Abstract
Renal cell carcinoma (RCC) is the most common malignant tumor of the urinary system, and it has a high frequency of local invasion and distant metastasis. Although multiple advances have been made in the diagnosis and therapy of RCC, the vast majority of patients with metastatic RCC remain incurable. In this study, an aptamer named SW-4 against RCC 786-O cells was identified from a known sequence pool. The identified aptamer exhibited high binding affinity for target cells with dissociation constants in the nanomolar range. Binding analysis revealed that SW-4 only bound to RCC 786-O cells, but not HEK293T cells or human proximal tubular HK-2 cells, indicating that SW-4 has excellent binding selectivity. By sequence optimization, the 26-nt truncated SW-4b demonstrated improved binding affinity, and it was internalized into target cells via caveolae-mediated endocytosis in a temperature-dependent manner. Furthermore, fluorescence imaging confirmed that SW-4b accumulated at tumor sites in 786-O xenograft nude mice models and specifically recognized clinical RCC tissues. Meanwhile, SW-4b inhibited proliferation of 786-O cells by arresting cell cycle progression at the S phase. Taken together, these results indicate that SW-4b is a potential candidate for development into a novel tool for diagnosis and targeted therapy of RCC.
Keywords: renal cell carcinoma, aptamer, binding affinity, internalization, proliferation
Introduction
Kidney cancer is a cancer that originates from kidney tissues. Renal cell carcinoma (RCC) is the most common type of kidney cancer, accounting for 90%–95% of neoplasms arising from the kidney and approximately 3% of all malignancies in adults.1 In recent years, the incidence of RCC has been steadily increasing by 2%–4% each year with a concomitant increase in the mortality rate.2 Currently, surgical resection remains the most effective treatment option for early-stage RCC. Unfortunately, approximately one-third of patients with renal cell carcinoma present with metastatic disease at the time of diagnosis, and a substantial proportion of patients with localized disease will recur and metastasize within 3 years following surgery. Renal cell carcinoma is not sensitive to traditional radiotherapy and chemotherapy, which results in a 5-year survival rate of less than 10% after diagnosis of metastatic disease.3, 4 Therefore, novel approaches for diagnosis, management, and treatment of RCC are urgently needed.
Aptamers are short, single-stranded DNA (ssDNA) or RNA oligonucleotides that can bind to targets by folding into distinct secondary and tertiary structures.5, 6 Much like protein antibodies, aptamers show high specificity and affinity to their targets. Typically, the dissociation constants of aptamer-target complexes are in the nanomolar to picomolar range.7, 8, 9 Meanwhile, aptamers have unique properties that differ from conventional antibodies, including ease of chemical synthesis, high chemical stability, low molecular weight, lack of immunogenicity, rapid tissue penetration, low toxicity, and ease of modification and manipulation.7, 10 Based on these features, aptamers have substantial potential for molecular imaging, targeted drug development, and biomarker discovery in the cancer field.
Aptamers are typically generated from libraries of random ssDNA or RNA sequences through an in vitro iterative selection process known as SELEX (systematic evolution of ligands by exponential enrichment), which involves repetitive rounds of alternating steps of partitioning candidate oligonucleotides and their PCR amplification.11 According to different types of targets, some variants of the SELEX procedure have been developed, such as cell/tissue SELEX,12, 13, 14, 15 protein SELEX,16 and in vivo SELEX.17 However, the traditional SELEX process is usually time consuming and labor intensive. To rapidly obtain target-specific aptamers, non-SELEX-based methods for the selection of aptamers have recently been put into practice. For example, Aptamer AS1411 is a 16-base G-rich DNA oligonucleotide with anticancer activity that operates through binding of nucleolin, and its development was based on the observation that guanosine-rich oligonucleotides (GROs) possess antiproliferative properties against cancer cells.18
In this study, we obtained an ssDNA aptamer named SW-4 from a known sequence pool that specifically bound to RCC 786-O cells with high affinity, but not HEK293T cells or human proximal tubular HK-2 cells. By sequence optimization, the 26-nt truncated SW-4b demonstrated improved binding affinity for 786-O cells. Fluorescence imaging confirmed that SW-4b accumulated at tumor sites in 786-O xenograft nude mice models and showed excellent recognition ability in clinical RCC tissues. Furthermore, SW-4b inhibited the proliferation of 786-O cells by arresting cell cycle progression at the S phase. Taken together, these results indicated that SW-4b is a potential candidate for development into a novel tool for diagnosis and targeted therapy of RCC.
Results and Discussion
Identification of ssDNA Aptamers against RCC Cell Line 786-O
In general, target-specific aptamers are typically generated by an in vitro selection process known as SELEX.12, 13, 19 The SELEX process starts with a chemically synthesized random DNA oligonucleotide library containing 1013–1016 ssDNA molecules. Through iterative rounds of selection and amplification, specific aptamers are enriched and identified by high-throughput sequencing analysis. However, to rapidly and accurately obtain specific aptamers against human renal cell carcinoma, we adopted a strategy to select specific aptamers from a known sequence pool. Based on the predicting secondary structures of ssDNAs with unique stem and loop, we designed and synthesized an aptamer library, termed a swan library, consisting of approximately 50 aptamers with identified sequences but unidentified functional activity (Table S1). Human renal cancer cell line 786-O cells were used as the target, and epithelial HEK293T cells, as well as human proximal tubular epithelial HK-2 cells, were used as negative control cell lines.
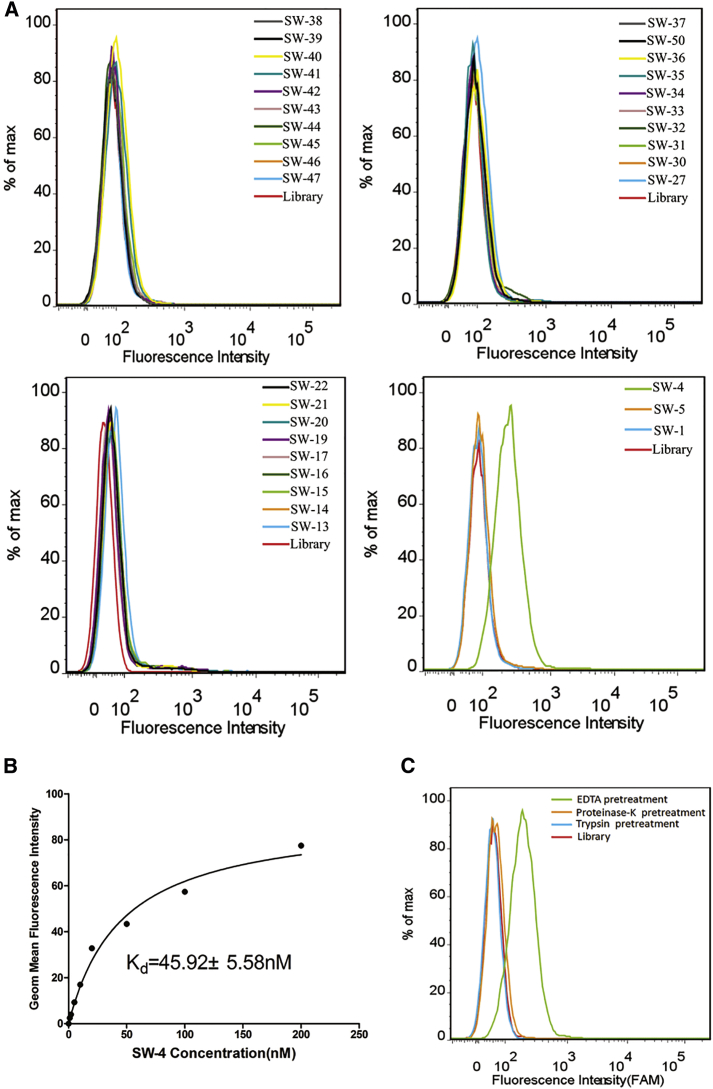
The binding abilities of the aptamers from the swan library to the target cells were tested with flow cytometry. Interestingly, one of them, termed SW-4, showed significant binding to 786-O cells with a strong fluorescence shift compared to the ssDNA library (Figure 1A). To further determine the binding affinity of SW-4 to 786-O, the equilibrium dissociation constant (Kd) was evaluated. 786-O cells were incubated with different concentrations of FAM-labeled aptamers or the initial library at 4°C, and the fluorescence intensity was monitored by flow cytometry. After the geometric mean fluorescence intensity of cells treated with the library was subtracted from that of cells treated with SW-4, the equilibrium dissociation constants (Kd) of SW-4 for 786-O cells were obtained by fitting the dependence of the fluorescence intensity of specific binding on the concentration of the aptamers to the equation Y∖B max X/(Kd + X). As shown in Figure 1B, the Kd of aptamer SW-4 for 786-O cells was approximately 45.92 ± 5.58 nM, indicating that aptamer SW-4 bound with high affinity to the target 786-O cells.
Figure 1.
Selection of ssDNA Aptamers against RCC Cell Line 786-O
(A) The binding abilities of aptamers from the swan library against 786-O target cells were analyzed by flow cytometry. (B) The dissociation constant of SW-4 for 786-O target cells was determined by flow cytometry. (C) After treatment with proteinase K, trypsin, or EDTA, the binding of SW-4 to 786-O was analyzed by flow cytometry.
To investigate what type of target molecules are bound by SW-4, 786-O cells were treated with proteinase K or trypsin for a short time before incubating the aptamer with treated cells. Intriguingly, SW-4 completely lost its ability to bind 786-O cells after enzyme treatment (Figure 1C). This result indicated that the enzymes removed the SW-4-binding target molecule from the cell surface, implying that the binding targets of SW-4 are probably cell surface-bound proteins.
Determination of the Binding Selectivity
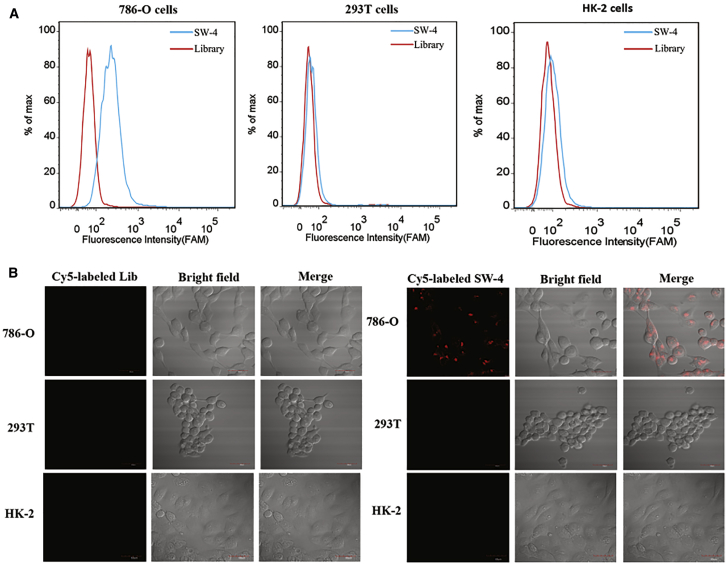
Clinical suitability of an aptamer depends on its ability to distinguish cancer cells from normal cells. Therefore, we further examined the binding selectivity of SW-4. Normal human embryonic renal cell line HEK293T and human renal proximal tubular epithelial HK-2 cells were selected as control cells. FAM-labeled SW-4 was incubated with 786-O, HEK293T, or HK-2 cells. Aptamer SW-4 showed significantly higher fluorescence intensity than that of the unselected library against 786-O, but not against HEK293T or HK-2 cells, implying that SW-4 was specific to human renal cancer cells, especially the 786-O cell line. Selective cell recognition of SW-4 was further confirmed by confocal imaging using Cy5-labeled SW-4. As shown in Figure 2B, intense fluorescence was observed in 786-O cells after incubation with SW-4, with no obvious fluorescence observed from HEK293T and HK-2 cells. Moreover, all of the above cells did not display a significant fluorescence signal when incubated with the control ssDNA library. These results reveal that aptamer SW-4 could selectively recognize 786-O cells.
Figure 2.
Determination of Binding Selectivity
(A) FAM-labeled SW-4 was incubated with 786-O, HEK293T, and HK-2 cells. The binding ability was analyzed by flow cytometry. (B) 786-O, HEK293T, and HK-2 cells were incubated with Cy5-labeled SW-4 and library at 4°C. Fluorescence, bright-field, or merged confocal images are shown.
Sequence Optimization of Aptamer SW-4
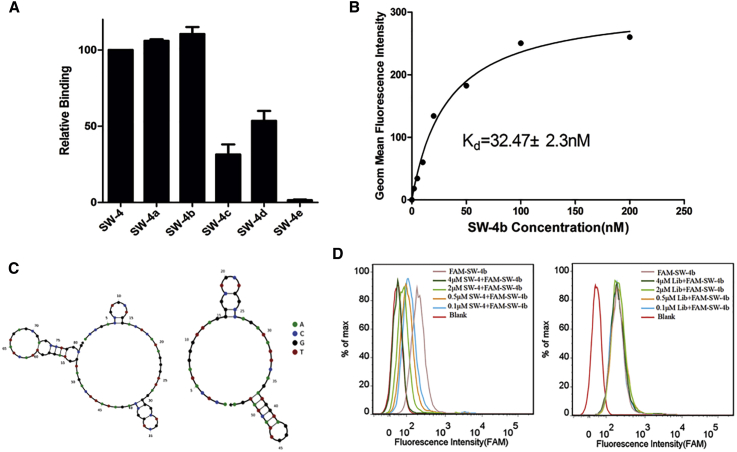
Previous studies have revealed that short aptamers reduce the cost of chemical synthesis, increase tissue penetration, and maintain structural stability.20 Thus, the full-length aptamer SW-4 (80 nt, 1–80) was further optimized by gradually removing nucleotides at the 5′ and 3′ termini. Five truncated sequences, SW-4a, SW-4b, SW-4c, SW-4d, and SW-4e, are listed in Table S2. Their binding affinities to 786-O cells were evaluated by flow cytometry. As shown in Figure 3A, SW-4a and SW-4b, but not SW-4c, SW-4d, and SW-4e, retained their binding ability to 786-O cells, and SW-4b exhibited a higher binding affinity with a Kd value of 32.47 ± 2.3 nM compared with full-length SW-4 (Figure 3B). The structure predicted by Nupack indicated that truncated SW-4b had a structure with two hairpins on a big loop, which was different from full-length SW-4 with three hairpins on the loop (Figure 3C). To identify whether truncated SW-4b has the same binding sites as SW-4, a competitive assay was performed. 786-O cells were incubated with increasing amounts of unlabeled SW-4, followed by FAM-labeled SW-4b. Interestingly, the binding ability of SW-4b was gradually abolished, accompanied by an increasing concentration of SW-4. By contrast, an increasing amount of library did not affect the SW-4b binding (Figure 3D), suggesting that the binding sites of truncated SW-4b are identical to those of SW-4.
Figure 3.
Sequence Optimization of SW-4
(A) Binding of different truncated SW-4 sequences to 786-O cells was assayed by flow cytometry. (B) The dissociation constant of SW-4b for 786-O cells was determined by flow cytometry. (C) Secondary structure of full-length SW-4 (left) and truncated SW-4b (right) was predicted by NUPACK. (D) 786-O cells were pre-incubated with the indicated concentration of SW-4 before being incubated with FAM-labeled SW-4b (200 nM). Competition binding was analyzed by flow cytometry.
Internalization of SW-4b into 786-O Cells via Caveolae-Mediated Endocytosis
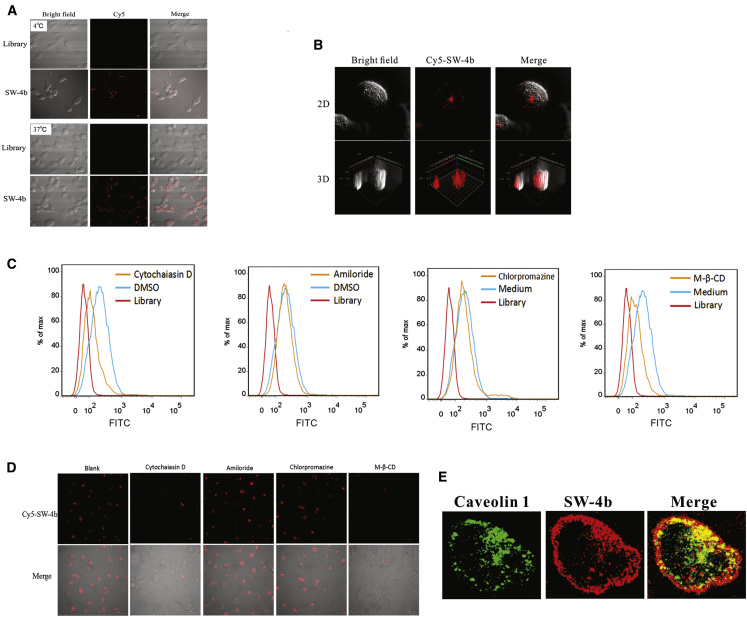
To investigate SW-4b internalization properties, 786-O cells were incubated with Cy5-labeled SW-4b at 4°C and 37°C for 1 hr. Confocal imaging showed that strong fluorescence was observed on the periphery of 786-O cells after incubation at 4°C. However, after incubation at 37°C, a strong fluorescence signal originated from inside cells, which was confirmed by constructing three-dimensional images using Z-scan (Figures 4A and 4B). These results suggest that SW-4b could be rapidly internalized into 786-O cells in a temperature-dependent manner.
Figure 4.
Internalization of SW-4b into 786-O Cells
(A) 786-O cells were incubated with Cy5-labeled SW-4b at 4°C or 37°C. Fluorescence image, bright-field, or merged confocal images are shown. (B) 786-O cells were incubated with Cy5-labeled SW-4b at 37°C. The two-dimensional and three-dimensional images were analyzed by confocal microscopy. (C and D) After 786-O cells were treated with cytochalasin D, amiloride, M-β-CD, or chlorpromazine, binding of SW-4b to 786-O was analyzed by flow cytometry (C) or confocal microscopy (D). (E) The colocalization of the Cy5-SW-4b (red) with caveolin 1 (green) is shown by confocal microscopy.
Endocytosis is the main pathway by which aptamers are internalized into cells. To confirm that the internalization of SW-4b is mediated by endocytosis, 786-O cells were pretreated with an actin polymerization inhibitor, cytochalasin D, at 37°C and assessed for SW-4b uptake by flow cytometry. Compared with untreated cells, cytochalasin D-treated cells showed a decrease in SW-4b uptake, consistent with endocytosis. In general, endocytic pathways can be divided into classic clathrin- and caveolae-mediated endocytosis, clathrin- and caveolae-independent endocytosis, and micropinocytosis.21, 22 To explore the possible endocytic pathway of SW-4b, specific inhibitors for different pathways were used, including methyl-β-cyclodextrin (M-β-CD, caveolae), amiloride (macropinocytosis), and chlorpromazine (clathrin). As shown in Figure 4C, pretreatment of 786-O cells with M-β-CD caused a significant decrease in the fluorescence signal compared with control untreated cells, indicating that the blockage of caveolae function inhibited cellular uptake of SW-4b. By contrast, no significant change in the fluorescence signal was detected after 786-O cells were pretreated with amiloride or chlorpromazine, suggesting that endocytosis might not be mediated by either micropinocytosis or the clathrin pathway. These findings were also confirmed by confocal images (Figure 4D). Furthermore, after 786-O cells were incubated with Cy5-SW-4b, endogenous caveolin 1 protein, a caveolae marker, was labeled with its corresponding antibodies conjugated to DyLight 488. An intense yellow color was observed in the merged image, indicating that SW-4b colocalized with caveolin 1 protein (Figure 4E).
Collectively, we conclude that the internalization of SW-4b mainly occurs through caveolae-mediated endocytosis. It has been reported that internalization via caveolae-mediated endocytosis may avoid the lysosome and, hence, subsequent degradation,23 suggesting that this is a nonacidic and nondigestive internalization pathway. Thus, internalization of SW-4b via caveolae-mediated endocytosis makes this aptamer an ideal candidate for development in aptamer-based drug delivery systems.
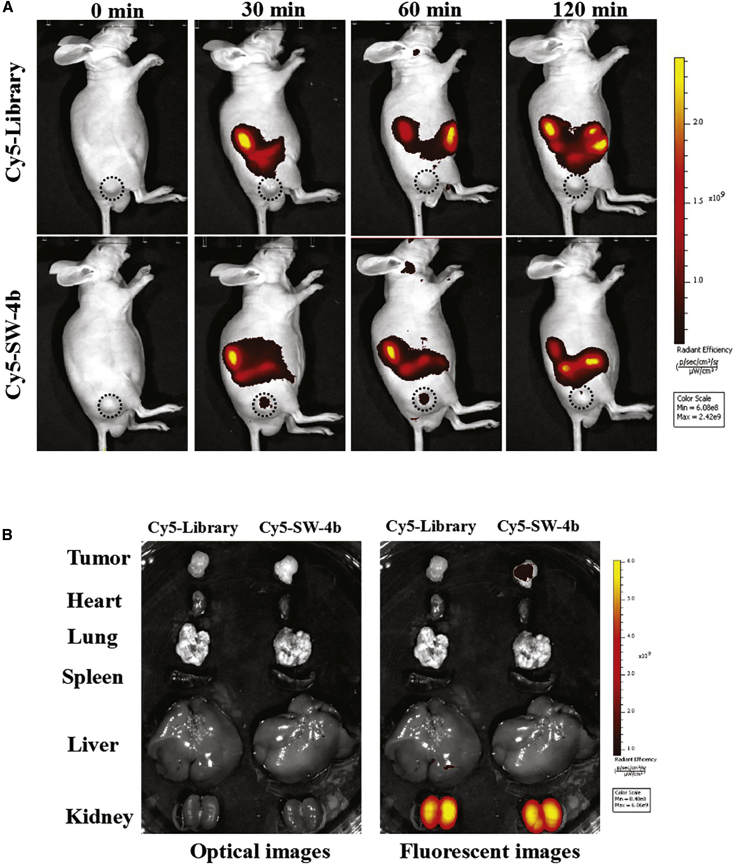
In Vivo and Ex Vivo Fluorescence Imaging
To determine whether aptamer SW-4b retains its recognition ability in vivo, a systematic comparative investigation was performed to validate the selectivity of SW-4b in vivo. Cy5-labeled SW-4b and library were respectively injected into the 786-O xenograft through the tail vein. As expected, the fluorescence signal gradually increased at the tumor site 60 min post-injection, indicating that SW-4b held the ability to rapidly accumulate at the tumor site. After this, the fluorescence signal significantly decreased and almost disappeared at 2 hr post-injection, suggesting that SW-4b can be cleared rapidly by metabolism in vivo (Figure 5A, lower). By contrast, no fluorescence signal was observed at the tumor site during the entire procedure after injection of the Cy5-labeled library into RCC tumor-bearing mice (Figure 5A, upper), revealing that the control library could not specifically target tumor sites in vivo. Furthermore, the biodistribution of Cy5-labeled SW-4b in mice was also examined after 2 hr post-injection. Compared to the control, the mice injected by Cy5-labeled SW-4b exhibited more accumulation in tumor than heart, lung, liver, or spleen. Meanwhile, a strong fluorescence signal was observed in the kidney, which is due to non-specific accumulation of aptamers, because the small size of aptamers is susceptible to kidney filtration (Figure 5B). These results demonstrate that aptamer SW-4b holds the ability to target RCC in vivo.
Figure 5.
In Vivo and Ex Vivo Fluorescence Imaging of SW-4b
(A and B) RCC tumor-bearing mice were intravenously injected with Cy5-labeled SW-4b (lower) or the control ssDNA library (upper). Fluorescence images of live mice were collected by an in vivo imaging system (A), and optical and fluorescent images of organs isolated from RCC tumor-bearing mice are shown (B).
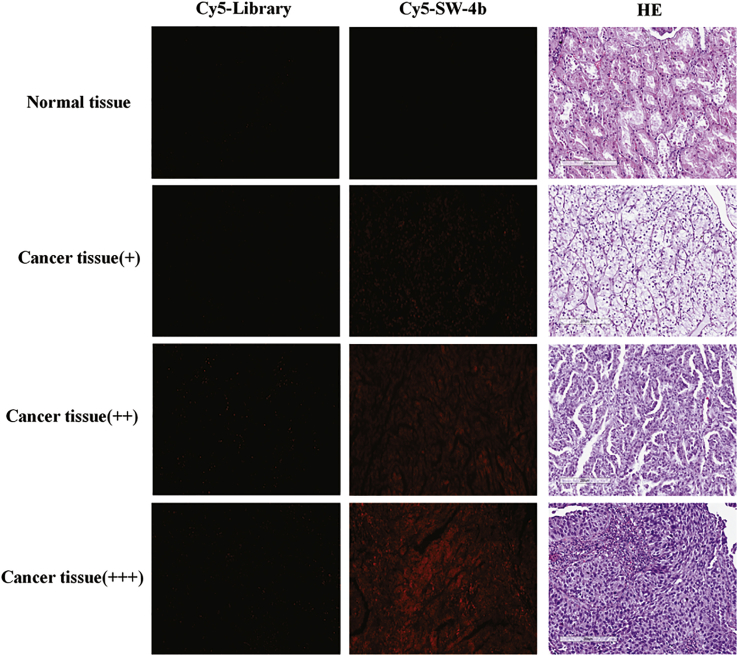
Imaging RCC Clinical Tissues with Aptamer
The above results show that SW-4b has excellent specificity to 786-O cells in vitro and in vivo. Thus, we speculate that SW-4b may also hold the ability to recognize clinical RCC tissues. To test this hypothesis, Cy5-labeled SW-4b was incubated with an RCC tissue microarray containing 110 RCC tissues and 10 normal renal tissues. As shown in Figure 6 and Table 1, 81.8% of the RCC tissues had a positive fluorescence signal. By contrast, only a 30% positive rating was observed among normal renal tissues. These results clearly illustrate that SW-4b had stronger binding and recognition ability for RCC tissues compared to normal renal tissue. Therefore, SW-4b has potential for use in clinical applications for the diagnosis and treatment of RCC.
Figure 6.
Fluorescence Images of Tissue Sections Stained with Cy5-Labeled SW-4b
Cy5-labeled SW-4b or library was incubated with an RCC tissue microarray containing 110 RCC tissues and 10 normal renal tissues. Typical fluorescence images are shown.
Table 1.
Assessment of the Clinical Tissues Stained with Cy5-Labeled SW-4b
| Fluorescence Intensity | Normal Tissues | Cancer Tissues Stage I | Cancer Tissues Stage II | Cancer Tissues Stage III |
|---|---|---|---|---|
| − | 7 | 13 | 5 | 2 |
| + | 2 | 34 | 12 | 2 |
| ++ | 1 | 19 | 5 | 1 |
| +++ | 0 | 13 | 3 | 1 |
−, no fluorescence; +, weak fluorescence intensity; ++, moderate fluorescence intensity; +++, strong fluorescence intensity.
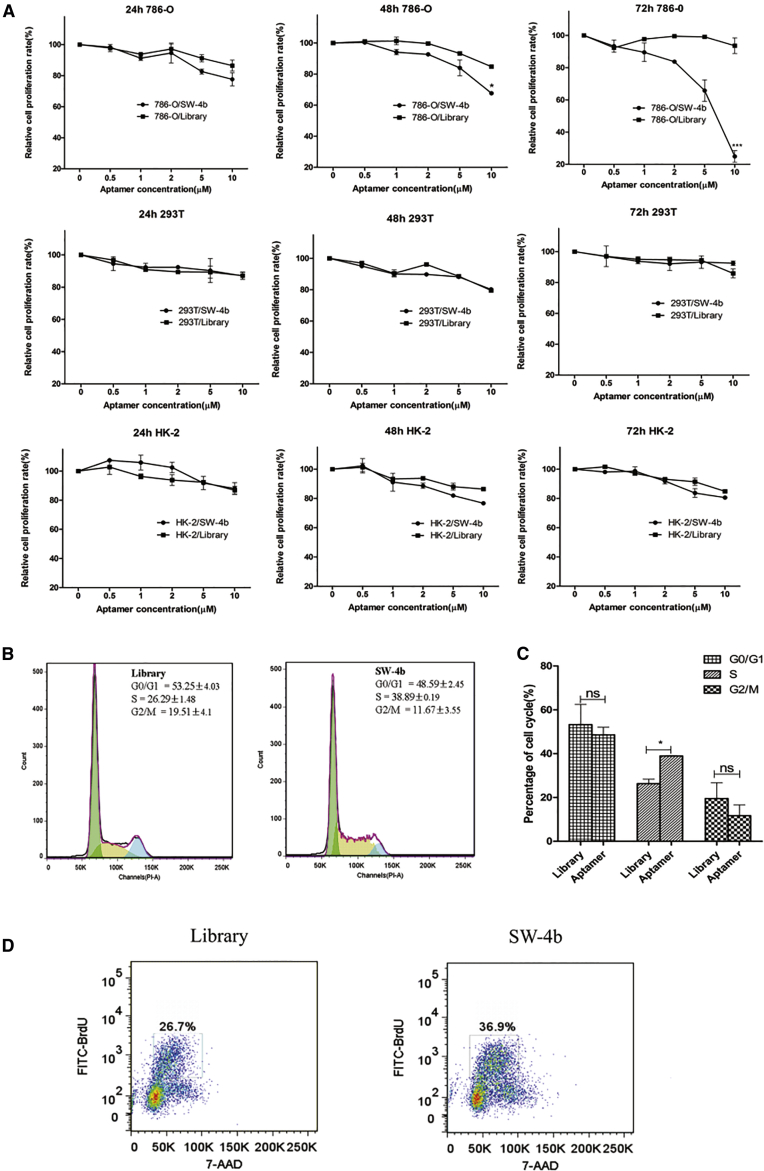
Aptamer SW-4b Caused Growth Inhibition of Target Cells and Induced S Phase Arrest
SW-4b could specifically recognize RCC cells in vitro and in vivo. We therefore investigated whether SW-4b is able to inhibit the growth of RCC cells. To this end, 786-O, HEK293T, and HK-2 cells were exposed to different concentrations of SW-4b for 24, 48, and 72 hr, and cell viability was measured using a Cell Counting Kit-8 (CCK-8) assay. Interestingly, SW-4b inhibited proliferation of 786-O cells in a dose-dependent manner, and maximum inhibitory effect was found at 10 μM for 72 hr (Figure 7A). The concentration of SW-4b needed to inhibit 50% of the proliferation (IC50) of 786-O was 4.7 μM. By contrast, SW-4b did not exhibit a significant inhibitory effect on the proliferation of non-cancerous cells, including HEK293T and HK-2 cells, at different concentrations and different time points (Figure 7A). Taken together, these data indicate that SW-4b can selectively inhibit the proliferation of RCC cells.
Figure 7.
SW-4b Inhibited the Proliferation of 786-O Cells by Inducing S Phase Arrest of Cell Cycle
(A) 786-O, HEK293T, and HK-2 cells were treated with the indicated concentration of SW-4b or the control ssDNA library. Cell proliferation at different time points was evaluated by the CCK-8 assay. (B and C) 786-O cells were treated with 10 μmol/L SW-4b or the control ssDNA library. Cell cycle distribution was analyzed by flow cytometry (B), and the percentage of cell cycle phase is shown (C). (D) 786-O cells treated with library or SW-4b were labeled with BrdU for 60 min before harvesting. BrdU incorporation was analyzed by flow cytometry.
Cell proliferation depends on an orderly cell cycle process. To explore the underlying mechanism by which SW-4b affects proliferation of RCC cells, we analyzed the cell cycle distribution by flow cytometry. After 786-O cells were treated with SW-4b and the library at 10 μM for 72 hr, the cells were harvested and stained with propidium iodide (PI) for flow cytometry analysis. Compared with control cells treated with the library, the percentage of cells in the S phase significantly increased from 26.29% ± 1.48% to 38.89% ± 0.19% (Figures 7B and 7C). These increases were accompanied by concomitant decreases in the percentage of cells in the G0/G1 and G2/M phases. Furthermore, we measured the bromodeoxyuridine (BrdU) incorporation by flow cytometry. Consistent with cell cycle analysis, we found that 786-O cells treated with SW-4b resulted in a significant increase of BrdU incorporation (Figure 7C), suggesting that SW-4b inhibits proliferation of 786-O cells by inducing cell-cycle arrest at the S phase.
Conclusions
In summary, our study successfully identified the aptamer SW-4 that showed high binding affinity and specificity against RCC 786-O cells. By sequence optimization, the 26-nt truncated SW-4b demonstrated improved binding affinity, and it was internalized into target cells via caveolae-mediated endocytosis in a temperature-dependent manner. Fluorescence imaging from in vivo and clinical samples confirmed the excellent recognition ability of SW-4b. Most importantly, SW-4b showed significant antiproliferation activity against 786-O cells, which functions by arresting cell cycle progression at the S phase. Thus, SW-4b is a potential candidate for development into a novel tool for RCC diagnosis and targeted therapy.
Materials and Methods
Cell Lines and Cell Culture
Human clear cell carcinoma of the kidney cell line 786-O, epithelial cell line HEK293T, and human proximal tubular epithelial line HK-2 were obtained from the Chinese Academy of Sciences (CAS) cell bank (Shanghai, China). All cell lines were cultured at 37°C in a humid atmosphere with 5% CO2. 786-O cells and HK-2 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin streptomycin (pen-strep). The 293T cells were cultured in DMEM supplemented with 10% FBS (HyClone) and 100 U/mL pen-strep.
Buffer
Washing buffer was prepared by adding 5 mM Mgcl2 and 4.5 g/L glucose to Dulbecco’s PBS (D-PBS). Binding buffer was prepared by adding 0.1 mg/mL yeast tRNA and 1 mg/L BSA to the washing buffer.
DNA Synthesis and Labeling
DNA used in all experiments was synthesized and labeled with FAM or Cy5 at the 5′ terminus by Sangon Biotech (Shanghai, China).
Flow Cytometric Analysis
Target and negative cells were cultured in a 10-mm dish at 90% confluence, and cells were dissociated by 0.002 EDTA after washing with D-PBS twice. Then, 2 × 105 786-O, HEK293T, or HK-2 cells were incubated with 100 nM FAM-labeled control library or aptamer in 200 μL binding buffer at 4°C for 30 min. Before flow cytometric analysis (BD FACSVerse flow cytometer), each sample was washed two times with washing buffer and then filtered with 400 μL D-PBS. All experiments were repeated three times.
Confocal Microscopy Imaging
Confocal microscopy imaging was employed to monitor the binding of the aptamer to live cells. Then, 1 × 105cells were seeded in a 35-mm glass-bottom dish and cultured for approximately 24 hr. The cells were then incubated with Cy5-labeled aptamer or Cy5-labeled library at 4°C or 37°C for 1 hr after washing twice with washing buffer. After again washing twice with washing buffer, cells were imaged by an FV1000-X81 confocal microscope (Olympus, Japan). The images were analyzed by FV10-ASW version 3.1 software.
Target Type Analysis
786-O cells cultured to 90% confluence were washed twice with D-PBS and then treated with 200 μL 0.25% trypsin or 0.1 mg/mL proteinase K for 3 min, followed by adding complete culture medium to inhibit the proteinase activity. Cells were then washed with washing buffer and the detached cells were collected. The cells were incubated with 100 nM aptamers at 4°C for 30 min. After washing with washing buffer, all cell samples were tested by flow cytometry.
Affinity Analysis
To test the affinity of aptamers SW-4 and SW-4b, 786-O cells (2 × 105) were incubated with different concentrations of aptamers and then analyzed by flow cytometry. The dissociation constants (Kd) were calculated from the cell-binding results, as detected by flow cytometry, by measuring the geometric mean fluorescence intensity of the specific binding by different aptamer concentration. Afterward, the graphs of the results were plotted using GraphPad Prism 5.
In Vivo and Ex Vivo Fluorescence Imaging
Male athymic BALB/c (BALB/c-nude) mice were purchased from Shanghai SLAC Laboratory Animal. They were 5–6 weeks old at the start of each experiment. All animal operations were in accord with institutional animal use and care regulations, approved by the Laboratory Animal Center of Hunan. Nude mice received a subcutaneous injection of 5 × 106 786-O cells into the backside. Tumors were then allowed to grow over a period of 40–45 days until they reached 0.5–1.5 cm in diameter. Once the mice (3 mice/group) were anesthetized and motionless, a 100-μL volume of PBS containing 5.5 nM Cy5-labeled aptamer and library were injected intravenously via the tail vein. At certain time points, fluorescence images of live mice were collected by an IVIS Lumina II in vivo imaging system (Caliper LifeScience, USA). For the ex vivo fluorescence experiment, tumor-bearing mice intravenously injected with Cy5-labeled aptamer or library were sacrificed by cervical dislocation under narcosis after imaging. After anatomization, the dissected organs, including liver, kidney, spleen, lung, heart, and tumor tissue, were imaged with the IVIS Lumina II in vivo imaging system, as described above.
Using Aptamer SW-4b for Human Tissue Staining
A kidney tissue array was preheated at 60°C for 2 hr and then deparaffinized in xylene (15 min, twice). Tissue sections were then immersed in decreasing ethanol concentrations (100%, 95%, 90%, 80%, and 70%) at 5-min intervals. The hydrated tissues were pretreated in 0.01 M citrate buffer at pH 6.0 and heated in a pressure cooker for 20 min. Afterward, tissue sections were blocked with precooled binding buffer, 20% FBS, and 0.1 mg/mL salmon sperm DNA for 1 hr at room temperature, and they were incubated with a 200 nM Cy5-labeled library or aptamer SW-4b for 1 hr. The fluorescence signals were detected using a Panoramic SCAN (3DHISTECH). Meanwhile, the fluorescence intensity of every case was calculated and evaluated as negative (−, <30,000), weak (+, 30,000–60,000), moderate (++, 60,000–100,000), or strong (+++, >100,000).
Analyzing Aptamer Internalization and Investigating the Uptake Mechanism
The 786-O cells were incubated with 200 nM Cy5-labeled aptamer at 4°C or 37°C for 1 hr. Then, cells were washed two times with washing buffer before imaging. To investigate the uptake mechanism, 786-O cells were pretreated with 3 mM amiloride for 1 hr, 10 mM M-β-CD for 30 min, 10 μg/mL chlorpromazine for 30 min, and 5 μM cytochalasin D for 30 min. After incubation with aptamer, the cell samples were analyzed by flow cytometry and confocal microscopy. All experiments were repeated at least three times.
Cell Function Assays
To evaluate cell proliferation, 4,000 cells were seeded in 96-well plates and cultured for approximately 24 hr. Then, the cells were treated daily with SW-4b or the 54-nt library as a control with 0, 0.5, 1, 2, 5, and 10 μM in RPMI 1640 medium containing 10% serum for up to 24, 48, or 72 hr. A CCK-8 (DOJINDO Laboratories, Tabaru, Japan) assay was performed. Briefly, at each set time point, CCK-8 solution was added to each well and incubated for 1 hr. The absorbance at 405 nm was measured with a microplate reader.
For the cell cycle study, cells (5 × 104) were seeded in 6-well plates, cultured for approximately 24 hr, and treated with 10 μM aptamer SW-4b or the 54-nt control library for 72 hr in RPMI 1640 medium containing 10% serum. Cells were then collected and washed with cold PBS and fixed in 70% ethanol at −20°C for 20 hr. Subsequently, the cells were incubated with PI solution (10 μg/mL PI and 0.2 mg/mL RNase A; Beyotime Institute of Biotechnology, Jiangsu, China) for 30 min at 37°C. After washing with cold PBS, the cells were suspended in 500 μL cold PBS, and cell cycle analysis was performed by flow cytometry.
Statistical Analysis
The statistical analysis was performed using GraphPad Prism 7.0. Statistical significance was determined using the t tests; p ≤ 0.05 was considered significant.
Author Contributions
H.Z., M.Y., and W.T. conceived the project and designed the experiments. H.Z., Z.W., L.X., Y.Z., Lin Zhang., Lei Zhang., and J.Li. carried out the main experiments and collected the data. H.Z., M.Y., W.T., J.Liu., W.X., B.P., L.H., T.D., and X.H. analyzed the data. H.Z., M.Y., and W.T. wrote the manuscript.
Conflicts of Interest
The authors have declared that no competing interest exists.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81171950, 81272220, 81672760 and 31701249), the Hunan Provincial Natural Science Foundation of China (2016JJ3048 and 2018JJ3037) and the Hunan Provincial Key Research and Development Plan (2018SK2128).
Footnotes
Supplemental Information includes two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.07.015.
Contributor Information
Mao Ye, Email: goldleaf@hnu.edu.cn.
Xiaoxiao Hu, Email: xxhu@hnu.edu.cn.
Weihong Tan, Email: tan@chem.ufl.edu.
Supplemental Information
References
- 1.Cairns P. Renal cell carcinoma. Cancer Biomark. 2010;9:461–473. doi: 10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Znaor A., Lortet-Tieulent J., Laversanne M., Jemal A., Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur. Urol. 2015;67:519–530. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Bukowski R.M. Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin-2. Cancer. 1997;80:1198–1220. doi: 10.1002/(sici)1097-0142(19971001)80:7<1198::aid-cncr3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 4.Zisman A., Pantuck A.J., Wieder J., Chao D.H., Dorey F., Said J.W., deKernion J.B., Figlin R.A., Belldegrun A.S. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J. Clin. Oncol. 2002;20:4559–4566. doi: 10.1200/JCO.2002.05.111. [DOI] [PubMed] [Google Scholar]
- 5.Ye M., Hu J., Peng M., Liu J., Liu J., Liu H., Zhao X., Tan W. Generating aptamers by cell-SELEX for applications in molecular medicine. Int. J. Mol. Sci. 2012;13:3341–3353. doi: 10.3390/ijms13033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:440. doi: 10.1038/nrd.2017.86. [DOI] [PubMed] [Google Scholar]
- 7.Duan M., Long Y., Yang C., Wu X., Sun Y., Li J., Hu X., Lin W., Han D., Zhao Y. Selection and characterization of DNA aptamer for metastatic prostate cancer recognition and tissue imaging. Oncotarget. 2016;7:36436–36446. doi: 10.18632/oncotarget.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X., Zhao Z., Bai H., Fu T., Yang C., Hu X., Liu Q., Champanhac C., Teng I.T., Ye M., Tan W. DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long Y., Qin Z., Duan M., Li S., Wu X., Lin W., Li J., Zhao Z., Liu J., Xiong D. Screening and identification of DNA aptamers toward Schistosoma japonicum eggs via SELEX. Sci. Rep. 2016;6:24986. doi: 10.1038/srep24986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang X., Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yüce M., Ullah N., Budak H. Trends in aptamer selection methods and applications. Analyst (Lond.) 2015;140:5379–5399. doi: 10.1039/c5an00954e. [DOI] [PubMed] [Google Scholar]
- 12.Li X., Zhang W., Liu L., Zhu Z., Ouyang G., An Y., Zhao C., Yang C.J. In vitro selection of DNA aptamers for metastatic breast cancer cell recognition and tissue imaging. Anal. Chem. 2014;86:6596–6603. doi: 10.1021/ac501205q. [DOI] [PubMed] [Google Scholar]
- 13.Sefah K., Shangguan D., Xiong X., O’Donoghue M.B., Tan W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 14.Yuan B., Jiang X., Chen Y., Guo Q., Wang K., Meng X., Huang Z., Wen X. Metastatic cancer cell and tissue-specific fluorescence imaging using a new DNA aptamer developed by Cell-SELEX. Talanta. 2017;170:56–62. doi: 10.1016/j.talanta.2017.03.094. [DOI] [PubMed] [Google Scholar]
- 15.Mercier M.C., Dontenwill M., Choulier L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers (Basel) 2017;9:E69. doi: 10.3390/cancers9060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y., Zhu Z., An Y., Zhang W., Zhang H., Liu D., Yu C., Duan W., Yang C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- 17.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ireson C.R., Kelland L.R. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Zhang J., Ma Y., Pei X., Liu Q., Lu B., Jin L., Wang J., Liu J. A cell-based single-stranded DNA aptamer specifically targets gastric cancer. Int. J. Biochem. Cell Biol. 2014;46:1–8. [PubMed] [Google Scholar]
- 20.Karlsen K.K., Wengel J. Locked nucleic acid and aptamers. Nucleic Acid Ther. 2012;22:366–370. doi: 10.1089/nat.2012.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 22.Hansen C.G., Nichols B.J. Molecular mechanisms of clathrin-independent endocytosis. J. Cell Sci. 2009;122:1713–1721. doi: 10.1242/jcs.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bengali Z., Rea J.C., Shea L.D. Gene expression and internalization following vector adsorption to immobilized proteins: dependence on protein identity and density. J. Gene Med. 2007;9:668–678. doi: 10.1002/jgm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.