Abstract
Background
Circular RNAs (circRNAs) are widely expressed in mammals and can regulate the development and progression of human tumors. has_circ_0015758 (circ-CFH) is an exon circRNA transcript from the GRCh37/hg19 fragment of chromosome 1 and is homologous to the protein-coding gene complement factor H (CFH). Currently, the function of circ-CFH in glioma remains unclear.
Material/Methods
In our study, circ-CFH, miR-149, and Akt1 mRNA expression levels were analyzed by qRT-PCR assays. To investigate the function of circ-CFH in cell proliferation, circ-CFH knockdown models were established by using circ-CFH siRNAs. Cell proliferation abilities were measured by CCK-8 and colony formation assays and in vivo experiments. In addition, the interaction between circ-CFH and miR-149 was assessed by luciferase reporter assays.
Results
Circ-CFH expression was significantly upregulated in glioma tissue and was correlated with tumor grade. Circ-CFH expression levels were also markedly higher in U251 and U373 glioma cell lines. Circ-CFH knockdown inhibited cell proliferation and colony formation abilities. Luciferase assays indicated that circ-CFH functions as a miR-149 sponge and inhibits its function in U251 and U373 cells. Subsequently, AKT1 was identified as a direct target of the circ-CFH/miR-149 axis.
Conclusions
Circ-CFH promotes glioma progression by sponging miR-149 and regulating the AKT1 signaling pathway. The circ-CFH/miR-149/AKT1 regulation axis may be a potential target for glioma therapy.
MeSH Keywords: Astrocytoma; MicroRNAs; Proto-Oncogene Proteins c-akt; RNA, Small Untranslated
Background
Glioma, the most prevalent and aggressive tumor of the brain, arises from transformed glial cells [1]. The yearly incidence of gliomas is approximately 6 in 100 000 cases, and the disease is incurable [2]. Furthermore, the high recurrence rate of glioma is mainly because of its extensive growth, malignant behavior, and diffuse invasive characters [3]. Despite many clinical and pathological subtype studies, there remain few effective treatments for this disease. Therefore, there is an urgent need to find sensitive biomarkers and therapeutic targets for glioma, and this process requires insights into the precise mechanisms underlying glioma pathology [4].
Circular RNA (circRNA) is a novel type of noncoding RNA (ncRNA) [5–7]. CircRNAs form a covalently closed continuous loop structure without 5′ or 3′ tails, and their circular structure makes them resistant to RNase R activity and more stable [3,8,9]. They are derived from exons due to the process of back-splicing [10,11]. CircRNAs are abundant and conserved in mammalian cells and have certain levels of tissue and cell specificity [12–14]. Generally, they can function as miRNA sponges and regulate gene transcription and expression [15,16]. In particular, the circRNA-miRNA-mRNA signaling axis has been established to explore their functions [17]. Recent studies confirmed that circRNAs can regulate the development and progression of diseases, including cancer [18,19]. An in-depth study revealed the function of circRNAs in tumor cell proliferation, migration, and invasion [15,18,20,21]. Taken together, these findings indicate that circRNAs could be used as new diagnostic or predictive biomarkers and provide new effective disease treatments.
MicroRNAs, which are endogenous, noncoding, single-stranded RNAs of ~22 nucleotides, are involved in post-transcriptional gene regulation [22–25]. MiRNAs can bind to the 3′UTR of the target mRNA to inhibit translation or stimulate mRNA degradation [26–31]. In mammals, up to 2% of the genome codes for miRNA genes and ~60% of human protein-coding genes are targeted by miRNAs [32,33]. Many studies have found that miRNAs can act as oncogenes or tumor suppressor genes by regulating cell proliferation, differentiation, angiogenesis, migration, and apoptosis [34–38]. MiRNA expression levels are usually altered and associated with pathological conditions [39,40]. There are more distinct miRNAs expressed in the brain than in any other organ, implying a complex regulation network [41–43].
In this study, we found a new circRNA, circ-CFH, that is derived from CFH gene exons and was upregulated in glioma tissue. Subsequently, the function and mechanism of circ-CFH were investigated. The antagonistic effect of circ-CFH on miR-149 was also confirmed. We revealed that circ-CFH plays an oncogenic role in glioma by functioning as a sponge of miR-149 to promote AKT1 expression.
Material and Methods
Tissue specimens
Thirty-one glioma tumor samples and paired adjacent normal tissues were obtained from Huai’an First People’s Hospital. The study protocol was approved by the Research Ethics Committee of Huai’an First People’s Hospital. In addition, written informed consent was obtained from all patients. The samples were immediately snap-frozen in liquid nitrogen and stored at −70°C before use.
Cell lines and cell culture
Primary normal human astrocytes (NHAs) were purchased from Sciencell Research Laboratories (Carlsbad, CA, USA). Human U87, A172, U251, TJ905, and U373 glioblastoma cell lines were purchased from the ATCC (Manassas, VA, USA). NHAs were cultured in Dulbecco’s modified Eagle’s medium (DMEM), but all glioma cell lines were cultured in DMEM-F12; both mediums were supplemented with 10% fetal bovine serum (FBS). The mediums and FBS were purchased from Invitrogen (Carlsbad, CA). All cells were maintained in a 37°C, 5% CO2 incubator.
Cell transfections
Circ-CFH siRNA (SiCirc-CFH), negative control siRNA (SiNC), and circ-CFH mimic (circCFH) were obtained from RiboBio (Guangzhou, China). miRNA mimics (miR-149, miR-145, miR-155, and miR-183), a negative control mimic (NC), and an antisense oligonucleotide (miR-149 ASO) were obtained from GenePharma Co., Ltd. (Shanghai, China). The sequences of the mimics were as follows: miR-149, 5′-UCU GGC UCC GUG UCU UCA CUC CC-3′; miR-145, 5′-GUC CAG UUU UCC CAG GAA UCC CU-3′; miR-183, 5′-UGA AUU CUA CCA GUG CCA UAU U-3′; and miR-155, 5′-UUAAUGCUAAUCGUGAUAGGGGUU-3′. U373 and U251 cells were seeded in plates to 80% confluence and transfected with the constructs (50 nM) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA from the glioma samples and cultured cells was extracted using Trizol reagent (#9109, Takara, China). After quantitation, 5 μg of RNA was transcribed into cDNA using a cDNA synthesis kit (#6130, Takara, China). The reverse transcription products were mixed with TaqMan universal PCR master mix II, and real-time PCR was performed on the Applied Biosystems® 7500 Real-Time PCR System. Relative expression levels were determined based on the 2−ΔΔCt method [44]. The mRNA expression of miR-145, miR-149, miR-155, and miR-183 was normalized to U6, and the mRNA expression of circ-CFH, AKT1, FOXM1, and LRIG2 was normalized to GAPDH. The specific primers used are listed as follows: Forward, 5′-CTC GCT TCG GCA CAT A-3′, Reverse, 5′-AAC GAT TCA CGA ATT TGC GT-3′ for U6; Forward, 5′-TGT GAG GGT TTC AGG AT-3′, Reverse, 5′-CCA TGA GAA ATC TCA GGT GGA-3′ for circ-CFH; Forward, 5′-CTG AAC GGG AAG CTC ACT GG-3′, Reverse, 5′-AAA GTG GTC GTT GAG GGC AA-3′ for GAPDH; Forward, 5′-CGC GCT CGA GCC CAG AGC AAT AAG CCA CAT-3′, Reverse, 5′-GGT GTC GTG GAG TCG GCA ATT CAG TTG AG-3′ for miR-145; Forward, 5′-TCT GGC TCC GTG TCT TCA CTC CC-3′, Reverse, 5′-AGT GGT TGT TCT GCT CTC TGT GTC-3′ for miR-149; Forward, 5′-CAG TCC CGG GTG CAG GCT GGA GAG TGT GAC-3′, Reverse, 5′-GAT CGA TAT CCC TCA GGC AGT GAA AGG TGA TC-3′ for miR-183; Forward, 5′-AGC AAG CGC GGG GAA CCA AGG-3′, Reverse, 5′-TCC ATT GGG TGG GAG AGC CAA GG-3′ for miR-155; Forward, 5′-CTG AGA TTG TGT CAG CCC TGG A-3′, Reverse, 5′-CAC AGC CCG AAG TCT GTG ATC TTA-3′ for AKT1; Forward, 5′-GGG CGC ACG GCG GAA GAT GAA-3′, Reverse, 5′-CCA CTC TTC CAA GGG AGG GCT C-3′ for FOXM1; and Forward, 5′-CAT GTG CCC TCA CTA CCA-3′, Reverse, 5′-CTC CAG GAC CCG AGA ATA-3′ for LRIG2. All primers were designed and purchased from Sangon (Shanghai, China).
CCK-8 assays
U373 and U251 cells were seeded onto 96-well plates and transfected. At 0 h, 12 h, 24 h, 48 h, and 72 h after transfection, 10 μl of CCK8 solution (CK04, Dojindo, Japan) was added to each well; 2 h later, the optical density value of each well was measured at 450 nm using a microplate reader (S28629, Fisher Scientific).
Colony formation assays
At 24 h after transfection, 5×103 U251 or U373 cells were evenly plated on 6-well plates and incubated for 10 days. The colonies were fixed in 10% formaldehyde and stained with 1% crystal violet solution for 5 min. After washing with PBS 5 times, visible colonies were imaged and counted.
Tumor xenograft growth assays
At 24 h after transfection, 1×106 U251 cells were injected into the right cerebral hemisphere of 5-week-old BALBc nude mice. The growth curves and tumor volumes were measured every week. The following equation was used to calculate tumor volume: tumor volume=length×width×0.5. At 5 weeks after injection, the tumors were harvested and weighed.
Ki-67 staining assays
Xenograft tumors were fixed in 10% formalin and paraffin-embedded. Next, 2-μm sections were deparaffinized and subjected to antigen retrieval according to standard procedures. The sections were incubated with Ki-67 (clone MIB-1; Dako; dilution 1: 100) for 30 min. Then, the sections were incubated with secondary antibodies and visualized with 3, 3′-diaminobenzidine. Ki-67 immunoreactivity was evaluated using a labeling index (% of positive cells).
Plasmid construction and dual-luciferase activity assay
The fragments of the circ-CFH sequence containing the miR binding site or mutant binding sites were amplified by PCR. Then, the fragments were cloned into a luciferase vector psi-CHECK (Promega, Madison, USA) and named circCFH wild-type or circCFH mutant. For dual-luciferase activity assays, U251 or U373 cells were seeded in 24-well plates and co-transfected with circCFH wild-type or circCFH mutant and miRNA mimics. Luciferase activity was detected 48 h after transfections by the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocols.
Statistical analysis
The data from this study are all presented as the mean ±SEM, and GraphPad (Ver. Prism 7, GraphPad Prism Software, La Jolla, CA, USA) was used for statistical analyses. The data were subjected to one-way analysis of variance, followed by comparison using the t test to evaluate the differences between the means. A *p value less than 0.05 was considered as statistically significant.
Results
The expression of circCFH was upregulated in glioma
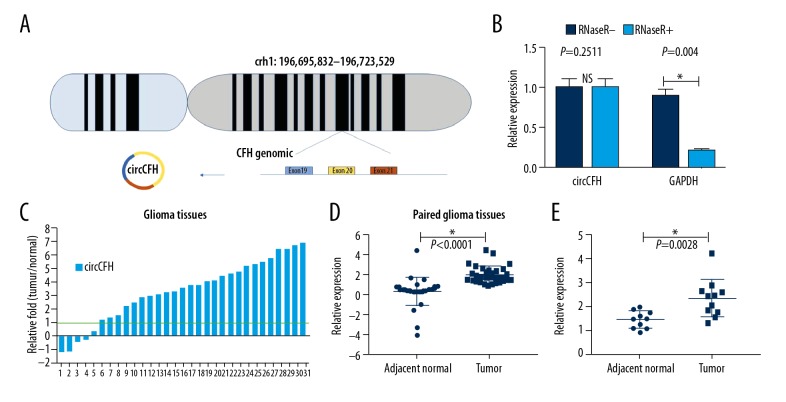
has_circ_0015758 (circ-CFH) is encoded by the sequence located at chr1: 196,695,832–196,723,529. Circ-CFH spliced by 3 exons of the CFH genome contains the following: Exon 19, Exon 20, and Exon 21 (Figure 1A). Total RNA extracted from the cultured cells was treated with or without RNase R and then subjected to qRT-PCR. Circ-CFH was not degraded (1.00±0.11 vs. 0.89±0.09), whereas GAPDH (a linear RNA) was reduced by RNase R treatment (1.00±0.12 vs. 0.21±0.03, Figure 1B). These results validated the circular structure of circ-CFH. Moreover, we analyzed the expression of circ-CFH in 31 pairs of human tumors compared to their adjacent normal tissues; qRT-PCR results showed that the expression in 80% of the tumor tissues was notably upregulated (Figure 1C). A scatter map indicated that the different expression levels of circ-CFH in the gliomas tissues and paired adjacent normal tissue were statistically significant (2.03±0.85 vs. 0.34±1.41, Figure 1D). In addition, the circ-CFH expression was significantly upregulated in grade I–II gliomas vs. grade III–IV gliomas (2.32±0.79 vs. 1.41±0.36, Figure 1E); therefore, circ-CFH expression may be correlated with tumor grade (* P<0.05, Figure 1B, 1D, 1E).
Figure 1.
Circ-CFH expression in normal tissues and glioma. (A) Chromosome localization of the CFH gene and its structure. (B) qRT-PCR results showed that CFH was resistant to RNase R, and GAPDH, a linear mRNA, was used as a control. (C) Circ-CFH expression in 31 pairs of human tumor tissues compared to their adjacent normal tissues was analyzed by qRT-PCR. (D) Scatter map of circ-CFH expression in 31 paired glioma tissues. (E) Circ-CFH expression in grade I–II and grade III–IV gliomas. * P<0.05.
Circular RNA circ-CFH promoted glioma progression
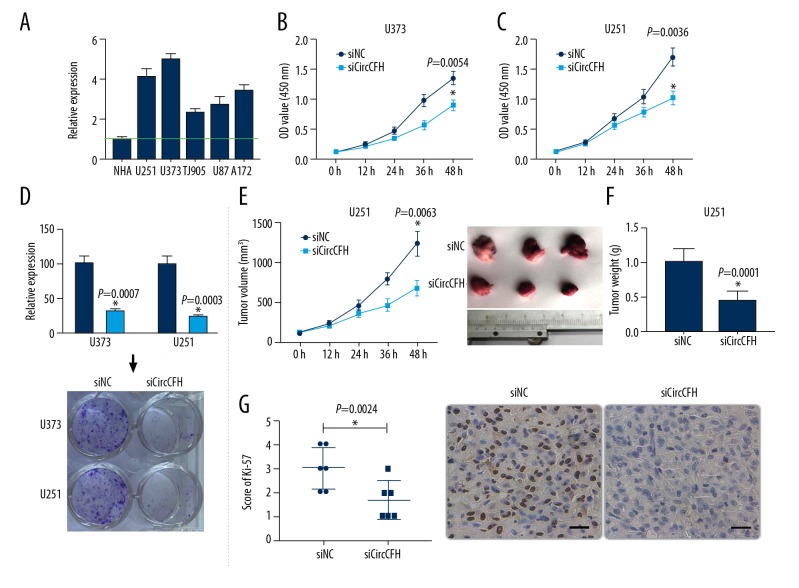
Because circ-CFH was upregulated in gliomas, we next investigated the effect of circ-CFH on glioma cells. The expression of circCFH in 6 glioma cell lines (NHA, 0.78±0.09; U251, 3.19±0.36; U373, 3.89±0.22; TJ905, 1.82±0.15; U87, 2.14±0.31; and A172, 2.64±0.25) was evaluated; it was relatively higher in U251 and U373 cells than in the others (Figure 2A). Therefore, U251 and U373 cells were used for further study. siCircCFH was used to knock down the expression of circ-CFH and siNC was used as a control. CCK-8 assays indicated that U251 and U373 cells transfected with siCircCFH showed decreased cell proliferation (U251, 1.34±0.11 vs. 0.89±0.09; U373, 1.69±0.15 vs. 1.01±0.12, * P<0.05, Figure 2B, 2C). Similarly, transfection with siCircCFH also significantly inhibited the colony formation ability and decreased the numbers of colonies formed on the plates (* P<0.05, Figure 2D). To explore the effect of circ-CFH in vivo, U251 cells transfected with siNC or siCircCFH were injected subcutaneously into nude mice. The volume growth curves and final weights of the xenograft tumors were measured. The results showed that knocking down the expression of circ-CFH markedly decreased the growth of the tumors in vivo (Tumor volume, 1235±154 vs. 678±101 mm3; Tumor weight, 1.03±0.19 vs. 0.45±0.14 g, Figure 2E, 2F). Then, the xenograft tumors in the 2 groups were immunohistochemically stained for Ki-67; as the results show, Ki-67 immuno-positive staining in the siCircCFH group was significantly lower than that in the siNC group (* P<0.05, Figure 2G). Taken together, these findings indicate that circ-CFH promoted glioma progression.
Figure 2.
Effects of circCFH on glioma cells and tissue. (A) Expression of circ-CFH in different glioma cell lines; the expression was higher in U251 and U373 cell lines. (B, C) After U251 and U373 cells were transfected with siNC or siCircCFH, CCK-8 assays were used to measure cell proliferation. (D) Effect of circCFH on U251 and U373 cell colony formation. (E, F) U251 cells transfected with siNC or siCircCFH were injected in nude mice, and the growth of tumor xenografts was determined in the 2 groups; the growth curves of the tumors were measured once a week for 5 weeks, and the final weights of the tumors were compared. (G) Immunohistochemical staining of Ki-67 in xenograft tumor slices; a summarized graphical representation and representative images are shown. The scale bar is 20 μm. * P<0.05
Circ-CFH functions as a miRNA sponge for miR-149 in glioma cells
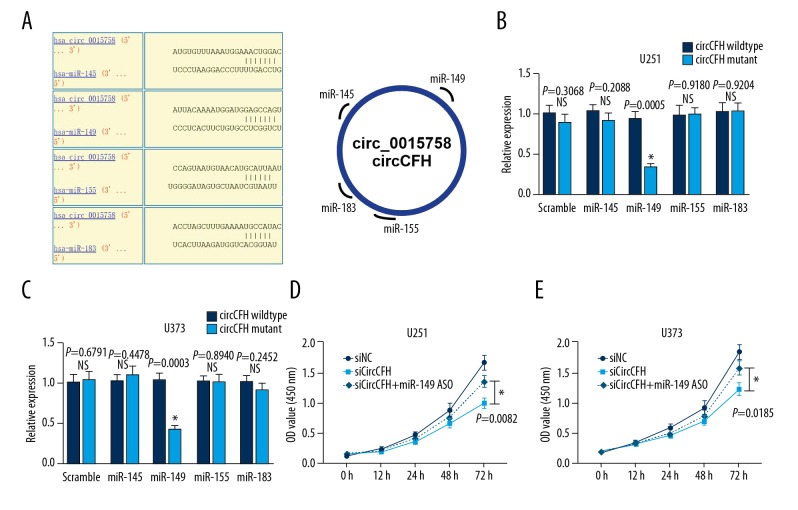
Because circRNA could bind to miRNA and act as a miRNA sponge, we next studied whether circ-CFH could bind miRNA. According to a circular RNA interactome bio-informatic analysis, 4 miRNAs (miR-145, miR-149, miR-155, and miR-183) shared a binding site with circ-CFH (Figure 3A). To confirm whether these miRNAs can directly bind with circRNA-CFH, dual-luciferase assays were performed. U251 and U373 cells were co-transfected with each miRNA mimic and the circ-CFH wild-type reporter or circ-CFH mutant reporter. The data showed that luciferase intensity was not significantly different between the circ-CFH wild-type group and the circ-CFH mutant group when the miR-145, miR-155, and miR-183 mimics were transfected. However, the co-transfection of miR-149 mimics and the circ-CFH wild-type reporter inhibited the luciferase activity, and this reduction was not present in the circ-CFH mutant reporter group (* P<0.05, Figure 3B, 3C). These results indicated that circRNA-CFH specifically interacts with miR-149 and may serve as a sponge in U251 and U373 cells.
Figure 3.
Circ-CFH binding with miR-149 in glioma cells. (A) According to the circular RNA interactome prediction, 4 miRNAs could bind to circ-CFH; the putative complementary site is shown in the schematic. (B, C) For luciferase analysis, U251 and U373 cells were co-transfected with each miRNA mimic and the circCFH wild-type reporter or circCFH mutant reporter, and the luciferase intensity was measured. The scrambled mimic was used as a control. (D, E) After U251 and U373 cells were transfected with siNC, siCircCFH, or siCircCFH + miR-149 ASO, CCK-8 assays were used to measure cell proliferation. * P<0.05.
To determine whether circ-CFH functions as a miR-149 sponge and inhibits its function, U251 and U373 cells were transfected with siNC, siCirc-CFH, or siCirc-CFH + miR-149 ASO; then, CCK-8 assays were used to measure cell proliferation. Cell proliferation was significantly higher in the siCircCFH+miR-149 ASO group than in the siCirc-CFH group (0.98±0.08 vs. 1.34±0.10; 1.21±0.11 vs. 1.57±0.12,* P<0.05, Figure 3D, 3E). These results indicate that circ-CFH promotes glioma progression by sponging miR-149.
AKT1 is a direct target of miR-149 and is positively regulated by circ-CFH
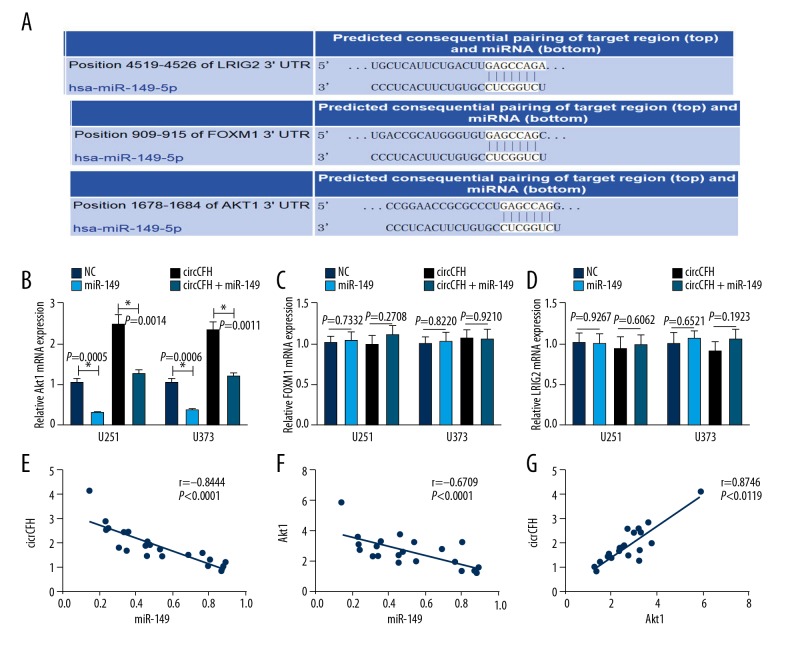
We have shown that circ-CFH can directly bind to miR-149 and act as a sponge, so we next determined whether circ-CFH could regulate miR-149 target genes. A TargetScan analysis indicated that AKT1, FOXM1, and LRIG2 were the predicted targets of miR-149, and the putative binding sites between them are shown (Figure 4A). To investigate the miR-149 target genes in glioma cells and their correlation with circ-CFH, U251 and U373 cells were transfected with NC mimics, miR-149 mimics, or circ-CFH or co-transfected with circ-CFH and miR-149 mimics. The data show that the overexpression of miR-149 had no obvious effect on FOXM1 and LRIG expression, but the mRNA level of AKT1 was decreased (U251, 1.13±0.14 vs. 0.31±0.03; U373, 1.22±0.13 vs. 0.41±0.05); thus, AKT1 was the miR-124 target gene in glioma cells. Furthermore, circ-CFH increased the mRNA level of AKT1, and the upregulation of AKT1 by circ-CFH could be abrogated by miR-149 mimics (U251, 2.77±0.27 vs. 1.38±0.14; U373, 2.82 vs. ±0.26 vs. 1.42±0.13, * P<0.05, Figure 4B–4D). A correlation analysis confirmed that circ-CFH and AKT1 expression were inversely correlated with miR-149 (r=−0.8444, Figure 4E; r=−0.6709, Figure 4F), while circ-CFH expression was positively correlated with AKT1 expression (r=0.8746, Figure 4G). These results indicated that circ-CFH can regulate AKT1 by sponging miR-149.
Figure 4.
AKT1 is a direct target of miR-149 and is positively regulated by circ-CFH. (A) The TargetScan predictions for the putative complementary site of miR-149 and AKT1, FOXM1, and LRIG2 are shown in the schematic. (B–D) U251 and U373 cells were transfected with NC mimics, miR-149 mimics or circCFH or co-transfected with siCircCFH and miR-149 mimics; then, the mRNA expression of Akt1, FOXM1 and LRIG2 was measured by qRT-PCR. * P<0.05. (E) Correlation between circ-CFH and miR-149 expression (P<0.05). (F) Correlation between Akt1 and miR-149 expression (P<0.05). (G) Correlation between circ-CFH and Akt1 expression (P<0.05).
Discussion
In the past decade, RNA sequencing and mapping have revealed that thousands of expressed genes also differentially generate circular RNAs by back-splicing or lariat introns [17]. These RNAs may not translate into proteins, but are stable and contain exons. Many of these circRNAs are dysregulated in various diseases, suggesting their involvement in disease development and progression [46]. Regarding their high conservation, stability, and relatively tissue-specific expression patterns, they could act as novel biomarkers for cancer [46]. Glioma is the most common malignant nervous system tumor and is characterized by high fatality rates and poor prognosis. Some endogenous circRNAs involved in glioma have been researched; these include TTBK2 circular RNA and cZNF292 circular RNA [47,48]. In our study, we identified a novel circRNA in glioma; our findings may provide important evidence for circular RNA research in glioma carcinoma.
The complement factor H (CFH) gene encodes for complement factor H, which is a complement regulator and has an important role in inhibiting complement activation and inflammation [49]. In our study, we found that a CFH circRNA could arise from CFH exons and be generated by back-splicing. Circ-CFH contains 3 exons and is resistant to RNase R. However, the function of circ-CFH in glioma was unknown. We found that circ-CFH was upregulated in glioma tissues and cell lines, and its expression level was also correlated with tumor grade. Then, we explored the specific role of circ-CFH in glioma. The results indicated that circ-CFH functions as an oncogene in glioma tumorigenesis. To the best of our knowledge, this is the first study to identify the expression and functional role of circ-CFH in glioma.
Previous studies have revealed that miR-149 increased apoptosis in neuroblastoma cells, and the blockage of miR-149 promoted tumors in prostate carcinoma [50,51]. In addition, other studies found that miR-149 decreased cell proliferation by targeting FOXM1 [52]. In glioma cells, it was also reported that miR-149 can inhibit invasiveness through blocking AKT1 signaling [53]. In our study, we confirmed a new regulation aids, circ-CFH/miR-149/Akt1, in glioma cell tumorigenesis. These results are an extension of miR-149 research in glioma. This axis also serves as solid evidence that circRNA/miRNA/mRNA regulatory systems play important physiological functions in various diseases.
Conclusions
In summary, our data indicate that circ-CFH expression levels are upregulated in glioma tissues and cell lines. Additionally, the mechanistic analysis showed that circ-CFH increased the proliferative and colony formation abilities of glioma cells by specifically sponging miR-149 and releasing AKT1. Our study uncovers a novel circ-CFH-miR-149-AKT1 signaling pathway involved in glioma cell progression (Figure 5). Therefore, circ-CFH may serve as a therapeutic target for glioma treatment.
Figure 5.
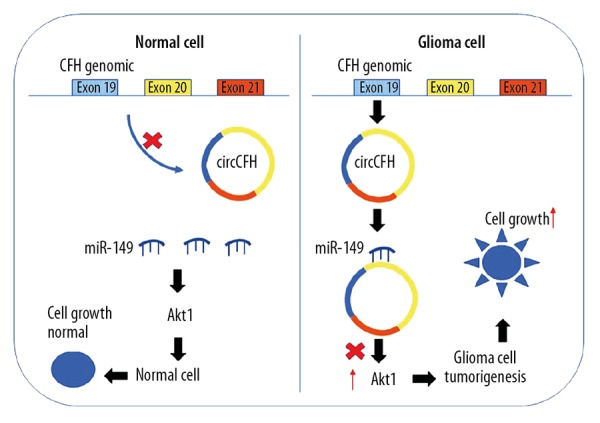
Schematic representation of the function and mechanism of circ-CFH glioma cells. Circ-CFH expression was significantly upregulated in glioma cells compared to that in normal cells. Circ-CFH functions as a miR-149 sponge to regulate the miR-149/Akt1 pathway in glioma cells. Increased circ-CFH expression levels in glioma decreased the activity of miR-449a; therefore, upregulated Akt1 expression ultimately promoted glioma progression.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neurooncology. 2012;14(Suppl 5):v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vannini E, Maltese F, Olimpico F, et al. Progression of motor deficits in glioma-bearing mice: Impact of CNF1 therapy at symptomatic stages. Oncotarget. 2017;8(14):23539–50. doi: 10.18632/oncotarget.15328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciaglia E, Torelli G, Pisanti S, et al. Cannabinoid receptor CB1 regulates STAT3 activity and its expression dictates the responsiveness to SR141716 treatment in human glioma patients’ cells. Oncotarget. 2015;6(17):15464–81. doi: 10.18632/oncotarget.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfonso JCL, Talkenberger K, Seifert M, et al. The biology and mathematical modelling of glioma invasion: A review. J R Soc Interface. 2017;14(136) doi: 10.1098/rsif.2017.0490. pii: 20170490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andres-Leon E, Nunez-Torres R, Rojas AM. Corrigendum: miARma-Seq: A comprehensive tool for miRNA, mRNA and circRNA analysis. Sci Rep. 2018;8:46928. doi: 10.1038/srep46928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Bartsch D, Zirkel A, Kurian L. Characterization of circular RNAs (circRNA) associated with the translation machinery. Methods Mol Biol. 2018;1724:159–66. doi: 10.1007/978-1-4939-7562-4_13. [DOI] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–88. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 10.Guo XY, Chen JN, Sun F, et al. circRNA_0046367 prevents hepatoxicity of lipid peroxidation: An inhibitory role against hepatic steatosis. Oxid Med Cell Longev. 2017;2017:3960197. doi: 10.1155/2017/3960197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo XY, Sun F, Chen JN, et al. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol. 2018;24(3):323–37. doi: 10.3748/wjg.v24.i3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Xie F, Zheng FX, et al. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J Huazhong Univ Sci Technolog Med Sci. 2017;37(6):886–90. doi: 10.1007/s11596-017-1822-9. [DOI] [PubMed] [Google Scholar]
- 13.Li JL, Zheng WY, Li XH, et al. [Research on circRNA in regulatory mechanism of diseases]. Zhonghua Bing Li Xue Za Zhi. 2017;46(8):578–81. doi: 10.3760/cma.j.issn.0529-5807.2017.08.015. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 14.Li L, Guo J, Chen Y, et al. Comprehensive CircRNA expression profile and selection of key CircRNAs during priming phase of rat liver regeneration. BMC Genomics. 2017;18(1):80. doi: 10.1186/s12864-016-3476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–81. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider T, Hung LH, Schreiner S, et al. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. 2016;6:31313. doi: 10.1038/srep31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XQ, Yang JH. Discovering circRNA-microRNA Interactions from CLIP-Seq Data. Methods Mol Biol. 2018;1724:193–207. doi: 10.1007/978-1-4939-7562-4_16. [DOI] [PubMed] [Google Scholar]
- 18.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–91. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou M, Huang C, Li X, He X, et al. Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget. 2017;8(47):81825–37. doi: 10.18632/oncotarget.18998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Liu CX, Xue W, et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67(2):214–27. doi: 10.1016/j.molcel.2017.05.023. e7. [DOI] [PubMed] [Google Scholar]
- 21.Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol. 2018;9:9. doi: 10.3389/fimmu.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abreu FB, Liu X, Tsongalis GJ. miRNA analysis in pancreatic cancer: The Dartmouth experience. Clin Chem Lab Med. 2017;55(5):755–62. doi: 10.1515/cclm-2017-0046. [DOI] [PubMed] [Google Scholar]
- 23.Acharya S, Saha S. Importance of proximity measures in clustering of cancer and miRNA datasets: proposal of an automated framework. Mol Biosyst. 2016;12(11):3478–501. doi: 10.1039/c6mb00609d. [DOI] [PubMed] [Google Scholar]
- 24.Adams BD, Arem H, Hubal MJ, et al. Exercise and weight loss interventions and miRNA expression in women with breast cancer. Breast Cancer Res Treat. 2018;170(1):55–67. doi: 10.1007/s10549-018-4738-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Advani J, Subbannayya Y, Patel K, et al. Long-term cigarette smoke exposure and changes in MiRNA expression and proteome in non-small-cell lung cancer. OMICS. 2017;21(7):390–403. doi: 10.1089/omi.2017.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baltruskeviciene E, Schveigert D, Stankevicius V, et al. Down-regulation of miRNA-148a and miRNA-625-3p in colorectal cancer is associated with tumor budding. BMC Cancer. 2017;17(1):607. doi: 10.1186/s12885-017-3575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bash-Imam Z, Therizols G, Vincent A, et al. Translational reprogramming of colorectal cancer cells induced by 5-fluorouracil through a miRNA-dependent mechanism. Oncotarget. 2017;8(28):46219–33. doi: 10.18632/oncotarget.17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37(1):33–44. doi: 10.1007/s10555-017-9724-7. [DOI] [PubMed] [Google Scholar]
- 29.Bhardwaj A, Singh H, Rajapakshe K, et al. Regulation of miRNA-29c and its downstream pathways in preneoplastic progression of triple-negative breast cancer. Oncotarget. 2017;8(12):19645–60. doi: 10.18632/oncotarget.14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanca A, Cheng L, Montironi R, et al. Mirna expression in bladder cancer and their potential role in clinical practice. Curr Drug Metab. 2017;18(8):712–22. doi: 10.2174/1389200218666170518164507. [DOI] [PubMed] [Google Scholar]
- 31.Braicu OL, Budisan L, Buiga R, et al. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. Onco Targets Ther. 2017;10:4225–38. doi: 10.2147/OTT.S137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du F, Yuan P, Zhao ZT, et al. A miRNA-based signature predicts development of disease recurrence in HER2 positive breast cancer after adjuvant trastuzumab-based treatment. Sci Rep. 2016;6:33825. doi: 10.1038/srep33825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farina NH, Ramsey JE, Cuke ME, et al. Development of a predictive miRNA signature for breast cancer risk among high-risk women. Oncotarget. 2017;8(68):112170–83. doi: 10.18632/oncotarget.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavrilas LI, Ionescu C, Tudoran O, et al. The role of bioactive dietary components in modulating miRNA expression in colorectal cancer. Nutrients. 2016;8(10) doi: 10.3390/nu8100590. pii: E590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilot D, Galibert MD. miRNA displacement as a promising approach for cancer therapy. Mol Cell Oncol. 2017;5(1):e1406432. doi: 10.1080/23723556.2017.1406432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gov E, Kori M, Arga KY. Multiomics analysis of tumor microenvironment reveals Gata2 and miRNA-124-3p as potential novel biomarkers in ovarian cancer. OMICS. 2017;21(10):603–15. doi: 10.1089/omi.2017.0115. [DOI] [PubMed] [Google Scholar]
- 37.Granados-Lopez AJ, Ruiz-Carrillo JL, Servin-Gonzalez LS, et al. Use of mature miRNA strand selection in miRNAs families in cervical cancer development. Int J Mol Sci. 2017;18(2) doi: 10.3390/ijms18020407. pii: E407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu L, Zhang J, Shi M, Peng C. The effects of miRNA-1180 on suppression of pancreatic cancer. Am J Transl Res. 2017;9(6):2798–806. [PMC free article] [PubMed] [Google Scholar]
- 39.Mandujano-Tinoco EA, Garcia-Venzor A, Munoz-Galindo L, et al. miRNA expression profile in multicellular breast cancer spheroids. Biochim Biophys Acta. 2017;1864(10):1642–55. doi: 10.1016/j.bbamcr.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 40.Mao Y, Liu R, Zhou H, et al. Transcriptome analysis of miRNA-lncRNA-mRNA interactions in the malignant transformation process of gastric cancer initiation. Cancer Gene Ther. 2017;24(6):267–75. doi: 10.1038/cgt.2017.14. [DOI] [PubMed] [Google Scholar]
- 41.Ohzawa H, Miki A, Teratani T, et al. Usefulness of miRNA profiles for predicting pathological responses to neoadjuvant chemotherapy in patients with human epidermal growth factor receptor 2-positive breast cancer. Oncol Lett. 2017;13(3):1731–40. doi: 10.3892/ol.2017.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osipov ID, Zaporozhchenko IA, Bondar AA, et al. Cell-free miRNA-141 and miRNA-205 as prostate cancer biomarkers. Adv Exp Med Biol. 2016;924:9–12. doi: 10.1007/978-3-319-42044-8_2. [DOI] [PubMed] [Google Scholar]
- 43.Osip’yants AI, Knyazev EN, Galatenko AV, et al. Changes in the level of circulating hsa-miR-297 and hsa-miR-19b-3p miRNA are associated with generalization of prostate cancer. Bull Exp Biol Med. 2017;162(3):379–82. doi: 10.1007/s10517-017-3620-6. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–32. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 46.Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 47.Yang P, Qiu Z, Jiang Y, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7(39):63449–55. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng J, Liu X, Xue Y, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10(1):52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Lv Q, Fan W, et al. Influence of CFH gene on symptom severity of schizophrenia. Neuropsychiatr Dis Treat. 2017;13:697–706. doi: 10.2147/NDT.S132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He DX, Gu XT, Li YR, et al. Methylation-regulated miR-149 modulates chemoresistance by targeting GlcNAc N-deacetylase/N-sulfotransferase-1 in human breast cancer. FEBS J. 2014;281(20):4718–30. doi: 10.1111/febs.13012. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Rivas LG, Jerez JM, Carmona R, et al. A microRNA signature associated with early recurrence in breast cancer. PLoS One. 2014;9(3):e91884. doi: 10.1371/journal.pone.0091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu K, Liu X, Mao X, et al. MicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem. 2015;35(2):499–515. doi: 10.1159/000369715. [DOI] [PubMed] [Google Scholar]
- 53.Pan SJ, Zhan SK, Pei BG, et al. MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. Int J Immunopathol Pharmacol. 2012;25(4):871–81. doi: 10.1177/039463201202500405. [DOI] [PubMed] [Google Scholar]