Abstract
Introduction
Terminal differentiation-induced non-coding RNA (TINCR) has been suggested to have aberrant expression in multiple human cancers, and functions as tumor suppressor or promoter in various types of human tumors depending on the specific cancer types. The expression status and biological function of TINCR in prostate cancer is still unknown.
Materials and methods
In our study, we detected TINCR expression in prostate cancer tissue samples and cell lines, and analyzed the association between TINCR expression and clinical parameters in 160 prostate cancer patients. Moreover, we conducted gain-of-function and loss-of-function studies in prostate cancer cell to explore the biological function and molecular mechanism of TINCR.
Results
In our results, low-expression TINCR was observed in prostate cancer, and correlated with advanced clinical T stage, lymph node involvement, distant metastasis, high Gleason score and poor prognosis in prostate cancer patients. Moreover, levels of TINCR expression were negatively associated with TRIP13 mRNA and protein expressions in prostate cancer tissues, and negatively regulated the TRIP13 mRNA and protein expressions in prostate cancer cell lines. TINCR inhibits prostate cancer cell proliferation, migration and invasion via suppressing TRIP13 expression.
Conclusion
TINCR plays a tumor suppressive role in regulating prostate cancer cell proliferation, migration and invasion through modulating TRIP13 expression.
Keywords: lncRNA, TINCR, biomarker, TRIP13, prostate cancer
Introduction
Prostate cancer is the second common malignancy in man and the fifth most common cause of cancer-related death among males worldwide.1 There were about 1,111,700 newly diagnosed prostate cancer patients and 307,500 prostate cancer deaths all over the world in 2012.1 In China, a significantly increasing incidence and mortality trend was observed in prostate cancer patients from 2000 to 2011.2 The China Cancer Statistics suggested that prostate cancer is the seventh most common malignant cancer in males with an estimated 60,300 new cases and more than 26,000 deaths in 2015.2 Although the growth of prostate cancer is slow, distant metastasis has often occurred at diagnosis.3,4 Owing to the poor understanding of the molecular mechanisms underlying prostate cancer progression, the rate of long-term survival in prostate cancer patients is still low.5,6 Therefore, it is urgently needed to investigate the mechanism underlying the development of prostate cancer for developing new therapies.
Terminal differentiation-induced non-coding RNA (TINCR, also known as LINC00036) is a 3.7 kb long non-coding RNA (lncRNA) and located in the human chromosome 19p13.3.7 Originally, TINCR was detected in a genomic screen for lncRNA expression change during differentiation, and has been found to control human epidermal differentiation.8 Recently, TINCR has been suggested to have aberrant expression in multiple human cancers and be involved in tumorigenesis.9,10 However, the expression status and biological function of TINCR in prostate cancer are still unknown. At the initial stage, we analyzed The Cancer Genome Atlas (TCGA) RNA-seq datasets, which contain 52 normal prostatic tissue samples and 374 prostate cancer tissue samples, and observed that TINCR expression was decreased in prostate cancer tissues compared with normal prostatic tissues and associated with favorable prognosis in prostate cancer patients. Thus, we supposed that TINCR acts as a tumor suppressor in prostate cancer progression. In order to identify the role in prostate cancer, we analyzed the association between TINCR expression and clinicopathological characteristics to explore the clinical value in prostate cancer patients, and conducted gain-of-function and loss-of-function studies to investigate the effect of TINCR on prostate cancer cell proliferation, migration and invasion.
Materials and methods
TCGA database analysis
To determine the aberrant expression of TINCR, we investigated TINCR expression in 52 pairs of normal prostatic tissues and prostate cancer tissues from TCGA (http://cancergenome.nih.gov). Meanwhile, a total of 374 prostate cancer cases were available for the survival analysis.
Clinical tissue samples
The study methodologies conformed to the standards set by the Declaration of Helsinki, and the study methodologies were approved by the Ethics Committee of The Fourth Affiliated Hospital of China Medical University and The Affiliated Hospital of Chifeng Medical College. Written informed consent was obtained from each patient. Fresh 160 prostate cancer tissue specimens and 30 paired normal prostatic tissue specimens from 160 patients were collected from The Fourth Affiliated Hospital of China Medical University and The Affiliated Hospital of Chifeng Medical College. Fresh clinical tissue samples were immediately stored in liquid nitrogen for quantitative (qRT-PCR). None of the patients had received anti-tumor therapy before diagnose.
qRT-PCR
RNA isolation and TINCR expression determination were carried out according to previous description.11 All the primers were listed as follows: TINCR forward, 5′-TGTG-GCCCAAACTCAGGGATACAT-3′; reverse, 5′-AGATGA-CAGTGGCTGGAGTTGTCA-3′ and GAPDH forward, 5′-GCTCTCTGCTCCTCCTGTTC-3′; reverse, 5′-ACGAC-CAAATCCGTTGACTC-3′. The GAPDH gene was used as the internal control gene.
Immunohistochemistry staining and evaluation of staining
Immunohistochemistry was performed as described previously11 with a rabbit anti-TRIP13 antibody at the concentration of 1:100 (Abcam, Cambridge, UK). Five random fields in each slide were selected and evaluated independently by two pathologists. Any disagreements were arbitrated by the third pathologists. A semi-quantitative scoring system was used to evaluate the staining results based on the percentage and intensity of positively stained cells.12 The intensity was scored as 0–3 (0: negative; 1: weak; 2: moderate; 3: strong). The staining extent was scored as 1–4 (1: 0–25%; 2: 26–50%; 3: 51–75%; 4: 76–100%). The final score was calculated by multiplying intensity and extent score. Low-expression of TRIP13 was defined as 0–7 scores; high expression of TRIP13 was defined as 8–12 scores.
Western blotting
Western blotting was carried out accordingly as described previously11 with a rabbit anti-TRIP13 antibody at the concentration of 1:1000 (Abcam). An horseradish peroxidase (HRP) conjugated anti-rabbit IgG antibody was used as the secondary antibody (CWBio, Beijing, China). Signals were detected using enhanced chemiluminescence (ECL) kit (CWBio), and analyzed by Quantity One Software (Bio-Rad Laboratories Inc., Hercules, CA, USA). Each experiment was performed in triplicate.
Cell culture and transfection
The human prostate cancer cell lines (LNCaP, PC3, DU145 and 22Rv1) and normal human prostate epithelial cell lines (RWPE-1 and P69) were purchased from the Chinese Academy of Sciences. P69, LNCaP, PC3, DU145 and 22Rv1 cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum (Gibco, Waltham, MA, USA). RWPE-1 cell was cultured in Keratinocyte Serum Free Medium (Invitrogen, Waltham, MA, USA) supplemented with bovine pituitary extract and human recombinant epidermal growth factor. All cell lines were cultured at 37°C in a humidified atmosphere of 5% CO2.
The short-hairpin RNA (5′-AATACCTGCTACTTC-ATGC-3′) targeting the TINCR gene was cloned into the pSilencer 3.1 (pSilencer-TINCR) according to the manufacturer’s protocol. The coding sequence region of human TINCR was amplified and cloned into pcDNA3.1 express vector (pcDNA-TINCR). Cells were transfected using Lipofectamine™ 3000 reagent (Invitrogen) in Opti-MEM (Gibco), according to the manufacturer’s instructions. Transfection efficiencies were tested by qRT-PCR.
Cell proliferation analysis
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich Co., St Louis, MO, USA) assay was used for detecting cell proliferation according to previous description.11 After transfection, cells were grown in a 96-well plate for 24, 48, 72 and 96 h. Each experiment was performed in triplicate.
Cell migration and invasion assays
In vitro cell migration and invasion assays were conducted according to a previous description.11 Each experiment was performed in triplicate.
Statistical analysis
The SPSS 17.0 software was used for statistical analysis. The GraphPad prism 5.0 software was used to make diagrams. The difference in TINCR expression between normal prostatic tissues and prostate cancer tissues was detected by Wilcoxon signed rank test. Student’s t-test was used for comparisons of two independent groups. The correlation between prostate cancer characteristics and TINCR expression was examined by chi-square test. Log-rank test and univariate Cox’s regression model were used for survival analysis. The association between TINCR expression and TRIP13 mRNA level was detected by using Spearman’s correlation coefficient analysis. P-values of 0.05 or less were considered to be statistically significant.
Results
Levels of TINCR expression are decreased in prostate cancer tissues and cell lines
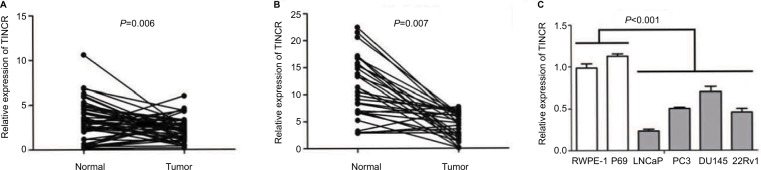
In order to investigate the expression of TINCR in prostate cancer, we analyzed the expression of TINCR in 52 pairs of normal prostatic tissues and prostate cancer tissues from TCGA database, and found that TINCR expression was significantly decreased in prostate cancer tissues compared with normal prostatic tissues (P=0.006, Figure 1A). Furthermore, we conducted qRT-PCR to confirm TINCR expression status in prostate cancer tissues and cell lines. Compared with paired normal prostatic tissues, TINCR levels were obviously reduced in prostate cancer tissues (P<0.001, Figure 1B). Moreover, TINCR showed a dramatically lower expression level in prostate cancer cell lines (LNCaP, PC3, DU145 and 22Rv1) compared with normal human prostate epithelial cell lines (RWPE-1 and P69) (P<0.001, Figure 1C).
Figure 1.
Levels of TINCR expression are decreased in prostate cancer tissues and cell lines.
Notes: (A) TINCR expression is significantly decreased in prostate cancer tissues compared with paired normal prostatic tissues (clinical samples from TCGA database). (B) TINCR levels are obviously reduced in prostate cancer tissues compared with paired normal prostatic tissues (clinical samples from our study). (C) TINCR has a lower expression level in prostate cancer cell lines (LNCaP, PC3, DU145 and 22Rv1) compared with normal human prostate epithelial cell lines (RWPE-1 and P69).
Abbreviations: TINCR, terminal differentiation-induced non-coding RNA; TCGA, The Cancer Genome Atlas.
Low expression of TINCR correlates with the malignant status of prostate cancer patients
We measured TINCR expression in 160 prostate cancer specimens by using qRT-PCR, and explored the relationship between TINCR expression and clinicopathological characteristics of prostate cancer patients. According to a published study,13 we divided all cases into two groups according to TINCR expression: TINCR high-expression group (n=80) and TINCR low-expression group (n=80). As summarized in Table 1, TINCR low expression was strikingly associated with advanced clinical T stage (T1–T2 vs. T3–T4; P=0.039), lymph node involvement (no vs. yes; P=0.001), distant metastasis (no vs. yes; P<0.001) and high Gleason score (≤8 vs. >8, P=0.003). However, TINCR expression was not associated with age (P=0.746), smoking (P=0.738) and levels of serum prostate-specific antigen (P=0.418).
Table 1.
Correlations between TINCR expression and clinicopathological characteristics in prostate cancer
| Characteristics | n | Low expression (%) | High expression (%) | P-value |
|---|---|---|---|---|
| Age (years) | ||||
| ≤65 | 62 | 30 (48.4) | 32 (51.6) | 0.746 |
| >65 | 98 | 50 (51.0) | 48 (49.0) | |
| Smoking | ||||
| No | 54 | 26 (48.1) | 28 (51.9) | 0.738 |
| Yes | 106 | 54 (50.9) | 52 (49.1) | |
| Clinical T stage | ||||
| T1–T2 | 73 | 30 (41.1) | 43 (58.9) | 0.039 |
| T3–T4 | 87 | 50 (57.5) | 37 (42.5) | |
| Lymph node involvement | ||||
| No | 130 | 57 (43.8) | 73 (56.2) | 0.001 |
| Yes | 30 | 23 (76.7) | 7 (23.3) | |
| Distant metastasis | ||||
| No | 135 | 58 (43.0) | 77 (57.0) | <0.001 |
| Yes | 25 | 22 (88.0) | 3 (12.0) | |
| Gleason score | ||||
| ≤8 | 70 | 27 (37.0) | 46 (63.0) | 0.003 |
| >8 | 90 | 53 (60.9) | 34 (39.1) | |
| Serum PSA (ng/mL | ) | |||
| ≤20 | 63 | 34 (54.0) | 29 (46.0) | 0.418 |
| >20 | 97 | 46 (47.4) | 51 (52.6) |
Abbreviations: PSA, prostate-specific antigen; TINCR, terminal differentiation-induced non-coding RNA.
Low expression of TINCR predicts a poor prognosis in prostate cancer patients
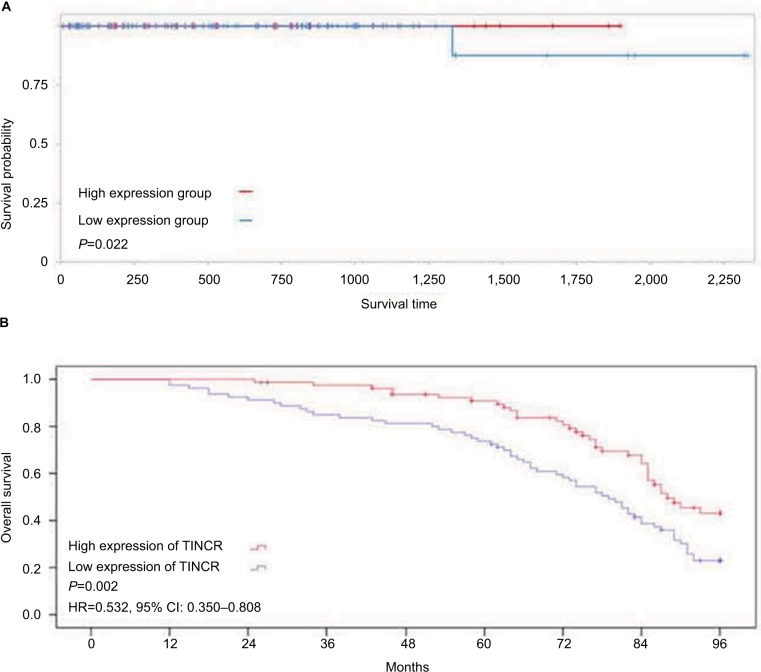
The prognostic value of TINCR was further investigated in prostate cancer patients. We analyzed a cohort of 374 prostate cancer cases from TCGA database, and found that patients with low expression of TINCR had a shorter survival time than those with high expression of TINCR (P=0.022, Figure 2A). Similar result was found in our study: level of TINCR was positively correlated with the overall survival of prostate cancer cases (P=0.002, Figure 2B). Meanwhile, the univariate analysis indicated that the TINCR low expression was an unfavorable prognostic factor for prostate cancer patients (HR=0.532, 95% CI: 0.350–0.808, P=0.002, Figure 2B).
Figure 2.
Low expression of TINCR predicts a poor prognosis in prostate cancer patients.
Notes: (A) Prostate cancer patients with low expression of TINCR have a shorter survival time than those with high expression of TINCR (clinical samples from TCGA database). (B) Level of TINCR is positively correlated with the overall survival of prostate cancer cases (clinical samples from our study).
Abbreviations: TINCR, terminal differentiation-induced non-coding RNA; TCGA, The Cancer Genome Atlas.
The relationship between TINCR and TRIP13 in prostate cancer
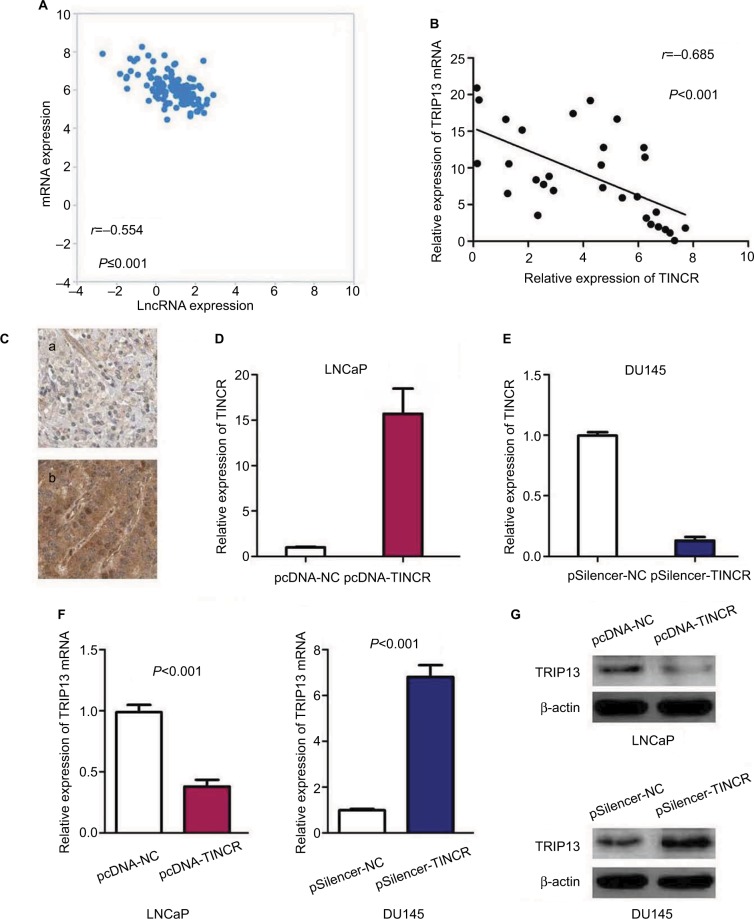
TRIP13 has been suggested to be overexpressed in multiple tumors and function as oncogene in carcinogenesis.14 Interestingly, we observed that TINCR expression was markedly negatively correlated with TRIP13 mRNA expression in 374 prostate cancer tissue samples from TCGA database (r=−0.554, P<0.001, Figure 3A). Furthermore, the relationship of TINCR expression with TRIP13 mRNA and protein was explored in prostate cancer tissues. We found that levels of TINCR expression were negatively associated with TRIP13 mRNA expression (r=−0.685, P<0.001, Figure 3B) and TRIP13 protein expression (r=−0.554, P=0.002, Figure 3C and Table 2). In order to explore the effect of TINCR on TRIP13 expression, we induced upregulation of TINCR expression in LNCaP cell by pcDNA-TINCR (Figure 3D) and downregulation of TINCR expression in DU145 cell by pSilencer-TINCR (Figure 3E). We observed that upregulation of TINCR expression dramatically reduced TRIP13 mRNA and protein expressions in LNCaP cell, and downregulation of TINCR expression markedly increased TRIP13 mRNA and protein expressions in DU145 cell (Figure 3F and G).
Figure 3.
The relationship between TINCR and TRIP13 in prostate cancer.
Notes: (A) TINCR expression is markedly negatively correlated with TRIP13 mRNA expression in prostate cancer tissue samples from TCGA database. (B) Levels of TINCR expression are negatively associated with TRIP13 mRNA expression in prostate cancer tissue samples from our study. (C) Immunohistochemical staining of TRIP13 in prostate cancer tissues: a, low-expression of TRIP13; b, high-expression of TRIP13. (D) The efficiency of pcDNA-TINCR is confirmed by qRT-PCR in LNCaP. (E) The efficiency of pSilencer-TINCR is confirmed by qRT-PCR in DU145. (F) Upregulation of TINCR expression reduces TRIP13 mRNA expression in LNCaP cell, and downregulation of TINCR expression increases TRIP13 mRNA expression in DU145 cell. (G) Upregulation of TINCR expression decreases TRIP13 mRNA expression in LNCaP cell, and downregulation of TINCR expression elevates TRIP13 mRNA expression in DU145 cell.
Abbreviations: qRT-PCR, quantitative RT-PCR; TINCR, terminal differentiation-induced non-coding RNA; TCGA, The Cancer Genome Atlas.
Table 2.
The association between TINCR and TRIP13 protein in prostate cancer
| Group | n | TRIP13 protein
|
r
|
P-value | ||
|---|---|---|---|---|---|---|
| High expression | Low expression | |||||
| TINCR | High expression | 15 | 5 | 10 | −0.55 | 0.002 |
| Low expression | 15 | 13 | 2 | |||
Abbreviation: TINCR, terminal differentiation-induced non-coding RNA.
TINCR regulates prostate cancer cell proliferation, migration and invasion
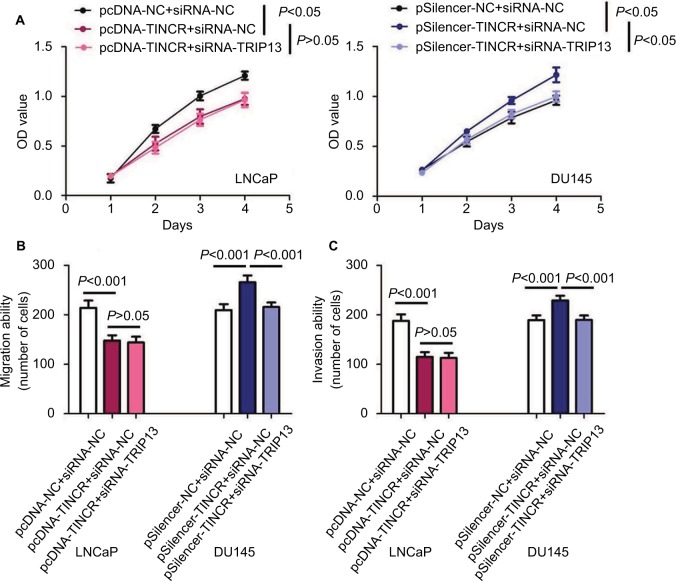
The MTT assay was performed to evaluate the effect of TINCR on prostate cancer cell proliferation. The results of MTT assay indicated that upregulation of TINCR expression dramatically inhibited cell proliferation, and downregulation of TINCR expression strikingly promoted cell proliferation (P<0.05, Figure 4A). The cell migration and invasion assays were conducted to evaluate the effect of TINCR on prostate cancer cell motility. Upregulation of TINCR expression significantly accelerated LNCaP cell migration, and down-regulation of TINCR expression obviously depressed DU145 cell migration (P<0.001, Figure 4B). Similar to results of cell migration assay, invasion assay demonstrated that upregulation of TINCR expression notably promoted LNCaP cell invasion, and downregulation of TINCR expression markedly repressed DU145 cell invasion (P<0.001, Figure 4C and Figure S1).
Figure 4.
The biological function of TINCR and TRIP13 regulates cell proliferation, migration and invasion in prostate cancer.
Notes: (A) The effect of TINCR and/or TRIP13 on prostate cancer cell proliferation. (B) The effect of TINCR and/or TRIP13 on prostate cancer cell migration. (C) The effect of TINCR and/or TRIP13 on prostate cancer cell invasion.
Abbreviations: OD, optical density; TINCR, terminal differentiation-induced non-coding RNA.
TINCR modulates TRIP13 to control prostate cancer cell proliferation, migration and invasion
Abovementioned results suggested that TINCR expression was negatively associated with TRIP13 expression in prostate cancer tissues, and negatively modulated TRIP13 expression in prostate cancer cells. Meanwhile, we found that TRIP13 functioned as an oncogene in regulating prostate cancer cell proliferation, migration and invasion (article in press). In order to elucidate the relationship between TINCR and TRIP13 in regulating prostate cancer cell proliferation, migration and invasion, pcDNA-TINCR and siRNA-TRIP13 were co-transfected into LNCaP cell, and pSilencer-TINCR and siRNA-TRIP13 were co-transfected into DU145 cell. We observed that co-transfection of pcDNA-TINCR and siRNA-TRIP13 did not further suppress LNCaP cell proliferation, migration and invasion in comparison to pcDNA-TINCR (all P>0.05, Figure 4A–C), and co-transfection of pSilencer-TINCR and siRNA-TRIP13 strikingly rescued facilitation of pSilencer-TINCR in DU145 cell proliferation, migration and invasion (all P<0.05, Figure 4A–C). Thus, TRIP13 was a functional protein for TINCR in regulating prostate cancer cell proliferation, migration and invasion.
Discussion
TINCR (also known as LINC00036) is a 3.7 kb lncRNA and plays a key role in stabilizing mRNA for key differentiation genes.8 Recently, it was found that TINCR is aberrantly expressed (high or low) in various types of human tumors depending on the specific cancer types. Kretz et al reported that TINCR expression was declined in squamous cell carcinoma tissues, consistent with decreased differentiation seen in squamous cell carcinomas.8 Moreover, Zhang et al suggested that TINCR expression was obviously reduced in colorectal cancer tissues and cell lines compared with their counterparts.15 However, there was more evidence suggesting that TINCR was overexpressed in human cancers including hepatocellular carcinoma,16 bladder cancer,17 breast cancer,18 esophageal cancer19 and gastric cancer.20,21 In addition, the expression status of TINCR in lung cancer is still controversial. Liu et al indicated that TINCR expression was decreased in lung cancer tissues and cells compared with adjacent normal tissues and human bronchial epithelial cells.22 On the contrary, Zhu and He showed that TINCR was overexpressed in non-small cell lung cancer tissues and cell lines compared with normal adjacent tissues and normal bronchial epithelial cell lines.23 Unfortunately, both studies did not show details of lung cancer classification (lung adenocarcinoma, lung squamous cell carcinoma, small cell lung cancer and large cell lung cancer). Thus, the difference between Liu et al’s results and Zhu and He’s results may be most likely due to heterogeneity of lung cancer. The expression of TINCR in prostate cancer is still unknown. In order to explore TINCR expression in prostate cancer, we firstly analyzed the expression of TINCR in 52 pairs of normal prostatic tissues and prostate cancer tissues from TCGA database, and found that TINCR expression was significantly decreased in prostate cancer tissues compared with normal prostatic tissues. Furthermore, we confirmed the expression of TINCR in prostate cancer tissues and cell lines, and found that levels of TINCR expression were reduced in prostate cancer tissues and cell lines compared with paired normal prostatic tissues and normal human prostate epithelial cell lines, respectively. Afterwards, we analyzed the relationship between TINCR expression and clinical parameters in 160 prostate cancer patients, and found that low expression of TINCR was significantly associated with advanced clinical T stage, lymph node involvement, distant metastasis and high Gleason score. However, high expression of TINCR was associated with clinical progression in several human malignancies. Tian et al demonstrated that high expression of TINCR was significantly correlated with tumor size, tumor differentiation status, TNM stage and vascular invasion in hepatocellular carcinoma patients.16 Similarly, TINCR overexpression had a positive association with advanced TNM stage in bladder cancer and gastric cancer patients.17,24
In recent years, TINCR expression has been found to be a prognostic predictor in most human cancers, which has a good or poor prognostic value depending on cancer types. In non-small cell lung cancer patients, high levels of TINCR were markedly associated with poor overall survival.23 Meanwhile, Xu et al reported that TINCR overexpression was correlated with short survival time in breast cancer patients.18 Instead, low expression of TINCR was obviously associated with unfavorable outcome in patients with head and neck squamous cell carcinoma.25 Interestingly, Li et al suggested that low levels of TINCR dramatically correlated with poor clinical outcome in gastric cancer patients.26 However, Xu et al revealed the high-expression TINCR patients had higher recurrence rates than the low-expression TINCR patients, but high-expression TINCR was not an independent prognostic factor for gastric cancer as found in univariate and multivariate analyses.24 Due to different results about the prognostic value of TINCR in gastric cancer, we tried to analyze the relationship between TINCR and survival time in gastric cancer patients from TCGA database, and found that TINCR low expression might associate with poor prognosis in gastric cancer patients, but P-value was at critical value. Therefore, more studies should be performed to confirm the prognostic value of TINCR in gastric cancer patients. In order to investigate the prognostic value of TINCR in prostate cancer, we analyzed a cohort included 374 prostate cancer cases from TCGA database, and found that patients with low expression of TINCR had a shorter survival time than those with high-expression of TINCR. Furthermore, we also found that low expression of TINCR predicted a poor outcome in Chinese prostate cancer patients.
Based on the clinical and prognostic significance of TINCR in prostate cancer, TINCR might have an anti-tumor effect on prostate cancer cell. In order to explore the biological function of TINCR in prostate cancer, we conducted gain-of-function and loss-of-function studies in prostate cancer cell, and found that upregulation of TINCR expression dramatically inhibited cell proliferation, migration and invasion, and downregulation of TINCR expression strikingly promoted cell proliferation, migration and invasion. Similarly, Hazawa et al indicated that TINCR suppression resulted in promotion of squamous carcinoma cell growth and migration.25 Moreover, silencing of TINCR expression obviously promoted colorectal cancer cell growth and metastasis in vivo and in vitro.15 In order to elucidate the molecular mechanism of TINCR involved in prostate cancer tumorigenesis, we observed that TINCR expression was obviously negatively correlated with TRIP13 expression in prostate cancer tissue samples from TCGA database. TRIP13 has been suggested to be overexpressed in multiple human cancers and to function as an oncogene in regulating tumor cell proliferation, migration and invasion.27–30 Therefore, we further confirmed the relationship of TINCR expression with TRIP13 mRNA and protein expressions in prostate cancer tissues, and found that TINCR expression was negatively correlated with TRIP13 mRNA and protein expressions. Moreover, upregulation of TINCR expression dramatically reduced TRIP13 mRNA and protein expressions, and downregulation of TINCR expression markedly increased TRIP13 mRNA and protein expressions in prostate cancer cell lines. We next probed whether TRIP13 was involved in TINCR-mediated functions in prostate cancer cell. Thus, we conducted the rescued-function studies and found that TRIP13 was a functional protein for TINCR in regulating prostate cancer cell proliferation, migration and invasion. Thus, TINCR has an anti-tumor effect on prostate cancer cell proliferation, migration and invasion via suppressing TRIP13 expression.
Conclusion
In conclusion, low-expression TINCR is observed in prostate cancer, and correlates with clinical progression and poor prognosis. TINCR negatively modulates TRIP13 expression to inhibit prostate cancer cell proliferation, migration and invasion.
Acknowledgments
This work was supported by funding from Shenyang Natural Science Foundation (grant no. 20158960239).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bandini M, Fossati N, Gandaglia G, et al. Neoadjuvant and adjuvant treatment in high-risk prostate cancer. Expert Rev Clin Pharmacol. 2018;11(4):425–438. doi: 10.1080/17512433.2018.1429265. [DOI] [PubMed] [Google Scholar]
- 4.Tilki D, Pompe RS, Bandini M, et al. Local treatment for metastatic prostate cancer: a systematic review. Int J Urol. 2018;25(5):390–403. doi: 10.1111/iju.13535. [DOI] [PubMed] [Google Scholar]
- 5.Cucchiara V, Cooperberg MR, Dall’Era M, et al. Genomic markers in prostate cancer decision making. Eur Urol. 2018;73(4):572–582. doi: 10.1016/j.eururo.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Wade CA, Kyprianou N. Profiling prostate cancer therapeutic resistance. Int J Mol Sci. 2018;19(3) doi: 10.3390/ijms19030904. pii: E904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwakiri J, Terai G, Hamada M. Computational prediction of lncRNA-mRNA interactions by integrating tissue specificity in human transcrip-tome. Biol Direct. 2017;12(1):15. doi: 10.1186/s13062-017-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long non-coding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612. doi: 10.18632/oncotarget.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10(2):103–121. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L, Li Y, Xue D, Liu Y. PCMT1 is an unfavorable predictor and functions as an oncogene in bladder cancer. IUBMB Life. 2018;70(4):291–299. doi: 10.1002/iub.1717. [DOI] [PubMed] [Google Scholar]
- 12.Sheng N, Yan L, Wu K, et al. TRIP 13 promotes tumor growth and is associated with poor prognosis in colorectal cancer. Cell Death Dis. 2018;9(3):402. doi: 10.1038/s41419-018-0434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37(5) doi: 10.1042/BSR20171019. pii: BSR20171019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dazhi W, Mengxi Z, Fufeng C, Meixing Y. Elevated expression of thyroid hormone receptor-interacting protein 13 drives tumorigenesis and affects clinical outcome. Biomark Med. 2017;11(1):19–31. doi: 10.2217/bmm-2016-0169. [DOI] [PubMed] [Google Scholar]
- 15.Zhang ZY, Lu YX, Zhang ZY, et al. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget. 2016;7(16):22639–22649. doi: 10.18632/oncotarget.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian F, Xu J, Xue F, Guan E, Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2017;37(4) doi: 10.1042/BSR20170301. pii: BSR20170301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Liu Y, He A, et al. Theophylline controllable RNAi-based genetic switches regulate expression of lncRNA TINCR and malignant phenotypes in bladder cancer cells. Sci Rep. 2016;6:30798. doi: 10.1038/srep30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16(1):129. doi: 10.1186/s12943-017-0696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Qiu M, Chen Y, et al. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29(8):950–958. doi: 10.1111/dote.12436. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Liu H, Yang H, Gao Y, Zhang G, Hu J. The long noncoding RNA, TINCR, functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in gastric cancer. Onco Targets Ther. 2017;10:3353–3362. doi: 10.2147/OTT.S137726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Shi H, Xi H, et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics. 2017;7(1):213–227. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Ma J, Xu F, Li L. TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. Biomed Pharmacother. 2018;99:9–17. doi: 10.1016/j.biopha.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 23.Zhu ZJ, He JK. TINCR facilitates non-small cell lung cancer progression through BRAF-activated MAPK pathway. Biochem Biophys Res Commun. 2018;497(4):971–977. doi: 10.1016/j.bbrc.2018.02.059. [DOI] [PubMed] [Google Scholar]
- 24.Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 25.Hazawa M, Lin DC, Handral H, et al. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36(16):2243–2254. doi: 10.1038/onc.2016.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CY, Liang GY, Yao WZ, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48(5):1965–1976. doi: 10.3892/ijo.2016.3407. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Zhang G, Li X, et al. Thyroid hormone receptor interactor 13 (TRIP13) overexpression associated with tumor progression and poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2018;499(3):416–424. doi: 10.1016/j.bbrc.2018.03.129. [DOI] [PubMed] [Google Scholar]
- 28.Zhou K, Zhang W, Zhang Q, et al. Loss of thyroid hormone receptor interactor 13 inhibits cell proliferation and survival in human chronic lymphocytic leukemia. Oncotarget. 2017;8(15):25469–25481. doi: 10.18632/oncotarget.16038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurita K, Maeda M, Mansour MA, et al. TRIP13 is expressed in colorectal cancer and promotes cancer cell invasion. Oncol Lett. 2016;12(6):5240–5246. doi: 10.3892/ol.2016.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee R, Russo N, Liu M, et al. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat Commun. 2014;5:4527. doi: 10.1038/ncomms5527. [DOI] [PMC free article] [PubMed] [Google Scholar]