, with lukM-lukF’ often co-occurring with tst, but not with pvl. Methicillin-resistance was not found, but resistance to penicillin/ampicillin (86%) and tetracycline (54%) was very common.
Conclusions
We found a high degree of relatedness among bovine S. aureus isolates in North-Western Ethiopia, suggesting contagious within and between farm transmission of strains that are often resistant to commonly used antimicrobials. This highlights the need for effective preventive measures that aim at limiting transmission of bacteria rather than using antimicrobials to control S. aureus mastitis in Ethiopia.
Electronic supplementary material
The online version of this article (10.1186/s12917-018-1558-1) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus aureus, Spa, MLST, Virulence factor, Antimicrobial resistance, Mastitis, Ethiopia
Background
Staphylococcus aureus is a contagious pathogen that causes mastitis in dairy cattle [1, 2], and is an opportunistic pathogen in humans and many other animal species [3–5]. Also in Ethiopia, S. aureus is frequently isolated from cows with mastitis [6–8], and from milk or milk products [9].
In dairy cows, S. aureus can be isolated from milk as well as from different other body sites [10, 11]. Transmission of S. aureus intramammary infections (IMI) is believed to mainly occur during the milking process [12], but this has been poorly researched in situations where hand milking is common, as is the case in Ethiopia. Additionally, hand milking may introduce possibilities for transmission between farmers and their cattle. Typing of S. aureus isolates may give insight in their likely origin, because MLST and spa-types and virulence factors often are host-associated [13, 14]. Genetic similarity of isolates within and between farms suggests contagious transmission in the spread of bacteria [4] whereas a greater variety of genotypes within herds or regions is more suggestive of environmental pathogens [15]. Genotyping does not only give information on modes of transmission, but can also be used to identify virulence factors of the bacteria. Depending on their clonal lineage, S. aureus may carry the phage-encoded leukocidin genes lukM-lukF’ [16–18]. LukMF’ is a potent toxin specifically killing bovine neutrophils [19] and likely to contribute to the clinical severity of mastitis [20]. The proportion of bovine S. aureus isolates encoding this toxin varies largely between countries [21], but to our knowledge this has not been estimated in isolates of IMI from African cattle.
Because the use of antimicrobial drugs, both in humans and animals, is poorly controlled, multidrug resistant S. aureus are frequently isolated from animals [6, 7] and humans in Ethiopia [22–24]. In S. aureus isolated from humans, there is a trend of increasing antimicrobial resistance (AMR) [25]. Describing AMR patterns in both humans and animals may contribute to the knowledge on the importance of the issue in Ethiopia and to a more prudent and effective antimicrobial use.
In smallholder dairy farming, effective control of S. aureus mastitis is important, because mastitis can have a substantial effect on family income. Since only a limited number of studies characterized bovine S. aureus isolated from Ethiopia [9, 26], the aim of the present study was to characterize S. aureus isolates from milk samples from hand milked dairy cows in North-Western Ethiopia by spa and MLST typing, virulence genes and antimicrobial susceptibility.
Methods
Isolate collection
A total of 135 phenotypically isolated S. aureus were available from an earlier cross-sectional study conducted from October 2014 to December 2016 in 167 urban and peri-urban smallholder dairy farms in the regions Gondar and Bahir Dar, in the North-West of Ethiopia [8]. Nine isolates were lost. Forty seven isolates were excluded from the study because data of isolates were lost (11), data mismatched (8), and genotypically non S. aureus (28). Seventy nine S. aureus isolates that had complete data records were available (two from clinical mastitis (CM), 53 from California mastitis test (CMT) positive and 24 from CMT negative quarters) for spa and MLST typing, virulence genes detection and antimicrobial susceptibility testing. The 79 isolates were derived from 60 dairy cows in 42 dairy herds and were kept in glycerol stocks at − 80 °C until further study.
DNA extraction
Single colonies from an overnight culture grown on blood agar plates were suspended in 1 mL of distilled water, homogenized using Vortex and centrifuged for 1 min at 13,000 rpm. Supernatant was removed and the bacteria were re-suspended in 200 μL of distilled water, boiled for 10 min at 100 °C and centrifuged at 13,000 rpm for 1 min. The extracted DNA was diluted 1:10 in distilled water and stored at − 20 °C until further analysis.
Virulence genes detection
Polymerase chain reactions were performed on 10 μL of the DNA of all 79 isolates to confirm the isolates were S. aureus (femA), to test for presence of the mecA gene, encoding resistance to methicillin, and to amplify the polymorphic X region of the surface protein A (spa) gene. We also used PCR to detect lukM-lukF’ and pvl genes encoding the bovine and human associated bi-component leukocidins, respectively, and the toxic shock syndrome toxin gene-1 (tst). Primers and protocols are summarized in Table 1. Amplifications were carried out in final volumes of 25 μL using a T100™ Thermal Cycler (Bio-Rad, USA). The presence of appropriate amplicon sizes was examined by electrophoresis of 10 μL PCR product on 1.5% agarose at 100 V for 40 min. PCR products were visualized with a molecular Imager Gel Doc XR+ Imaging system (Bio-Rad, USA).
Table 1.
Sequences of primer sets with their corresponding PCR protocols and product sizes
| Target gene | Primer sequence (5′-3′) | PCR-protocol | Amplicon size | Reference |
|---|---|---|---|---|
| femA | F: TGCCTTTACAGATAGCATGCCA | 1 | 142 bp | [52] |
| R: AGTAAGTAAGCAAGCTGCAATGACC | ||||
| mecA | F: GGCTATCGTGTCACAATCGTT | 2 | 689 bp | [53] |
| R: TCACCTTGTCCGTAACCTGA | ||||
| spa | F: AGACGATCCTTCGGTGAGC | 3 | Variable | [54] |
| R: GCTTTTGCAATGTCATTTACTG | ||||
| lukM | F: TGAGTGGGTATGGCATGAAAGA | 4 | 572 bp | [20] |
| R: TGGACATTTTGTGTTACACCCC | ||||
| lukF’ | F: ACTCAGGCTATACCAACCCA | 1 | 425 bp | [20] |
| R: CGAGCTACTCTGTCTGCCAC | ||||
| pvl | F: GCTGGACAAAACTTCTTGGAATAT | 5 | 85 bp | [55] |
| R: GATAGGACACCAATAAATTCTGGATTG | ||||
| tst | F: CAACATACTAGCGAAGGAACT | 6 | 277 bp | [56] |
| R: GATATGTGGATCCGTCATTCA |
1 95 °C for 2 min, 35× (95 °C for 30 s, 59.5 °C for 30 s, 72 °C for 35 s), 72 °C for 5 min and 12 °C for ∞
2 95 °C for 1.5 min, 30× (95 °C for 45 s, 55 °C for 30 s, 72 °C for 45 s), 72 °C for 5 min and 12 °C for ∞
3 95 °C for 2 min, 30× (95 °C for 45 s, 60 °C for 45 s, 72 °C for 1:30 min), 72 °C for 5 min and 12 °C for ∞
4 95 °C for 2 min, 33× (95 °C for 30 s, 58.2 °C for 30 s, 72 °C for 30 s), 72 °C for 5 min and 12 °C for ∞
5 95 °C for 2 min, 35× (95 °C for 30 s, 55.5 °C for 30 s, 72 °C for 35 s), 72 °C for 5 min and 12 °C for ∞
6 95 °C for 2 min, 35× (95 °C for 30 s, 59 °C for 30 s, 72 °C for 30 s), 72 °C for 5 min and 12 °C for ∞
Spa and MLST typing
The product of the spa gene PCR was cleaned up from unbound primers and nucleotides using ExoSAP-IT PCR Cleanup Reagent (Affymetrix, USA) according to manufacturer’s instructions and was submitted for Sanger sequencing (Baseclear BV, The Netherlands). The spa types were assigned using BioNumerics version 7 (Applied Maths, Belgium), using the spa-typing plugin and Ridom SpaServer (http://www.spaserver.ridom.de). A minimum spanning tree (MST) was constructed in BioNumerics using the spa clustering method of the spa typing plugin. Repeat successions were aligned using a gap creation cost of 100%, gap extension cost of 50%, duplicate creation and duplicate extension costs of both 25% and a maximum duplication number of 3 repeats. The MST was built using the MST cluster method and the default cost matrix with a 1% grouping distance bin. To save costs, multilocus sequence typing (MLST) as described by Enright et al. [27] was performed on a selection of ten of the 79 isolates: on six t042 isolates, and on 4 isolates of spa types t273, t17184, t488 and t409. These ten isolates were selected aiming to optimally represent the various spa types and the two regions (Gondar and Bahir Dar). Sequence types were assigned using BioNumerics.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by determining minimum inhibitory concentrations (MICs) using the MICRONAUT system (Merlin Diagnostika, Germany) in customized prepared 96-wells microtiter plates containing dilution series of 17 antimicrobials. Inoculum preparation, broth composition and incubation conditions were performed as recommended by the manufacturer. Reading of the plates after incubation was done with a photometer (Skan, Merlin Diagnostika, Germany). Staphylococcus aureus ATCC 29213 was used as quality control strain. Clinical breakpoints were used according to the Clinical and Laboratory Standards Institute (CLSI) standards [28, 29]. Veterinary breakpoints were used for ceftiofur (cattle), cephalotin (dogs) and enrofloxacin (dogs). Because veterinary breakpoints for several antimicrobials have not been described, human breakpoints were used for penicillin, ampicillin, tetracycline, clindamycin, erythromycin, trimethoprim sulfamethoxazole, rifampicin, amoxicillin/clavulanic acid, cefoxitin, chloramphenicol, gentamicin, kanamycin and neomycin. For fusidic acid, EUCAST-criteria were used (www.eucast.org), because no breakpoints were available in the CLSI standards.
Results
Genetic typing
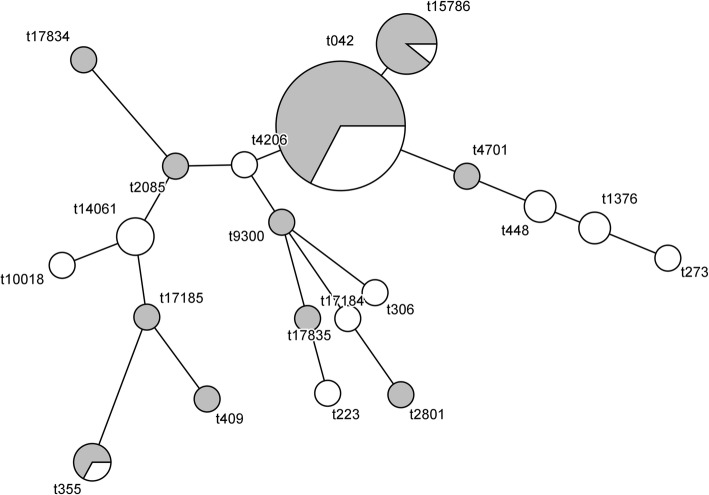
In total, twenty different spa types were found, the majority of isolates belonging to t042 (58%) and the closely related t15786 (11%) and the distribution of spa types was similar across the two regions (Fig. 1). The proportion of lukM-lukF’ positive S. aureus in our sample was low (3/79; 4%). Seven isolates carried pvl (7/79; 9%) and tst was detected in eight isolates (8/79, 10%). All t14061 isolates (3/79, 4%) were positive for tst; pvl was present in all t355 (3/79, 4%) and t1376 (2/79, 3%) isolates. None of the isolates carried the mecA gene. Table 2 summarizes the spa typing results and presents the isolates positive for virulence genes.
Fig. 1.
Minimum spanning tree based on spa types of 79 S. aureus isolates from Bahir Dar region (grey) and Gondar region (white) in North-Western Ethiopia. The size of the circles is proportional to the number of isolates found
Table 2.
Genetic characterization of 79 Staphylococcus aureus isolates from quarter milk samples of 60 cows in 42 dairy farms in North-Western Ethiopia
| spa type | Total (%) | N isolates positive for | Repeat sequence | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lukM-lukF’ | tst | pvl | |||||||||||||
| t042 | 46 (58) | 0 | 1 | 0 | 26 | 23 | 12 | 34 | 34 | 33 | 34 | ||||
| t15786 | 9 (11) | 0 | 0 | 0 | 26 | 23 | 12 | 34 | 34 | 34 | 33 | 34 | |||
| t355 | 3 (4) | 0 | 0 | 3 | 07 | 56 | 12 | 17 | 16 | 16 | 33 | 31 | 57 | 12 | |
| t14061 | 3 (4) | 2 | 3 | 0 | 04 | 31 | 17 | ||||||||
| t1376 | 2 (3) | 0 | 0 | 2 | 07 | 12 | 21 | 17 | 13 | 13 | 34 | 34 | 33 | 13 | |
| t488 | 2 (3) | 0 | 0 | 0 | 07 | 12 | 21 | 17 | 13 | 13 | 34 | 33 | 34 | ||
| t10018 | 1 (1) | 1 | 1 | 0 | 04 | 31 | 17 | 25 | 17 | 25 | 17 | ||||
| t306 | 1 (1) | 0 | 1 | 0 | 26 | 23 | 17 | 34 | 17 | 20 | 17 | 12 | 17 | 17 | 16 |
| t17835 | 1 (1) | 0 | 1 | 1 | 14 | 22 | 75 | 16 | 24 | 24 | 24 | 24 | 24 | ||
| t223 | 1 (1) | 0 | 1 | 0 | 26 | 23 | 13 | 23 | 05 | 17 | 25 | 17 | 25 | 16 | 28 |
| t273 | 1 (1) | 0 | 0 | 1 | 07 | 23 | 21 | 17 | 13 | 34 | 16 | 34 | 33 | 13 | |
| t2085 | 1 (1) | 0 | 0 | 0 | 26 | 33 | 34 | ||||||||
| t4206 | 1 (1) | 0 | 0 | 0 | 26 | 23 | 34 | 34 | 33 | 34 | |||||
| t9300 | 1 (1) | 0 | 0 | 0 | 26 | 23 | 21 | 34 | |||||||
| t4701 | 1 (1) | 0 | 0 | 0 | 26 | 12 | 21 | 17 | 34 | 34 | 34 | 33 | 34 | ||
| t17184 | 1 (1) | 0 | 0 | 0 | 26 | 23 | 13 | 17 | 25 | 17 | 28 | ||||
| t2801 | 1 (1) | 0 | 0 | 0 | 07 | 23 | 34 | 12 | 12 | 23 | 02 | 12 | 23 | ||
| t17834 | 1 (1) | 0 | 0 | 0 | 07 | 23 | 02 | 12 | 23 | 02 | 02 | 02 | 34 | ||
| t17185 | 1 (1) | 0 | 0 | 0 | 07 | 500 | 22 | 31 | 17 | ||||||
| t409 | 1 (1) | 0 | 0 | 0 | 60 | 61 | 34 | 22 | 34 | 17 | |||||
All six t042 isolates subjected to MLST typing belonged to a new ST, ST4550 which is a double locus variant of ST97. The other isolates subjected to MLST were ST1 (t273), ST22 (t17184), ST88 (t488) and ST848 (t409). All the data in this paper are available in Additional file 1.
Antimicrobial resistance
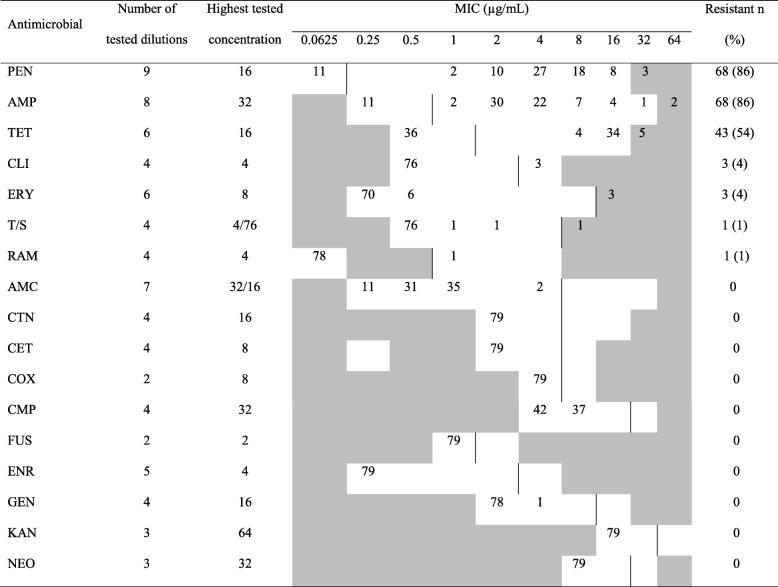
All 79 isolates were susceptible for cefoxitin and were therefore classified as methicillin-susceptible S. aureus. Resistance to penicillin/ampicillin (86%) and tetracycline (54%) was high, while the resistance to other antimicrobials was limited. The MIC values and numbers of isolates resistant to the antimicrobials included in the testing are presented in Table 3. Associations between spa-type and AMR patterns were not found. Percentages resistance to antimicrobials by spa-type for 79 S. aureus isolates is presented in Additional file 2.
Table 3.
Minimum inhibitory concentrations (MIC) for 79 S. aureus isolates from dairy farms in North-Western Ethiopia tested for susceptibility to 17 antimicrobials1. The dilution ranges tested are those contained in the white area. Values above this range indicate MICs higher than the highest concentration within the tested range. Vertical lines indicate clinical breakpoints, with values to the left of the line being susceptible or intermediate and those to the right being resistant
1PEN penicillin, AMP ampicillin, TET tetracycline, CLI clindamycin, ERY erythromycin, T/S Trimethoprim sulfamethoxazole, RAM rifampicin, AMC amoxicillin/clavulanic acid, CTN cephalothin (1st generation cephalosporin), CET ceftiofur (3rd generation cephalosporin), COX cefoxitin, CMP chloramphenicol, FUS fusidic acid, ENR enrofloxacin, GEN gentamicin, KAN kanamycin, NEO neomycin
Discussion
The aim of this study was to characterize S. aureus isolates cultured from milk samples of dairy farms in North-Western Ethiopia based on spa typing, virulence genes identification and AMR. The spa types t042 and the related t15786 were dominant, together representing 69% of the isolates. Such a high degree of relatedness suggests contagious transmission [13, 14]. Since our isolates were derived from different farms, contagious transmission is likely to occur not only within, but also between farms, possibly through cattle movements between farms, or from the government breeding and heifer distribution centers. Four of the spa types (t042, t223, t306 and t355) identified in the current study were previously isolated by Tarekgne et al. [9] in cattle in the Tigray region, Northern Ethiopia. Interestingly, in that study the dominant spa type was t314, which has a repeat sequence entirely different from the dominant types in our study. This suggests that, although contagious transmission seems to occur at a local scale, exchange of strains between regions seems to occur less, resulting in dominance of different clones in different regions. The Tigray region has a different cross-breeding and heifer distribution center, possibly explaining different dominant clones between regions. The six t042 isolates subjected to MLST belonged to the new ST4550, which is a double locus variant of ST97, a ST commonly found in bovine mastitis worldwide [30–32].
The proportion of isolates positive for lukM-lukF’ genes was low. As the majority of isolates in our study belonged to a ST related to ST97, this low lukM-lukF’ prevalence is in line with previous reports that found ST97 isolates to be negative for lukM-lukF’ [16, 17]. The isolates that were positive for lukM-lukF’ belonged to t14061 and t10018, the latter of which has previously been found in nasal swabs from Nigerian abattoir workers [33]. The bi-component leukocidin LukM-LukF’ is predominantly produced by strains isolated from bovine S. aureus mastitis [34] and specifically kills bovine neutrophils, whereas human neutrophils are not affected [18]. Based on an experimental study, Vrieling et al. [19] reported that LukM-LukF’ is produced during the course of infection and that a high levels in milk were associated with increased severity of mastitis. The low proportion of lukM-lukF’ positive isolates in our samples may be partly due to the fact that they came from a cross-sectional study that included only two cases of clinical mastitis [35]. In our collection the proportion of isolates positive for pvl was higher than that of lukM-lukF’, despite the fact that bovine neutrophils are insensitive to this human-associated leukocidin [21]. Although there are reports of pvl-positive cattle S. aureus [16, 35], pvl is mainly detected in S. aureus of human origin [19, 36, 37] whereas cattle isolates are often negative for pvl [38–40]. The finding of pvl in isolates cultured from cows may suggest human to cow transmission of S. aureus. Schmidt et al. [41] reported transmission of a human ST8 isolate to a dairy cow which subsequently caused mastitis and speculated that the close contact between dairy cows and humans, especially in the milking parlor, enables such transmission. Under the Ethiopian circumstances, where cows are hand milked, such transmission seems to be even more likely. Further characterization of the pvl-positive S. aureus in this study and of S. aureus isolates cultured from farmers and their families may help quantifying the importance of human to cattle transmission of S. aureus. Likewise, similar studies on bovine isolates that may be found in humans may also show sharing of S. aureus isolates between cattle and their farmers, which likely occurs along the same transmission routes. The tst gene encodes the toxic shock syndrome toxin, which S. aureus uses to facilitate colonization [42]. Although the tst gene is documented to be more prevalent in bovine than in human S. aureus strains [43], we only found it in a low number of isolates.
In line with the report of Tigabu et al. [44] from the Central highlands of Ethiopia, we did not find mecA positive S. aureus in our sample. The majority of our isolates were, however, resistant to penicillin, ampicillin and tetracycline. Resistance to penicillin and ampicillin is common among mastitis causing S. aureus, but varies largely between geographical regions [45–47]. In Ethiopia, penicillin and ampicillin are commonly used antimicrobials to treat mastitis, and tetracycline is often used to treat other bacterial infections. The high level of resistance to these antimicrobials is in line with previous Ethiopian reports on isolates from bovine mastitis [6, 7, 48]. This consistent picture of widespread AMR in S. aureus is alarming and likely results from the high usage of antimicrobials, possibly due to the fact that farmers can obtain antimicrobials over the counter without a prescription. Additionally, farmers are often not instructed on a proper treatment regimen, because guidance by veterinarians is lacking. A final reason for the high level of AMR may be that Ethiopian farmers are not inclined to cull animals, but rather continue treating their cows over and over again for an extended period of time. Treatment of cows with mastitis during lactation is an important component of a standard mastitis control program for contagious pathogens. Because AMR to penicillin and ampicillin is common, amoxicillin-clavulanic acid may be used for treatment of IMI without the need for tetracyclines, macrolides, lincosamides or third-generation cephalosporins. However, antimicrobial treatment is less than desirably effective in eliminating existing S. aureus infections [49] and treatment of intramammary infections with penicillin-resistant S. aureus strains generally results in a lower cure rate for treatment with either β-lactam or non-β-lactam antibiotics [50]. If cows do not respond favorably to treatment, separation or culling of the infected animals can be used to limit further transmission, but in Ethiopia, dairy cows are not easily culled because cross breed cows are very valuable for reproduction, even if they are chronically infected and have repeated CM cases. Therefore, encouraging dairy farmers to prevent transmission of S. aureus, rather than to rely on antimicrobial therapy is a preferable approach for mastitis control in North-Western Ethiopia. For instance, milking hygiene can be improved [8] and post milking teat disinfection [51] is not very expensive, is easy to perform, and is known to be very effective and can be practiced by Ethiopian dairy farmers. Implementation of these and other control measures will likely result in a lower prevalence of mastitis.
Conclusions
We showed a high degree of similarity between the bovine S. aureus isolates in North-Western Ethiopia, suggesting contagious transmission within and between farms. Moreover, there was a high level of resistance to antimicrobials that are commonly used to treat mastitis in Ethiopia. Therefore, implementing effective preventive measures aiming to limit transmission, rather than to use antimicrobials to control mastitis, along with guidelines for prudent use of antimicrobials, is important to decrease AMR.
Additional files
Dataset supporting the conclusions of this article. (XLS 79 kb)
Percentage resistance to antimicrobials by spa-type for 79 S. aureus isolates. (DOCX 21 kb)
Acknowledgements
We acknowledge the dairy farmers who participated in the study. We are thankful to the support of Merijn van den Hout (University of Applied Sciences, Den Bosch) and Lindert Benedictus (Division of Infection and Immunity, The Roslin Institute, The University of Edinburgh, Scotland, UK), for assistance in the lab work. Ato Teklu and Sisay, from Gondar artificial insemination service, are acknowledged for helping selecting participants.
Funding
This study was supported by the Netherlands organization for international cooperation in higher education (Nuffic) and the Gustav Rosenberger Memorial Fund.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article and its additional files.
Abbreviations
- AMC
Amoxicillin/clavulanic acid
- AMP
Ampicillin
- AMR
Antimicrobial resistance
- CET
Ceftiofur
- CLI
Clindamycin
- CLSI
Clinical and laboratory standards institute
- CM
Clinical mastitis
- CMP
Chloramphenicol
- CMT
California mastitis test
- COX
Cefoxitin
- CTN
Cephalothin
- ENR
Enrofloxacin
- ERY
Erythromycin
- FUS
Fusidic acid
- GEN
Gentamicin
- IMI
Intramammary infections
- KAN
Kanamycin
- MIC
Minimum inhibitory concentration
- MST
Minimum spanning tree
- NEO
Neomycin
- PEN
Penicillin
- RAM
Rifampicin
- ST
Sequence type
- T/S
Trimethoprim sulfamethoxazole
- TET
Tetracycline
Authors’ contributions
SAM, TST, GK and TJGML conceived and designed the study. SAM, JH, EMB, AER and MPS performed laboratory work. GK, TJGML, EMB, JH, VPMGR and SAM interpreted the results. SAM and GK: wrote the manuscript. All authors reviewed, edited and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cremonesi P, Pozzi F, Raschetti M, Bignoli G, Capra E, Graber HU, Vezzoli F, Piccinini R, Bertasi B, Biffani S, Castiglioni B, Luini M. Genomic characteristics of Staphylococcus aureus strains associated with high within-herd prevalence of intramammary infections in dairy cows. J Dairy Sci. 2015;98:6828–6838. doi: 10.3168/jds.2014-9074. [DOI] [PubMed] [Google Scholar]
- 2.Boss R, Cosandey A, Luini M, Artursson K, Bardiau M, Breitenwieser F, Hehenberger E, Lam T, Mansfeld M, Michel A, Mösslacher G, Naskova J, Nelson S, Podpečan O, Raemy A, Ryan E, Salat O, Zangerl P, Steiner A, Graber HU. Bovine Staphylococcus aureus: subtyping, evolution, and zoonotic transfer. J Dairy Sci. 2016;99:515–528. doi: 10.3168/jds.2015-9589. [DOI] [PubMed] [Google Scholar]
- 3.Nübel U, Strommenger B, Layer F, Witte W. From types to trees: reconstructing the spatial spread of Staphylococcus aureus based on DNA variation. Int J Medical Microbiol. 2011;301:614–618. doi: 10.1016/j.ijmm.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Zadoks RN, Middleton JR, McDougall S, Katholm J, Schukken YH. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J Mammary Gland Biol Neoplasia. 2011;16:357–372. doi: 10.1007/s10911-011-9236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sangvik M, Olsen RS, Olsen K, Simonsen GS, Furberg A, Ericson Sollid JU. Age- and gender-associated Staphylococcus aureus spa types found among nasal carriers in a general population: the Tromsø staph and skin study. J Cli Microbiol. 2011;49:4213–4218. doi: 10.1128/JCM.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Getahun K, Kelay B, Bekana M, Lobago F. Bovine mastitis and antibiotic resistance patterns in Selalle smallholder dairy farms, Central Ethiopia. Trop Anim Health Prod. 2008;40:261–268. doi: 10.1007/s11250-007-9090-5. [DOI] [PubMed] [Google Scholar]
- 7.Haftu R, Taddele H, Gugsa G, Kalayou S. Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large-scale dairy farms of northern Ethiopia. Trop Anim Health Prod. 2012;44:1765–1771. doi: 10.1007/s11250-012-0135-z. [DOI] [PubMed] [Google Scholar]
- 8.Mekonnen SA, Koop G, Melkie ST, Getahun CD, Hogeveen H, Lam TJGM. Prevalence of subclinical mastitis and associated risk factors at cow and herd level in dairy farms in north-West Ethiopia. Prev Vet Med. 2017;145:23–31. doi: 10.1016/j.prevetmed.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Tarekgne EK, Skjerdal T, Skeie S, Rudi K, Porcellato D, Félix B, Narvhus JA. Enterotoxin gene profile and molecular characterization of Staphylococcus aureus isolates from bovine bulk milk and milk products of Tigray region. Northern Ethiopia J Food Protection. 2016;79:1387–1395. doi: 10.4315/0362-028X.JFP-16-003. [DOI] [PubMed] [Google Scholar]
- 10.Joo YS, Fox LK, Davis WC, Bohach GA, Park YH. Staphylococcus aureus associated with mammary glands of cows: genotyping to distinguish different strains among herds. Vet Microbiol. 2001;80:131–138. doi: 10.1016/S0378-1135(00)00381-3. [DOI] [PubMed] [Google Scholar]
- 11.Middleton J, Fox L, Gay J, Tyler J, Besser T. Use of pulsed-field gel electrophoresis for detecting differences in Staphylococcus aureus strain populations between dairy herds with different cattle importation practices. Epidemiol Infect. 2002;129:387–395. doi: 10.1017/S095026880200746X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keefe G. Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet. Clin. North am. Food Anim. Pract. 2012;28:203–216. doi: 10.1016/j.cvfa.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Sommerhäuser J, Kloppert B, Wolter W, Zschöck M, Sobiraj A, Failing K. The epidemiology of Staphylococcus aureus infections from subclinical mastitis in dairy cows during a control programme. Vet Microbiol. 2003;96:91–102. doi: 10.1016/S0378-1135(03)00204-9. [DOI] [PubMed] [Google Scholar]
- 14.Gurjar A, Gioia G, Schukken Y, Welcome F, Zadoks R, Moroni P. Molecular diagnostics applied to mastitis problems on dairy farms. Vet Clin Food Anim. 2012;28:565–576. doi: 10.1016/j.cvfa.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Lam TJGM, Lipman LJA, Schukken YH, Gaastra W, Brand A. Epidemiological characteristics of bovine clinical mastitis caused by Staphylococcus aureus and Escherichia coli studied by DNA-fingerprinting. Am J Vet Res. 1997;57:39–42. [PubMed] [Google Scholar]
- 16.Hata E, Katsuda K, Kobayashi H, Uchida I, Tanaka K, Eguchi M. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Cli. Microbiol. 2010;48:2130–2139. doi: 10.1128/JCM.01940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlotter K, Ehricht R, Hotzel H, Monecke S, Pfeffer M, Donat K. Leukocidin genes lukF-P83 and lukM are associated with Staphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia. Germany Vet Res. 2012;43:42. doi: 10.1186/1297-9716-43-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrieling M, Koymans KJ, Heesterbeek DAC, Aerts PC, Rutten VPMG, de Haas CJC, van Kessel KPM, Koets AP, Nijland R, van Strijp JAG. Bovine Staphylococcus aureus secretes the leukocidin LukMF’ to kill migrating neutrophils through CCR1. mBio. 2015;6:3. doi: 10.1128/mBio.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vrieling M, Boerhout EM, van Wigcheren GF, Koymans KJ, Mols-Vorstermans TG, de Haas CJC, Aerts PC, Daemen IJJM, van Kessel KPM, Koets AP, Rutten VPMG, Nuijten PJM, van Strijp JAG, Benedictus L. LukMF’ is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci Rep. 2016;6:37759. doi: 10.1038/srep37759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoekstra J, Rutten V, Sommeling L, Werven T, Spaninks M, Duim B, Benedictus L, Koop G. High production of LukMF’ in Staphylococcus aureus field strains is associated with clinical bovine mastitis. Toxins. 2018;10:200. doi: 10.3390/toxins10050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koop G, Vrieling M, DML S, LSC L, Monie T, van Wigcheren G, Raisen C, Ba X, Gleadall N, Hadjirin N, Timmerman AJ, Wagenaar JA, Klunder HM, Ross Fitzgerald J, Zadoks R, Paterson GK, Torres C, Waller AS, Loeffler A, Loncaric I, Hoet AE, Bergström K, de Martino L, Pomba C, de Lencastre H, Ben Slama K, Gharsa H, Richardson EJ, Chilvers ER, de Haas C, van Kessel K, van Strijp JAG, Harrison EM, Holmes MA. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci. Rep. 2017;7:40660. doi: 10.1038/srep40660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemma MT, Zenebe Y, Tulu B, Mekonnen D, Mekonnen Z. Methicillin resistant Staphylococcus aureus among HIV infected pediatric patients in Northwest Ethiopia: carriage rates and antibiotic co-resistance profiles. PLoS One. 2015;10:9. doi: 10.1371/journal.pone.0137254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. Infect Diseases. 2016;16:689. doi: 10.1186/s12879-016-2014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dilnessa T, Bitew A. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus isolated from clinical samples at Yekatit 12 hospital medical college, Addis Ababa. Ethiopia BMC Infect Diseases. 2016;16:398. doi: 10.1186/s12879-016-1742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moges F, Endris M, Mulu A, Tessema B, Belyhun Y, Shiferaw Y, Huruy K, Unakal C, Kassu A. The growing challenges of antibacterial drug resistance in Ethiopia. J Global Antimicrob Resistance. 2014;2:148–154. doi: 10.1016/j.jgar.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Seyoum ET, Mekonene TK, Woldetsadik DA, Zewudie BM, Gebreyes WA. Enterotoxin gene profile of Staphylococcus aureus isolates recovered from bovine milk produced in Central Ethiopia. J Infect Dev Ctries. 2016;10:138–142. doi: 10.3855/jidc.6797. [DOI] [PubMed] [Google Scholar]
- 27.Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Cli. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CLSI, editor. Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute: Wayne, PA; 2015. [Google Scholar]
- 29.CLSI, editor. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 3rd ed. CLSI supplement VET01S. Clinical and Laboratory Standards Institute: Wayne, PA; 2017. [Google Scholar]
- 30.Smith EM, Green LE, Medley GF, Bird HE, Fox LK, Schukken YH, Kruze JV, Bradley AJ, Zadoks RN, Dowson CG. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Cli. Microbiol. 2005;43:4737–4743. doi: 10.1128/JCM.43.9.4737-4743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delgado S, García P, Fernández L, Jiméenez E, Mercedes Rodríguez-Baños M, del Campo R, Rodríguez JM. Characterization of Staphylococcus aureus strains involved in human and bovine mastitis. FEMS Immunol Med Microbiol. 2011;62:225–235. doi: 10.1111/j.1574-695X.2011.00806.x. [DOI] [PubMed] [Google Scholar]
- 32.Rabello RF, Moreira BM, Lopes RMM, Teixeira LM, Riley LW, Castro ACD. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J Medical Microbiol. 2007;56:1505–1511. doi: 10.1099/jmm.0.47357-0. [DOI] [PubMed] [Google Scholar]
- 33.Odetokun IA, Ballhausen B, Adetunji VO, Ghali-Mohammed I, Adelowo MT, Adetunji SA, Fetsch A. Staphylococcus aureus in two municipal abattoirs in Nigeria: risk perception, spread and public health implications. Vet Microbiol. 2018;216:52–59. doi: 10.1016/j.vetmic.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 34.Barrio MB, Rainard P, Prévost G. LukM/LukF0-PV is the most active Staphylococcus aureus leukotoxin on bovine neutrophils. Microbes and Infect. 2006;8:2068–2074. doi: 10.1016/j.micinf.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Haveri M, Roslöf A, Pyörälä S. Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J Appl Microbiol. 2007;103:993–1000. doi: 10.1111/j.1365-2672.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 36.Zecconi A, Cesaris L, Liandris E, Daprà V, Piccinini R. Role of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary gland. Microb Pathog. 2006;40:177–183. doi: 10.1016/j.micpath.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clinical Microbiol Reviews. 2000;13:16–34. doi: 10.1128/CMR.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rainard P, Corrales J, Barrio MB, Cochard T, Poutrel B. Leucotoxic activities of Staphylococcus aureus strains isolated from cows, ewes, and goats with mastitis: importance of LukM/LukF′-PV leukotoxin. Cli and Diagnostic Lab Immun. 2003;10:272–277. doi: 10.1128/CDLI.10.2.272-277.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada T, Tochimaru N, Nakasuji S, Hata E, Kobayashi H, Eguchi M, Kaneko J, Kamio Y, Kaidoh T, Takeuchi S. Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM-lukF-PV genes by bacteriophages in bovine isolates. Vet Microbiol. 2005;110:97–103. doi: 10.1016/j.vetmic.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Monecke S, Kuhnert P, Hotzel H, Slickers P, Ehricht R. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet Microbiol. 2007;125:128–140. doi: 10.1016/j.vetmic.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt T, Kock MM, Ehlers MM. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in south African dairy herds: genetic diversity and inter-species host transmission. Front Microbiol. 2017;8:511. doi: 10.3389/fmicb.2017.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. Identification and characterization of a new staphylococcal enterotoxin- related putative toxin encoded by two kinds of plasmids. Infect Immun. 2003;71:6088–6094. doi: 10.1128/IAI.71.10.6088-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Leeuwen W, Melles DC, Alaidan A, Al-Ahdal M, Boelens HAM, Snijders SV, Wertheim H, van Duijkeren E, Peeters JK, van der Spek PJ, Gorkink R, Simons G, Verbrugh HA, van Belkum A. Host- and tissue-specific pathogenic traits of Staphylococcus aureus. J Bacteriol. 2005;187:4584–4591. doi: 10.1128/JB.187.13.4584-4591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tigabu E, Kassa T, Asrat D, Alemayehu H, Sinmegn T, Adkins PRF, Gebreyes W. Phenotypic and genotypic characterization of Staphylococcus aureus isolates recovered from bovine milk in central highlands of Ethiopia. African J Microbiol Res. 2015;9:2209–2217. doi: 10.5897/AJMR2015.7562. [DOI] [Google Scholar]
- 45.Hendriksen RS, Mevius DJ, Schroeter A, Teale C, Meunier D, Butaye P, Franco A, Utinane A, Amado A, Moreno M, Greko C, Stärk K, Berghold C, Myllyniemi AL, Wasyl D, Sunde M, Aarestrup FM. Prevalence of antimicrobial resistance among bacterial pathogens isolated from cattle in different European countries: 2002-2004. Acta Vet Scand. 2008;8:50–28. doi: 10.1186/1751-0147-50-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jagielski T, Puacz E, Lisowski A, Siedlecki P, Dudziak W, Międzobrodzki J, Krukowski H. Short communication: antimicrobial susceptibility profiling and genotyping of Staphylococcus aureus isolates from bovine mastitis in Poland. J Dairy Sci. 2014;97:6122–6128. doi: 10.3168/jds.2014-8321. [DOI] [PubMed] [Google Scholar]
- 47.Wang D, Zhang L, Zhou X, He Y, Yong C, Shen M, Szenci O, Han B. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia. China J Dairy Sci. 2016;99:9560–9569. doi: 10.3168/jds.2016-11625. [DOI] [PubMed] [Google Scholar]
- 48.Mekonnen H, Workineh S, Bayleyegn M, Moges A, Tadele K. Antimicrobial susceptibility profiles of mastitis isolates from cows in three major Ethiopian dairies. Revue Méd Vét. 2005;156:391–394. [Google Scholar]
- 49.Philpot WN. Role of therapy in mastitis control. J Dairy Sci. 1969;52:708–713. doi: 10.3168/jds.S0022-0302(69)86633-6. [DOI] [PubMed] [Google Scholar]
- 50.Barkema HW, Schukken YH, Zadoks RN. Invited review: the role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J Dairy Sci. 2006;89:1877–1895. doi: 10.3168/jds.S0022-0302(06)72256-1. [DOI] [PubMed] [Google Scholar]
- 51.Lam TJ, Van Vliet JH, Schukken YH. Udder disinfection and mastitis in cattle: a literature review. Tijdschr Diergeneeskd. 1995;120:392–399. [PubMed] [Google Scholar]
- 52.Francois P, Pittet D, Bento M, Pepey B, Vaudaux P, Lew D, Schrenzel J. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Cli. Microbiol. 2003;41:254–260. doi: 10.1128/JCM.41.1.254-260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melo DA, Coelho IS, Motta CC, Rojas ACCM, Dubenczuk FC, Coelho SMO, Souza MMS. Impairments of mecA gene detection in bovine Staphylococcus spp. Brazilian J Microbiol. 2014;45:1075–1082. doi: 10.1590/S1517-83822014000300041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Voge U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Cli. Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pajić MJ, Rašić ZB, Velebit BM, Boboš SF, Mihajlović-Ukropina MM, Radinović MZ, Galfi AL, Petković JM, Trojačanec SI. The prevalence of methicillin resistance and Panton-valentine leukocidin synthesis genes in Staphylococcus aureus isolates of bovine and human origin. Vet Arhiv. 2014;84:205–214. [Google Scholar]
- 56.Cosandey A, Boss R, Luini M, Artursson K, Bardiau M, Breitenwieser F, Hehenberger E, Lam T, Mansfeld M, Michel A, Mösslacher G, Naskova J, Nelson S, Podpečan O, Raemy A, Ryan E, Salat O, Zangerl P, Steiner A, Graber HU. Staphylococcus aureus genotype B and other genotypes isolated from cow milk in european countries. J Dairy Sci. 2016;99:529–540. doi: 10.3168/jds.2015-9587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset supporting the conclusions of this article. (XLS 79 kb)
Percentage resistance to antimicrobials by spa-type for 79 S. aureus isolates. (DOCX 21 kb)
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article and its additional files.