Abstract
Background
Influenza A pandemics cause significant mortality and morbidity. H2N2 viruses have caused a prior pandemic, and are circulating in avian reservoirs. The age-related frequency of current population immunity to H2 viruses was evaluated.
Methods
Hemagglutinin inhibition (HAI) assays against historical human and recent avian influenza A(H2N2) viruses were performed across age groups in Rochester, New York, and Hong Kong, China. The impact of existing cross-reactive HAI immunity on the effective reproduction number was modeled.
Results
One hundred fifty individual sera from Rochester and 295 from Hong Kong were included. Eighty-five percent of patients born in Rochester and Hong Kong before 1968 had HAI titers ≥1:40 against A/Singapore/1/57, and >50% had titers ≥1:40 against A/Berkeley/1/68. The frequency of titers ≥1:40 to avian H2N2 A/mallard/England/727/06 and A/mallard/Netherlands/14/07 in subjects born before 1957 was 62% and 24%, respectively. There were no H2 HAI titers >1:40 in individuals born after 1968. These levels of seroprevalence reduce the initial reproduction number of A/Singapore/1/1957 or A/Berkeley/1/68 by 15%–20%. A basic reproduction number (R0) of the emerging transmissible virus <1.2 predicts a preventable pandemic.
Conclusions
Population immunity to H2 viruses is insufficient to block epidemic spread of H2 virus. An H2N2 pandemic would have lower impact in those born before 1968.
Keywords: H2, influenza, serology, effective reproduction number, pandemic risk assessment
Mathematical modeling was performed, using population immunity against candidate pandemic H2 influenza strains, to predict risk for pandemic infection. Population immunity is insufficient to block epidemic spread of H2 virus, but would have lower impact in those born before 1968.
Influenza pandemics may occur when influenza A viruses of animal origin with a novel hemagglutinin (HA, or H) with or without neuraminidase (NA, or N) subtypes to which the human population has little or no immunity infect humans and transmit efficiently from person to person. There were 3 influenza pandemics in the 20th century. In 1918, an influenza A virus of the H1N1 subtype emerged and caused widespread disease; subsequently, H1N1 viruses caused seasonal epidemics until 1957. In 1957 a new influenza A virus of the H2N2 subtype, sometimes referred to as the “Asian” influenza, emerged to cause a second pandemic, and subsequently H2N2 viruses replaced H1N1 viruses as the cause of seasonal influenza from 1957 to 1968. In 1968 a third pandemic was caused by an H3N2 virus, which replaced H2N2 viruses and continues to circulate in humans to the present day. Influenza A pandemics are associated with significant mortality, morbidity, and financial burden. For example, the pandemic of 1918–1919 resulted in at least 50 million influenza-related deaths [1], while the pandemics of 1957 (H2N2) and 1968 (H3N2) combined had estimated economic losses around US $32 billion (estimated in 1995 dollars) [2]. Although not designated a pandemic, the reemergence of H1N1 in 1977 also shared some characteristics with the other 3 pandemics. The 2009 H1N1 pandemic was caused by a virus subtype that was then circulating in humans and was unexpected because it was generally assumed that that population immunity would prevent emergence of a pandemic virus of a subtype currently endemic in humans. There continues to be concern regarding the potential for new influenza A viruses to be transmitted to humans, with documented severe zoonotic disease caused by influenza A H5 and H7 infections. However, because H2N2 virus has already caused a pandemic, and H2 subtype viruses are currently circulating in wild and domestic birds [3], reemergence of an H2N2 virus is one of the most likely scenarios for a new pandemic.
Anti-HA antibody, anti-NA antibody, and cell-mediated immunity have all been correlated with protective immunity in both experimental animals and in humans [4–7]. Anti-HA antibody in peripheral blood sera, as assessed by the hemagglutination inhibition (HAI) assay, has a strong correlation with protection against influenza infection and disease. Although complicated by significant interlaboratory variation, an HAI titer of 1:40 is generally accepted as a marker of reduced susceptibility [4]. Therefore, analysis of the population level of HAI antibody can be used to estimate the population susceptibility to infection [8]. For example, the seroprevalence of HAI antibody to pH1N1 in individuals older than 65 years correlated with a significantly decreased influenza-associated mortality for these individuals during 2009 [9]. It has been suggested that exposure to antigenically related H1N1 influenza virus 50–60 years earlier provided older adults with some degree of immunity against the H1N1pdm09 virus [10].
Similarly, individuals who were exposed to H2N2 viruses during the period from 1957 to 1968 may have persistent antibody to these viruses and be relatively protected from an emerging H2N2 virus. However, more than two-thirds of the world population in 2016 was born after 1968 [11], suggesting that there may be substantial susceptibility to these viruses. Because pandemics spread across the world within weeks after emergence [12], much faster than the process of developing and rolling out a vaccine to the newly emerged pandemic virus, which takes >6 months [12], attention has recently focused on developing systematic risk assessment algorithms for animal viruses of possible pandemic threat so that preemptive preparations including the development of vaccine seed strains can be initiated in advance. Examples of these include the Influenza Risk Assessment Tool and the Tool for Influenza Pandemic Risk Assessment [13]. An integral aspect of this risk assessment process is assessment of population immunity to the relevant virus.
In this study, we evaluated population immunity using HAI assays against human and avian H2N2 influenza strains in different age groups in the United States and Hong Kong. We then estimated the impact of existing cross-reactive HAI immunity on reducing the effective reproduction number (R) of a potentially pandemic H2 subtype virus and characterized the minimum basic reproduction number (R0) that such a virus must possess to cause a pandemic.
MATERIALS AND METHODS
Human Sera
Samples in Rochester were collected between 18 January 2010 and 14 March 2014 from nonimmunosuppressed individuals who were healthy or had stable medical conditions and were from 6 months to 80 years of age. In the United States, influenza activity typically peaks in January or February. According to the Centers for Disease Control and Prevention, in 2010–2011 influenza activity peaked in early February and in 2011–2012 it remained low through February and did not peak until mid-March. In 2012–2013, 2013–2014, and 2014–2015, influenza activity peaked in late December with some variability [14].
Sera from children and adults in Hong Kong were collected as part of a previous serological study between 24 August 24 and 19 December 2011, prior to the winter influenza season, which typically commences around February–March in Hong Kong [15]. The preceding influenza season peaked in February–March 2011 and the dominant virus subtype was pandemic H1N1. Two hundred ninety-five serum samples from this study were selected for testing in age strata.
Virus Antigens
Selection of viruses for HAI testing was based on 3 phylogenetic lineages of the H2 influenza virus subtypes: human and avian Eurasian lineages. Viruses were selected from each lineage to represent temporal and geographic diversity. A/Singapore/1/57(H2N2) and A/Berkeley/1/68(H2N2) represented the human lineage, whereas A/mallard/England/727/06(H2N2) and recent H2N2 virus isolate A/mallard/Netherland/14/07(H2N2) were of the Eurasian avian lineage [16]. The A/mallard/England/727/06 virus was generated by plasmid-based reverse genetics with HA and NA of A/mallard/England/727/06 and other internal virus genes of A/Puerto Rico/8/34 origin. Antigenic relatedness of the selected test viruses was determined by reciprocal HAI assays shown in Supplementary Table 1.
Viruses with pandemic potential were handled in a US Department of Agriculture–approved Animal Biosafety Level 3 (ABSL3)–enhanced facility. β-Propiolactone (BPL)–inactivated virus using standard procedures for use as antigen in the HAI test was prepared. The BPL-treated virus preparation was inoculated into 10-day-old hen’s eggs following standard virus culture procedures to confirm complete inactivation of the virus. The antigen was then removed from the ABSL3-enhanced laboratory for HAI analysis. In Rochester and Hong Kong, HAI studies were performed in Biosafety Level 2 conditions.
Serology
HAI tests were performed using turkey red blood cells in Rochester, and chicken red blood cells in Hong Kong, otherwise the 2 laboratories used the same procedure for the test. Sera were pretreated with receptor-destroying enzyme (Denka Seiken Co Ltd) and tested at a starting dilution of 1:10. HAI was performed using “V” bottom microtiter plates as previously described [17]. The positive control sera used were ferret hyperimmune sera against A/Kruitt/63, A/mallard/Netherlands/31/2006, A/Swine/Missouri/2124514/2006, A/mallard/Netherlands/14/2007, and A/Bakker/68 viruses, and goat hyperimmune sera against A/Japan/305/57 and A/Singapore/1/57. Negative controls consisted of antigen alone wells and the reagent control contained phosphate-buffered saline with red blood cells.
Statistical Analysis
Statistical significance was analyzed using GraphPad Prism software (GraphPad, San Diego, California) using 1-way analysis of variance, followed by Bonferroni post hoc analysis. P values <.05 were considered statistically significant.
Reproduction Number Modeling
The reproduction number of each virus in each population was calculated as follows. We partitioned the population into n = 8 age groups (0–10, 11–20, 21–30, 31–40, 41–50, 51–60, 61–70, ≥71) and m = 4 HAI titer levels (<1:20, 1:20, 1:40, ≥1:80). Let be the proportion of population in age group and be the proportion of age group i with the jth HAI titer. The age distribution was based on the most recent census data from the United States (2012) and Hong Kong (2011). To estimate for each age group, we used Bayesian inference with Dirichlet conjugates for multinomial likelihood , where was the number of individuals in age group i in our serologic study and was the number of subjects in age group i with the jth HAI titer level [18]. We assumed noninformative priors, that is, all priors were Dirichlet distributions with parameters for , and hence the joint posterior distributions of were Dirichlet distributions with parameters for . We assumed that an HAI titer of <1:20, 1:20, 1:40, and ≥1:80 reduced susceptibility by 0%, 25%, 50%, and 75%. As such, the proportion of the population that were immune was , where was the reduction in susceptibility conferred by the jth HAI titer level (ie, , , , and ). The basic reproduction number was the largest eigenvalue of the matrix , where was the average number of secondary cases in age group i generated by an infector in age group j when everyone in the population was susceptible [19]. We constructed the matrix using the United Kingdom social contact matrix [20] because analogous data are not available from the United States and Hong Kong. Because the immune proportion of age group i was , the effective reproduction number R was the largest eigenvalue of the matrix and the corresponding relative reduction in transmissibility was simply . The credible intervals for R were generated using 10000 samples randomly drawn from the joint posterior distribution of for each age group i.
RESULTS
A total of 150 individuals from Rochester and 295 individuals from Hong Kong were included in the analysis. The demographics of the study populations at the 2 sites are shown in Table 1. The demographics of the study population matched the demographics of the source population (data not shown).
Table 1.
Demographics of the United States and Hong Kong Study Populations
| Study Population | ||
|---|---|---|
| Characteristic | United States (n = 150) | Hong Kong (n = 295) |
| Median age, y | 27 | 43 |
| Sex | ||
| Male | 60 (40) | 125 (42) |
| Female | 90 (60) | 170 (58) |
| Ethnicity | ||
| White | 130 (87) | 0 |
| Native American | 1 (1) | 0 |
| Black | 14 (9) | 0 |
| Asian | 5 (3) | 295 (100) |
| Other | 0 | 0 |
| Birth year | ||
| 1930–1940 | 6 (4) | 24 (8) |
| 1941–1950 | 11 (7) | 38 (13) |
| 1951–1960 | 20 (13) | 39 (13) |
| 1961–1970 | 18 (12) | 37 (13) |
| 1971–1980 | 11 (7) | 38 (13) |
| 1981–1990 | 32 (21) | 40 (14) |
| 1991–2000 | 29 (19) | 40 (14) |
| 2001 or later | 23 (15) | 39 (13) |
Data are presented as No. (%).
The results of HAI testing of the sera from Rochester and Hong Kong gave very similar results, despite the 2 populations and 2 different laboratories. The geometric mean titers (GMTs) of antibody in the 2 populations against the test viruses by decade of birth are shown in Table 2. As expected, there were substantial levels of anti H2 HAI antibody in the sera of persons old enough to have been infected with H2N2 viruses between 1957 and 1968, and essentially no detectable antibody in persons born after 1968. Among persons born before 1957, the GMT of antibody against the early human A/Singapore/18/57 was significantly higher compared with titers against the later human A/Berkeley/1/68 virus, whereas in persons born from 1961 to 1970 there was a trend toward relatively higher titers against the A/Berkeley/1/68 virus. Titers against the avian H2N2 viruses were lower. There were no significant differences in the GMT against A/Singapore/1/57, A/Berkeley/1/68, or A/mallard/England/727/2006 in sera tested in Hong Kong and Rochester, but sera tested in Rochester had significantly higher titers against A/mallard/Netherlands/14/07 than the sera tested in Hong Kong.
Table 2.
Geometric Mean Titers of Hemagglutination Inhibition Antibodies to H2N2 Viruses Based on Birth Year and Location
| A/Singapore/1/1957 | A/Berkeley/1/1968 | A/Mallard/Eng/727/2006 | A/Mallard/Neth/14/2007 | |||||
|---|---|---|---|---|---|---|---|---|
| Birth Year | Hong Kong | Rochester | Hong Kong | Rochester | Hong Kong | Rochester | Hong Kong | Rochester |
| 1930–1940 | 129 (84–196) | 160 (68–378) | 19 (13–28) | 25 (7–88) | 20 (13–28) | 28 (8–94) | 6 (4–7) | 18 (6–49) |
| 1941–1950 | 320 (249–411) | 300 (189–479) | 35 (26–48) | 26 (13–53) | 42 (30–60) | 58 (40–86) | 13 (9–18) | 29 (22–39) |
| 1951–1960 | 202 (145–280) | 299 (197–453) | 37 (28–49) | 44 (26–75) | 33 23–46 | 41 (28–60) | 10 (7–13) | 25 (18–35) |
| 1961–1970 | 15 (9–24) | 27 (11–66) | 14 (10–20) | 34 (15–77) | 6 (5–8) | 10 (6–17) | 5 (5–6) | 8 (5–14) |
| 1971 or later | 5a | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
Data are presented as geometric mean titer (95% confidence interval).
aSamples in Hong Kong and Rochester were tested at a starting dilution of <1:10 and negative tests are given an imputed value of 5.
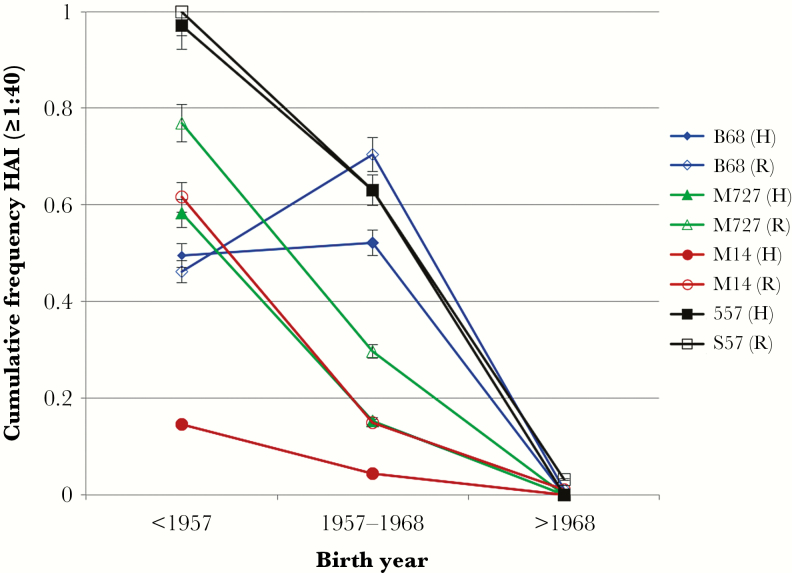
The prevalence of titers ≥1:40 against the test viruses is shown for sera from Rochester and Hong Kong in persons born before the 1957 H2N2 pandemic, during the years that H2N2 circulated (1957–1968), or after 1968 is shown in Figure 1. Ninety-eight percent of individuals from Rochester and Hong Kong born before 1957 had titers ≥1:40 against the A/Singapore/1/57 virus, whereas >63% of subjects born between 1957 and 1968 had titers ≥1:40. In contrast, none of those born after 1968 had titers >1:40 to A/Singapore/1/1957. Generally, persons born prior to 1957 had a lower prevalence of titers ≥1:40 to A/Berkley/1/1968, while the prevalence of titers ≥40 in persons born during circulation of these viruses was similar against A/Singapore/57 and A/Berkley/68. The prevalence of titers ≥1:40 against the 2 avian H2N2 viruses tested were significantly lower in these groups. No person born after 1968 had titer >1:40 against any of the H2 viruses.
Figure 1.
Frequency distribution sera with hemagglutination inhibition titers of ≥1:40 to human and avian H2N2 viruses in Hong Kong and Rochester, New York, against A/Singapore/1/57 (S57), A/Berkeley/1/68 (B68), A/mallard/England/727/2006 (M727), and A/mallard/Netherlands/14/07 (M14) among persons born before the circulation of H2N2 viruses (prior to 1957), born during the H2N2 epidemic period (1958–1968), and after H2N2 viruses circulated in humans (after 1968). Abbreviations: H, Hong Kong; HAI, hemagglutination inhibition; R, Rochester, New York.
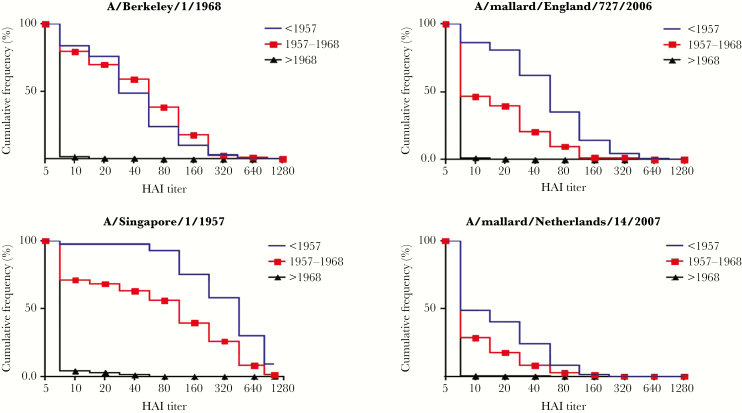
After combining the results from both populations, the cumulative distribution of antibody titers against the 4 test viruses in these 3 age groups is shown in Figure 2. Eighty-five percent of individuals born in both the United States and Hong Kong before 1968 had HAI titers ≥1:40 against A/Singapore/1/57 (Figure 2). More than 50% of subjects had HAI titers ≥1:40 to A/Berkeley/1/68 if born before 1968. The frequency of titers to avian H2N2 viruses was 62%, and 24% of subjects born before 1957 had titers of ≥1:40 to A/mallard/England/727/06 and A/mallard/Netherlands/14/07, respectively, with such titers seen in 21% and 8% of individuals born from 1957 to 1968.
Figure 2.
Reverse cumulative distribution of antibody titers against human and avian H2 viruses by period of birth. Data from Hong Kong and Rochester, New York, were combined. A hemagglutination inhibition titer of <1: 10 was considered a dilution of 5. Abbreviation: HAI, hemagglutination inhibition.
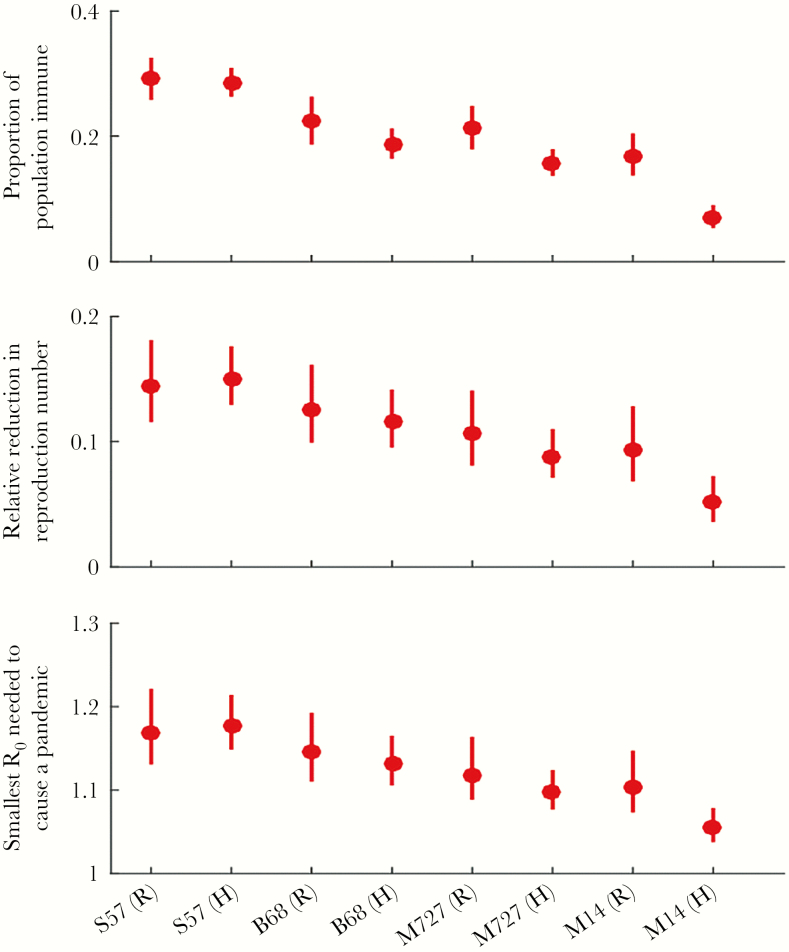
Successful pandemic emergence and spread of a virus depends on the proportion of the population that is immune and the initial R of the potentially pandemic strain. The median R for the 1957 A/H2N2 pandemic was 1.65 (interquartile range [IQR], 1.53–1.70) [21]. To assess the impact of these age-dependent population immunity profiles on the pandemic potential of each of these viruses if they were to emerge in the human population, we computed the impact of population immunity in reducing R (Figure 3). The current population immunity in the United States and Hong Kong would reduce the initial R of A/Singapore/1/1957 by around 15% (12%–18%) and that of A/Berkeley/1/1968 by around 12% (10%–17%). As such, a pandemic of A/Singapore/1/1957 and A/Berkeley/1/1968 would be prevented if initial R of the emerging virus was below 1.18 (1.14–1.22) and 1.14 (1.11–1.20), respectively. The threshold R below which a pandemic with the avian subtype H2 viruses A/mallard/England/727/2006 and A/mallard/Netherlands/14/2007 would be prevented was slightly lower.
Figure 3.
Estimations of overall population-level immunity against H2 viruses and the potential impact of population immunity on reproduction number, using 3 potential titer cutoffs for 100% immunity. Bars represent the 95% confidence intervals of the estimates. Data are shown from S57 A/Singapore/1/57, B68 A/Berkeley/1/68, M727 A/mallard/England/727/06, M14 A/mallard/Netherlands/14/07, Hong Kong, and Rochester, New York. Abbreviations: H, Hong Kong; R, Rochester, New York; R0, basic reproduction number.
DISCUSSION
The comparability of data from 2 geographically separated areas of the world, Rochester and Hong Kong, argues for the representativeness and generalizability of such studies that aim to assess population immunity to viruses of pandemic concern. Our study suggests that those individuals born prior to or during the period of H2N2 virus circulation were more likely to have higher HAI titers against the human H2N2 viruses than those born after 1968 when H2 infection in humans had been displaced by the H3N2 virus. In our study, we also confirmed evidence of cross-reactive HAI antibodies to unrelated avian H2 viruses, albeit at lower prevalence and titer. The prevalence of such cross-reactive antibodies was higher in those born prior 1957 and the GMT of these cross-reactive antibodies was higher in those born prior to 1960 than in the birth cohort of 1961–1970. This is possibly because those who were infected in the early pandemic waves of the H2N2 virus in 1957–1958 were reinfected some years later by antigenically drifted H2N2 viruses, possibly broadening cross-reactive immunity. As reported by others, we also observed low HAI titers <1:40 spanning across the age groups and this could be due to nonspecific inhibition by the sera. However, we checked the sera for nonspecific agglutinins where about 7% of the samples contained nonspecific inhibitors. Since the assays were run on sera which were confirmed not to contain nonspecific inhibitors, these low titers may be due to a certain type of antigen exposure that needs further investigation.
Sera from those born after 1963 had higher HAI titers to A/Berkeley/1/68 than to A/Singapore/1/57 whereas the converse was true in those born before 1963. The older group of individuals who were first infected by A/Singapore/1/1957-like viruses likely had a boost of these titers when they were subsequently infected by later drift variants (ie, A/Berkley/1/1968-like viruses), the phenomenon known as “original antigenic sin” [22]. The broader cross-reactivity may also occur due to targeting different antigenic sites, as a recent study points out that antibody response against H2 is mainly to the receptor binding domain resulting in a greater degree of cross-reactivity whereas for H1 or H3 viruses it is the hypervariable regions of HA resulting in a lesser cross-reactivity [23].
Soon after the H2N2 pandemic of 1957, studies done on sera collected prior to this pandemic were carried out and it was reported that people born prior to 1887 had detectable HAI antibody to H2 viruses. It was therefore suggested that the historical pandemic that was believed to have occurred in 1889–1890 was likely caused by an H2 subtype virus [24]. This prior exposure has also shown to elicit cross-reactive antibody for H2 strains that are currently circulating in animals, potential candidate pandemic stains [25].
Sera tested in Rochester yielded higher titers against A/mallard/ Netherlands/14/07 than the sera tested in Hong Kong. The differences between laboratories are not unexpected given the known laboratory-to-laboratory variation in the HAI test, and what remains remarkable is the closeness of results. One technical difference between the 2 laboratories was the source of erythrocytes for the HAI test; Rochester used turkey erythrocytes whereas Hong Kong used chicken erythrocytes. It is not known if this may contribute to this difference in HAI test results. An alternative reason for the discrepancy in titers between countries may be attributed to variation in vaccination rates. Vaccine uptake in Hong Kong is lower than in the United States. Potentially, repeated vaccination may increase cross-reactive antibody to this avian H2 strain.
Assuming that the H2 seroprevalence in Rochester and Hong Kong reflects global population seroprevalence, we modeled the impact of such immunity on possible emergence of an H2N2 pandemic. This model was based on the candidate pandemic H2 strain originating from escape of the human adapted H2N2 virus or from the avian gene pool that acquires transmissibility in humans. We estimate that the current levels of population immunity would reduce R of A/Singapore/1/1957- or A/Berkeley/1/1968-like viruses by around 15%–20% and the propagation of an epidemic would be prevented if the emerging virus had R0 of <1.2. The median reproduction number R for the 1918 A(H1N1), 1957 A(H2N2), 1968 A(H3N2), and 2009 A(H1N1) pandemics was 1.80, 1.65, 1.80, and 1.46 with IQRs of 1.47–2.27, 1.53–1.70, 1.56–1.85, and 1.30–1.70, respectively [21]. In particular, the smallest R0 estimate for A(H2N2) was 1.39 [21] and we know that the human H2N2 viruses are well adapted to transmission between humans. Thus, one can conclude that the laboratory escape of such a human-adapted virus would now very likely lead to a pandemic. The human population immunity to H2N2 continues to reduce because new birth cohorts born after 1968 continue to be added to the global population. The threshold R0 below which a pandemic with the avian subtype H2 viruses A/mallard/Netherlands/14/2007 and A/WWF/HK/MPN2606/2012 would be prevented is lower, and <1.1. This implies that human population immunity poses only a moderate brake on a human transmissible H2 virus if such a virus were to acquire transmissibility between humans. A recent risk assessment for viruses currently known to be circulating in wild birds has been carried out and the risk of these viruses acquiring transmissibility in humans was assessed to be low; most isolates replicated in human bronchial epithelial cells and ferrets. Several did transmit between ferrets by direct contact, but all HAs retained a preference for avian-like α2,3-linked sialic acid receptors [26]. These viruses still remain primarily in wild birds and have not been established in mammalian species including swine. Our study provides the systematic assessment of the impact of human population immunity that goes toward such an overall assessment of pandemic risk.
A limitation of the study is that other potential contributors to population immunity, such as cross-reactive NA-inhibiting antibody, stalk-specific antibody, cell-mediated immunity, and heterosubtype reactive HAI or neutralizing antibody [4–7, 27], were not assessed. Furthermore, in many parts of the world (especially in developing countries), the proportion of individuals born in 1968 or later might be larger than that in Rochester and Hong Kong. The level of immunity against A(H2N2) in these populations would be lower than that estimated here and the pandemic potential of A(H2N2) in these populations would thus be higher than estimated here.
In summary, we find that levels of population immunity to H2 subtype viruses are not substantial enough to block epidemic spread of an H2 virus that had acquired efficient transmissibility between humans. Furthermore, the existing levels of population immunity to H2 viruses will continue to decline with new birth cohorts being added to the human population. Given that human-adapted H2N2 viruses that arose subsequent to the 1957 pandemic are present in many laboratories worldwide, these findings support the need to have preparedness for H2 viruses as a credible pandemic threat. The approach used in the case study of H2 viruses is more broadly applicable in defining the impact of population immunity to viruses to which there is some level of cross-reactive HAI antibodies—for example, swine H1 and H3 viruses and avian H9N2 viruses [27]. Reliable methods for assessing population immunity are a key to reliable risk assessment of viruses for pandemic threat. It was the lack of such risk assessment of population immunity to the H1N2 triple reassortant swine viruses that led to these viruses not being recognized as potentially pandemic viruses prior to 2009; indeed, it was the triple reassortment swine HA that was the key protective antigen for the 2009 H1N1 pandemic virus.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are grateful to the members of the CEIRS H2N2 Working Group who collected and contributed influenza A H2 subtype viruses to this study.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences (NIGMS) or the National Institutes of Health (NIH).
Financial support. This study was supported by the National Institute of Allergy and Infectious Diseases, NIH (contract number HHSN272201400006C); the Theme-Based Research Scheme (project number T11-705/14N) from the government of the Hong Kong Special Administrative Region; and the NIGMS (award number U54GM088558).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med 2002; 76:105–15. [DOI] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention. Flu season 1999–2000: flu pandemics. https://www.cdc.gov/media/pressrel/r991007f.htm. Accessed 1 December 2016.
- 3. Schafer JR, Kawaoka Y, Bean WJ, Suss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology 1993; 194:781–8. [DOI] [PubMed] [Google Scholar]
- 4. Cox RJ. Correlates of protection to influenza virus: where do we go from here?Hum Vaccin Immunother 2013; 9:405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Monto AS, Petrie JG, Cross RT, et al. . Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis 2015; 212:1191–9. [DOI] [PubMed] [Google Scholar]
- 6. Wilkinson TM, Li CK, Chui CS, et al. . Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 2012; 18:274–80. [DOI] [PubMed] [Google Scholar]
- 7. Sridhar S, Begom S, Bermingham A, et al. . Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 2013; 19:1305–12. [DOI] [PubMed] [Google Scholar]
- 8. Broberg E, Nicoll A, Amato-Gauci A. Seroprevalence to influenza A(H1N1) 2009 virus—where are we?Clin Vaccine Immunol 2011; 18:1205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carter DM, Lu HR, Bloom CE, et al. . Complex patterns of human antisera reactivity to novel 2009 H1N1 and historical H1N1 influenza strains. PLoS One 2012; 7:e39435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hancock K, Veguilla V, Lu X, et al. . Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009; 361:1945–52. [DOI] [PubMed] [Google Scholar]
- 11. Nabel GJ, Wei CJ, Ledgerwood JE. Vaccinate for the next H2N2 pandemic now. Nature 2011; 471:157–8. [DOI] [PubMed] [Google Scholar]
- 12. Wu JT, Ma ES, Lee CK, et al. . The infection attack rate and severity of 2009 pandemic H1N1 influenza in Hong Kong. Clin Infect Dis 2010; 51:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox NJ, Trock SC, Burke SA. Pandemic preparedness and the influenza risk assessment tool (IRAT). Curr Top Microbiol Immunol 2014; 385:119–36. [DOI] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention. Past flu seasons. https://www.cdc.gov/flu/pastseasons/index.htm. Accessed 30 April 2018. [Google Scholar]
- 15. Kwok KO, Riley S, Perera R, et al. . Relative incidence and individual-level severity of seasonal influenza A H3N2 compared with 2009 pandemic H1N1. BMC Infect Dis 2017; 17:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joseph U, Linster M, Suzuki Y, et al. . CEIRS H2N2 Working Group Adaptation of pandemic H2N2 influenza A viruses in humans. J Virol 2015; 89:2442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. World Health Organization. Manual for the laboratory diagnosis and virological surveillance of influenza. Geneva, Switzerland: WHO, 2011. [Google Scholar]
- 18. Lunn D, Jackson C, Best N, Thomas A, Spiegelhalter D.. The BUGS book: a practical introduction to Bayesian analysis. Boca Raton, FL: CRC Press, 2012. [Google Scholar]
- 19. Wu JT, Leung K, Perera RA, et al. . Inferring influenza infection attack rate from seroprevalence data. PLoS Pathog 2014; 10:e1004054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mossong J, Hens N, Jit M, et al. . Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med 2008; 5:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biggerstaff M, Cauchemez S, Reed C, Gambhir M, Finelli L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis 2014; 14:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol 2009; 183:3294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brooke CB, Yewdell JW. Host versus flu: antibodies win a round?Nat Struct Mol Biol 2013; 20:245–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mulder J, Masurel N. Pre-epidemic antibody against 1957, strain of asiatic influenza in serum of older people living in the Netherlands. Lancet 1958; 810–14. [DOI] [PubMed] [Google Scholar]
- 25. Masurel N, Mulder J. Studies on the content of antibodies for equine influenza viruses in human sera. Bull World Health Organ 1966; 34:885–93. [PMC free article] [PubMed] [Google Scholar]
- 26. Jones JC, Baranovich T, Marathe BM, et al. . Risk assessment of H2N2 influenza viruses from the avian reservoir. J Virol 2013; JVI.02526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stephenson I, Nicholson KG, Glück R, et al. . Safety and antigenicity of whole virus and subunit influenza A/Hong Kong/1073/99 (H9N2) vaccine in healthy adults: phase I randomised trial. Lancet 2003; 362:1959–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.