Abstract
It was shown more than 40 years ago that the ability to perceive the bitterness of the fruit of the Antidesma bunius tree is inversely correlated with the ability to perceive the well-studied bitter tastant phenylthiocarbamide (PTC). To determine if variants of the TAS2R38 gene, which encodes the PTC taste receptor, or variants in any of the other TAS2R bitter or TAS1R sweet receptor genes account for Antidesma taste perception, we recruited an independent subject sample and examined associations between these taste receptor gene haplotypes and Antidesma perception. Consistent with previous findings, almost none of our subjects who reported Antidesma juice as bitter was a PTC “responder” by previous definitions (i.e. a PTC taster). In our study, of the 132 individuals who perceived PTC as bitter, 15 perceived Antidesma as bitter, although these 15 subjects had very weak bitterness perception scores. Examination of TAS2R38 gene haplotypes showed that, of the subjects who perceive Antidesma as bitter, all carried at least one copy of the TAS2R38 AVI (PTC non-taster) haplotype. However, 86 subjects carried at least one AVI haplotype and failed to perceive Antidesma as bitter. No other TAS2R or TAS1R gene variants showed an association with Antidesma bitter, sweet, or sour perception. Our results show that TAS2R38 haplotypes are associated with differential perception of Antidesma berry juice bitterness, and that all those who perceive this bitterness carry at least one AVI haplotype. This indicates that the AVI haplotype is necessary for this perception, but that additional variable factors are involved.
Keywords: Antidesma, balancing selection, fruit, TAS2R38, TAS2Rs, TAS1Rs
Introduction
The ability to taste phenylthiocarbamide (PTC) and the structurally related compound 6-e-propylthiouracil (PROP) is one of the best investigated human phenotypes. These compounds are well-known because of their ability to generate a bitter taste in some individuals but not in others, dividing individuals into those who cannot perceive any taste at all (“non-tasters”), versus others that perceive a strong bitter taste (“tasters”) (Fox 1932). Since then, many family, twin and population studies have shown that the inability to taste PTC is inherited in a nearly Mendelian recessive manner due to a locus on chromosome 7 now designated TAS2R38 (Drayna et al. 2003; Kim et al. 2003). Common variants in this gene explain all of the bimodality in the distribution of PTC taste perception thresholds, and approximately 75% of the total variation in PTC taste sensitivity (Blakeslee 1932; Drayna et al. 2003; Knaapila et al. 2012). TAS2R38 is a member of the TAS2R bitter taste receptor gene family (Adler et al. 2000) which in humans consists of 25 functional genes and 11 pseudogenes, many of which have undergone some form of natural selection (Wooding et al. 2004; Dong et al. 2009; Campbell et al. 2012; Risso et al. 2014; Risso et al. 2017). Like many other genes encoding G protein-coupled receptors, TAS2R38 has a single coding exon ~1000 bp in length, and within this coding sequence there are 3 single nucleotide polymorphisms (rs714598, rs1726866, rs10246939), at positions encoding amino acids 49, 262 and 296 respectively, which are the most common variant alleles. Two common haplotypes of these polymorphisms encode the “taster” PAV (Proline, Alanine, Valine) and “non-taster” AVI (Alanine, Valine, Isoleucine) forms of the receptor. Two uncommon haplotypes (frequency <5%, AAV and AAI) and 4 rare haplotypes (frequency < 1%, PAI, PVI, AVV, and PVV) carrying different combinations of these 3 variant amino acids have also been identified (Wooding et al. 2004; Carrai et al. 2011; Risso et al. 2016).
The fact that approximately 40% of individuals worldwide are AVI-carriers represents a paradox, since it is thought that bitter taste has evolved to prevent ingestion of potentially toxic or harmful compounds mostly found in plants (Wooding et al. 2004; Antinucci and Risso 2017). In addition, it has recently been suggested that an important function of the TAS2R38 receptor is to recognize and protect individuals from bacterial infections of the respiratory tract. For example, bacterial acyl-homoserine lactones (AHLs), quorum-sensing molecules produced by Pseudomonas aeruginosa and other gram negative bacteria, have been demonstrated to be ligands for the PAV form of T2R38 (Lee et al. 2012). Recently Verbeurgt et al. (2017) demonstrated that T2R38 is much more broadly tuned for bacterial compounds than previously thought, and that PAV is the functional allele of the receptor. Therefore, it is unclear how a presumably non-functional allele rose to a high frequency in the worldwide population.
Two major hypotheses have been presented to explain this paradox. One explanation is that genetic drift could be responsible for the present day distribution of both TAS2R38 forms, which reached their present population frequencies because of random fluctuations and therefore could be due to demographic events, rather than selective ones (Wang et al. 2004; Risso et al. 2016). However, most authors have hypothesized that both haplotypes have been maintained by balancing natural selection (Fisher et al. 1939; Wooding et al. 2004; Campbell et al. 2012). Some of these balancing selection hypotheses have also suggested that the AVI non-taster allele could encode a fully functional receptor for another, as yet unidentified bitter substance (Wooding et al. 2004).
A phenomenon that may shed light on this hypothesis occurs in the taste perception of the fruit of Antidesma bunius, a fruit tree of the Phyllanthaceae family native to Southeast Asia and northern Australia. The human taste perception of this fruit varies greatly among individuals, and it was reported more than 40 years ago that a perception of bitterness in this fruit is inversely correlated with the ability to taste PTC. In particular, individuals who perceive Antidesma fruit as bitter were reported to be uniformly PTC non-tasters and conversely, those who perceived PTC as bitter were uniformly Antidesma bitterness non-tasters (Henkin and Gillis 1977). A more recent abstract reported a similar finding in a smaller sample, and suggested that TAS2R38 gene haplotypes may also be related to Antidesma perception (Tharp et al. 2005).
To test this hypothesis and to shed more light on the evolutionary history of TAS2R38, we examined the associations between PTC perception, TAS2R38 haplotypes and Antidesma perception in an independent subject sample of 169 individuals. We also performed complete sequencing of the coding region of all the other 24 functional TAS2R bitter receptor genes in these individuals, and of the TAS1R2 and TAS1R3 sweet receptor genes, and we tested variants in these genes for association between PTC and Antidesma perception.
Materials and methods
Participants
Following pre-screening to ensure study eligibility, a total of 169 individuals were enrolled. Subjects were eligible if they were age 18 or older, reported themselves to not be pregnant, did not smoke, and had no medical defects related to their sense of taste. Written informed consent was obtained from all participants. The study was approved by the National Institutes of Health Combined Neurosciences/Blue Panel Institutional Review Board (National Institutes of Health protocol 01-DC-0230), and all procedures were performed in accordance with the Helsinki Declaration of 1975, as revised in 2000. Most of the participants (N = 114) were Caucasians, and the remaining subjects were Asian Americans (N = 36) or African Americans (N = 19). 57% of the subjects were females and the average age was 30.02 (± 10.27 SD). Additional details on enrolled subjects are shown in Supplementary Table 1.
DNA collection and sequencing
Saliva samples were collected from all participants using Oragene saliva collection kits (Genotek Inc., Kanata, Ontario, Canada) and genomic DNA was purified following the manufacturer’s protocol (http://www.dnagenotek.com/US/pdf/PD-PR-006.pdf). A dedicated set of primers was designed to completely sequence the single coding exon of the TAS2R38 gene, as previously published (Risso et al. 2015). In addition, PCR primers for all the human functional TAS2R bitter taste receptor genes and the TAS1R2 and TAS1R3 sweet taste receptor genes, were designed to fully sequence the coding regions of these genes (Supplementary Table 2) which were sequenced using dideoxy Sanger sequencing (Sanger et al. 1977). DNA chromatograms were individually analyzed with the Lasergene suite (DNASTAR, Madison, Wisconsin; http://www.dnastar.com/t-allproducts.aspx), to evaluate the presence of calling errors and document genotypes at variant sites in these genes.
Collection of taste phenotypes
A 15-kg sample of A. bunius berries was collected at The Kampong National Tropical Botanical Garden (Miami, FL) and subsequently stored at −20°C before preparing the taste solutions. Berries were pressed, skin and seeds were removed, and an aqueous extract was derived as previously described (Henkin and Gillis 1977). All subjects tasted an aliquot from the same aqueous extract sample. To measure PTC taste sensitivity, a single supra-threshold PTC concentration of 256 μmol/L was used because this concentration has been shown to provide the best discrimination between taster and non-taster status (Kim et al. 2003; Bufe et al. 2005). Volunteers were asked to refrain from eating and drinking for at least 3 h before the beginning of the session and to rinse their mouth with reverse osmosis water prior to the first and between the 2 samples. Subjects were first presented a 2-mL solution of Antidesma juice, followed by a 2-mL PTC solution, in 30 mL plastic cups. Subjects were asked to rate their perceptions after swishing them in their mouth for 10 s. The PTC solution was presented within an interval of 1–5 min. To collect the perceived taste intensities of both solutions, the labeled magnitude scale (LMS), which ranges between 0 (“Barely detectable”) to 100 (“Strongest imaginable”), was used (Green et al. 1993). A training session was performed to explain the main aims of the study and to orient the participants on how to use the LMS scale. Subjects were asked to report the taste modalities elicited by the Antidesma juice (including bitter, sweet, and sour), after being informed that they may perceive more than one taste. This was done in accordance with previous studies and because sweet tastants can mask the perception of bitter (Henkin and Gillis 1977; Ley 2008).
Statistical analyses
Statistical analyses were performed using the R statistical analysis software (R Development Core Team 2011). We used PLINK v.1.07 to perform an initial quality control of genotypes and excluded variants with a call rate <90% or a deviation from Hardy–Weinberg equilibrium (HWE) (P < 0.001) (Purcell et al. 2007). PHASE v.2.1. was used to statistically infer TAS2R and TAS1R haplotypes, using individuals from the 1000 Genomes Project as a reference (Stephens et al. 2001; Abecasis et al. 2012). Only haplotypes with posterior probability of 90% or above were considered for further analyses. Differences in TAS2R and TAS1R haplotype distributions between taste phenotypes were examined using logistic regression with adjustments for demographic variables such as age, sex and ethnicity, assuming an additive model for the effect of haplotypes. The significance levels of the association tests were adjusted using the Bonferroni correction (adjusted P = P value × number of individual tests) and P < 0.05 was considered statistically significant.
Results
Distribution of taste phenotypes
As expected, most of the studied subjects (N = 132, 78.1%) perceived the PTC solution to be bitter and were therefore classified as “PTC responders”, according to Henkin and Gillis 1977. The remaining individuals (N = 37, 21.9%) were unable to perceive any taste when tasting the PTC solution, reporting a score of “0” on the LMS, and were classified as “PTC non-responders”.
Of the 37 PTC non-responders in our sample, 15, (40.5%) perceived Antidesma as bitter (i.e. were responders). This was similar to previous results in which 25 of 45 PTC non-responders (45.4%) perceived Antidesma as bitter (Henkin and Gillis 1977). Conversely, of the 132 PTC responders in our sample, 15 (11.4%) were Antidesma responders (P < 0.001), although they reported extremely low LMS scores (ranging from 2 to 5) for the perceived intensity of PTC bitterness. No differences in Antidesma LMS bitterness scores were found between PTC responders and non-responders (mean of 26.93 vs. 25.86, P > 0.10), and no significant correlation was found in responsiveness to either PTC or Antidesma with age, sex or ethnicity (All P’s > 0.10).
Regarding other Antidesma taste perception modalities, the majority of subjects (N = 104, 61.5%) reported sour to be the main taste they perceived when tasting the juice, while 35 (20.7%) perceived it as “mostly sweet”. In agreement with previous findings (Henkin and Gillis 1977), 30 (17.8%) of the individuals tested perceived the Antidesma juice as “mostly bitter”. Of these 30 subjects (“Antidesma responders”), 28 (93.3%) of them also reported perception of a sour taste and 20 (66.7%) described it also as sweet. Of the 139 individuals who could not identify a bitter taste, 133 (95.7%) and 99 (71.2%) perceived it as sour or sweet, respectively. The difference in the distribution of either sour or sweet taste perception of Antidesma juice was not significantly different between individuals who perceived Antidesma taste as bitter or not, nor different between PTC responders and non-responders (P > 0.10).
Associations between TAS2R, TAS1R haplotypes and taste phenotypes
After phasing the genomic DNA sequence data, we identified 5 TAS2R38 haplotypes in our subjects. The most common haplotype was PAV (51.5%), followed by AVI (46.1%), AAV (1.5%), AAI (0.60%), and PAI (0.30%). A total of 54 single nucleotide polymorphisms (SNPs) with minor allele frequency (MAF) >0.01, were identified in the other 24 functional TAS2R genes. In addition, 35 SNPs were identified in TAS1R2 and TAS1R3 genes.
As expected, PTC perceived bitterness was highly associated with the TAS2R38 PAV taster haplotype (P < 10–5), confirming the validity of our taste phenotypes collection. A detailed distribution and summary of PTC LMS scores, categorized by TAS2R38 diplotype, are shown in Supplementary Table 3.
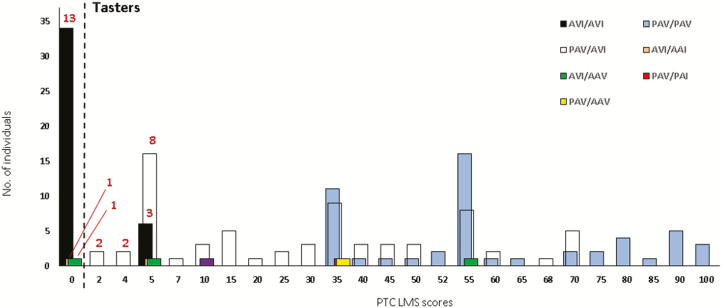
All the subjects who perceived Antidesma juice as bitter carried at least one AVI haplotype (N = 30, Figure 1, Supplementary Table 4). The majority of these Antidesma responders (N = 16, 53.3%) were AVI homozygotes, with the remaining being PAV/AVI (N = 12, 40%), AVI/AAI (N = 1, 3.3%), or AVI/AAV (N = 1, 3.3%) heterozygotes.
Figure 1.
Distribution of PTC LMS scores by TAS2R38 diplotypes for all 169 subjects tested. Numbers in red quantify the number of subjects, of a specific TAS2R38 diplotype, who recognized a bitter taste in the Antidesma juice. The dotted line sets the threshold between PTC tasters and non-tasters.
Overall, TAS2R38 haplotypes showed a strong and significant correlation with the reported LMS scores of Antidesma bitterness perception. The non-taster AVI haplotype was directly correlated and the taster PAV inversely correlated with the reported Antidesma bitterness LMS scores (P < 10–5, Table 1). No significant differences were found in the distribution of Antidesma bitterness LMS scores between AVI/AVI homozygotes and AVI/PAV heterozygotes (P > 0.10).
Table 1.
Distribution of bitterness LMS scores by TAS2R38 diplotypes in the 30 Antidesma responders.
| Diplotype | Antidesma bitter (%) | Antidesma bitterness LMS Score (SD) | Average PTC LMS score (SD) |
|---|---|---|---|
| AVI/AVI | 16 (53.4) | 26.81 (16.76) | 0.94 (2.01) |
| AVI/AAI | 1 (3.3) | 29 (0) | 0 (0) |
| AVI/AAV | 1 (3.3) | 20 (0) | 0 (0) |
| PAV/AVI | 12 (40) | 23.17 (11.67) | 4.33 (1.15) |
| PAV/PAI | / | / | / |
| PAV/AAV | / | / | / |
| PAV/PAV | / | / | / |
With similar testing, no significant correlations were found between any TAS2R or TAS1R SNPs with either Antidesma bitterness, sweetness, or sour perception (All P’s > 0.10). We additionally tested the hypothesis that variants in one of the other TAS2R or TAS1R genes could be present only in the AVI/AVI responders or AVI/AVI non-responders group. No variants were associated with Antidesma bitterness perception in either of these groups.
Discussion
The presence of 2 high-frequency TAS2R38 haplotypes in the population has presented a longstanding question in chemosensory and evolutionary biology. Since bitter taste perception is thought to protect organisms from ingesting toxic substances, it is not clear how the presumably non-functional AVI haplotype came to such a high frequency worldwide. To address this question, some studies have shown that TAS2R38 has undergone relaxation of selection in humans when compared with many other mammals, suggesting that the presence of both PAV and AVI haplotypes at high frequencies is due to demographic events, rather than selective ones (Wang et al. 2004; Risso et al. 2016). Other authors have noted the fact that bitter receptors are expressed in the respiratory and enteric systems and have suggested that pathogens may have been the real targets of natural selection. In support of this hypothesis, it has been shown that common polymorphisms in the TAS2R38 gene are linked to significant differences in the ability of the upper respiratory cells to kill and clear bacteria and regulate innate immune responses (Lee et al. 2012; Gillis et al. 2015).
It is notable that there is strong evidence that balancing selection maintained both PTC tasting and non-tasting haplotypes at high frequencies (Wooding et al. 2004; Campbell et al. 2012). Some of these balancing selection hypotheses have raised the possibility that the AVI non-taster allele encodes a fully functional receptor for other hypothetical bitter substances (Wooding et al. 2004), or for some not yet identified bacterial metabolites.
As one possible source of such a bitter substance, sensitivity to bitterness of the fruit of the A. bunius tree was previously reported to perfectly and inversely correlate with the ability to taste PTC bitterness: all subjects who perceived Antidesma extract as bitter found PTC not bitter, and none of the subjects who perceived PTC as bitter found Antidesma to be bitter. In that study, less than half of the PTC non-responders were able to detect Antidesma as bitter (45%) (Henkin and Gillis 1977). A more recent abstract reported a smaller study with a similar finding, again suggesting that TAS2R38 gene haplotypes may be related to Antidesma perception even though the active substance responsible for the bitter taste responsiveness of Antidesma has not been identified, although it is water soluble and heat stable (Henkin and Gillis 1977; Tharp et al. 2005).
A number of structural modeling and functional studies of the TAS2R38 receptor forms have been published that have shed light on possible receptor–ligand interactions in this system (Floriano et al. 2006; Miguet et al. 2006; Tan et al. 2011, 2012; Marchiori et al. 2013). Some of these studies have suggested that the AVI form presents steric hindrance or a lack of hydrogen bonding that would prevent ligand binding at the site used by PTC/PROP, while others have suggested that the variant amino acids in the AVI form disrupt signal transduction rather than ligand binding. Identification of potential ligands for the TAS2R38 AVI form in A. bunius followed by structural modeling studies could help clarify this issue.
We sought to expand on previous studies by sequencing the entire coding sequence and determining haplotypes of the TAS2R38 gene, as well as sequencing the entire coding sequence of all the other human bitter and sweet taste receptor genes, and by testing the association of variants in these genes with bitter, sweet and sour perception of Antidesma berry juice. Our results confirm that perception of the bitterness of Antidesma juice is highly correlated with the inability to perceive PTC as bitter. Similar to the previous results of Henkin and Gillis, who showed that approximately 15% of individuals are Antidesma responders, we found that approximately 18% of the participants of our study perceived Antidesma to be strongly bitter. Similarly, Henkin and Gillis reported that 45.4% of PTC non-tasters perceived Antidesma as bitter, which is consistent with our finding that 40.5% of PTC non-tasters perceived Antidesma as bitter.
However, our results differ from those of Henkin and Gillis in one aspect: while they found no individuals who were both PTC responders and Antidesma responders, we identified 15 PTC-responders who also perceived Antidesma to be bitter. However, it is important to note that these 15 subjects in our sample reported very low PTC bitterness LMS scores (ranging from 2 to 5). Thus, although not identical, our results are quite similar to those of Henkin and Gillis, and may be due to small differences in testing methodology or definition of cut-off values that define responders and non-responders.
We have also confirmed that TAS2R38 haplotypes strongly contribute to the bitterness perception of Antidesma, as suggested by previous authors (Tharp et al. 2005). In our sample, all subjects who perceived Antidesma bitterness carried at least one AVI haplotype and no subjects without this haplotype perceived this bitterness, indicating this haplotype is necessary for this perception. However, because a large fraction of AVI carriers fail to perceive this bitterness, another factor or factors must be involved. Such a factor could be represented by differences in saliva composition, and for example, variable components of saliva could affect the binding of the bitter compounds present in Antidesma juice. Of the many possible candidates, our results show that differences in other bitter taste receptor genes, or differences in sweet taste receptor genes, are unlikely to represent this additional contributor/s. Moreover, we showed that no coding sequence variants in any other bitter or sweet taste receptor gene contributed to differences in either Antidesma bitter, sweet or sour perceptions in our subject group. Our failure to identify any variants in any of the other TAS2R or TAS1R genes that could account for a lack of Antidesma bitterness perception in some carriers of an AVI haplotype could be due to other genetic differences in these individuals, or to the presence of components in saliva that vary between individuals that sequester the bitter component of Antidesma.
While our results strongly support previous hypotheses suggesting that TAS2R38 variants contribute to variation in Antidesma bitter perception, they argue against the simple hypothesis that the AVI haplotype of this gene encodes a receptor that acts by itself to specify this perception of bitterness. Our data also indicate that another factor or factors, which may or may not be heritable, is clearly involved in Antidesma bitterness perception. Of the many possible candidates, our results show that differences in other bitter taste receptor genes, or differences in sweet taste receptor genes, are unlikely to represent this additional contributor/s.
Supplementary material
Supplementary material can be found at Chemical Senses online.
Funding
This work was supported by the National Institute on Deafness and Other Communication Disorders Intramural grant Z1A-000046-16, by the University of Gastronomic Sciences, and by the U.S. Food and Drug Administration through funds obtained under the Family Smoking Prevention and Tobacco Control Act (https://www.fda.gov/TobaccoProducts/) (DD, SW, DR). The content was not reviewed by the Food and Drug Administration, but underwent the standard manuscript clearance process for scientific papers published from the NIH intramural research program. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be interpreted as a potential conflict of interest.
Acknowledgments
The authors would like to thank The Kampong National Tropical Botanical Garden (Miami, FL) for providing the Antidesma berries, Raffaello Verardi for help with berries lyophilization, Joanne Gutierrez for technical assistance, Thomas Friedman and Robert Morell for helpful comments on the manuscript, and Prof. Roberto Barale for helpful discussions.
References
- Abecasis GR, Auton A et al. 1000 Genomes Project Consortium 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. 2000. A novel family of mammalian taste receptors. Cell. 100:693–702. [DOI] [PubMed] [Google Scholar]
- Antinucci M, Risso D. 2017. A matter of taste: lineage-specific loss of function of taste receptor genes in vertebrates. Front Mol Biosci. 4:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF. 1932. Genetics of sensory thresholds: taste for phenyl thio carbamide. Proc Natl Acad Sci USA. 18:120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. 2005. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 15:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Ranciaro A, Froment A, Hirbo J, Omar S, Bodo JM, Nyambo T, Lema G, Zinshteyn D, Drayna D, et al. 2012. Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa. Mol Biol Evol. 29:1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrai M, Steinke V, Vodicka P, Pardini B, Rahner N, Holinski-Feder E, Morak M, Schackert HK, Görgens H, Stemmler S, et al. 2011. Association between TAS2R38 gene polymorphisms and colorectal cancer risk: a case-control study in two independent populations of Caucasian origin. PLoS One. 6:e20464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Jones G, Zhang S. 2009. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayna D, Coon H, Kim UK, Elsner T, Cromer K, Otterud B, Baird L, Peiffer AP, Leppert M; Utah Genetic Reference Project 2003. Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum Genet. 112:567–572. [DOI] [PubMed] [Google Scholar]
- Fisher R, Ford E, Huxley J. 1939. Taste-testing in anthropoid apes. Nature. 144:750. [Google Scholar]
- Floriano WB, Hall S, Vaidehi N, Kim U, Drayna D, Goddard WA 3rd. 2006. Modeling the human PTC bitter-taste receptor interactions with bitter tastants. J Mol Model. 12:931–941. [DOI] [PubMed] [Google Scholar]
- Fox AL. 1932. The relationship between chemical constitution and taste. Proc Natl Acad Sci USA. 18:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis S, Coldwell S, Drury JL, Arroyo F, Phi T, Saadat S, Kwong D, Chung WO. 2015. Genotype-specific regulation of oral innate immunity by T2R38 taste receptor. Mol Immunol. 68:663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BG, Shaffer GS, Gilmore MM. 1993. Derivation and evaluation of a semantic scale of oral sensation magnitude with apparent ratio properties. Chem Senses. 18:683–702. [Google Scholar]
- Henkin RI, Gillis WT. 1977. Divergent taste responsiveness to fruit of the tree Antidesma bunius. Nature. 265:536–537. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 299:1221–1225. [DOI] [PubMed] [Google Scholar]
- Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, James RS, Wysocki CJ, Rhyu M, Tordoff MG, et al. 2012. Genetic analysis of chemosensory traits in human twins. Chem Senses. 37:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RJ, Xiong G, Kofonow JM, Chen B, Lysenko A, Jiang P, Abraham V, Doghramji L, Adappa ND, Palmer JN, et al 2012. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 122(11):4145–4159. [DOI] [PMC free article] [PubMed]
- Ley JP. 2008. Masking bitter taste by molecules. Chem Percept. 1:58–77. [Google Scholar]
- Marchiori A, Capece L, Giorgetti A, Gasparini P, Behrens M, Carloni P, Meyerhof W. 2013. Coarse-grained/molecular mechanics of the TAS2R38 bitter taste receptor: experimentally-validated detailed structural prediction of agonist binding. PLoS One. 8:e64675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguet L, Zhang Z, Grigorov MG. 2006. Computational studies of ligand-receptor interactions in bitter taste receptors. J Recept Signal Transduct Res. 26:611–630. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: The R Foundation for Statistical Computing; ISBN: 3-900051-07-0 Available from http://www.R-project.org/ [Google Scholar]
- Reed DR, Cohen NA. 2012. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 122:4145–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Behrens M, Sainz E, Meyerhof W, Drayna D. 2017. Probing the evolutionary history of human bitter taste receptor pseudogenes by restoring their function. Mol Biol Evol. 34(7):1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso DS, Howard L, VanWaes C, Drayna D. 2015. A potential trigger for pine mouth: a case of a homozygous phenylthiocarbamide taster. Nutr Res. 35:1122–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso DS, Mezzavilla M, Pagani L, Robino A, Morini G, Tofanelli S, Carrai M, Campa D, Barale R, Caradonna F, et al. 2016. Global diversity in the TAS2R38 bitter taste receptor: revisiting a classic evolutionary PROPosal. Sci Rep. 6:25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Tofanelli S, Morini G, Luiselli D, Drayna D. 2014. Genetic variation in taste receptor pseudogenes provides evidence for a dynamic role in human evolution. BMC Evol Biol. 14:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Abrol R, Trzaskowski B, Goddard WA. 2011. The predicted 3D structure of bitter taste receptors, TAS2R38 based on a BiHelix and SuperBiHelix methodologies. Procedia Envir Sci. 8:543–548. [Google Scholar]
- Tan J, Abrol R, Trzaskowski B, Goddard WA 3rd. 2012. 3D structure prediction of TAS2R38 bitter receptors bound to agonists phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP). J Chem Inf Model. 52:1875–1885. [DOI] [PubMed] [Google Scholar]
- Tharp CD, Tharp A, Alarco SM, Reed DR, Breslin PA. 2005. PTC non-tasters find the fruit of Antidesma bunius bitter, while PTC tasters find it sweet. Chem Senses. 30:A126–A258. [Google Scholar]
- Verbeurgt C, Veithen A, Carlot S, Tarabichi M, Dumont JE, Hassid S, Chatelain P. 2017. The human bitter taste receptor T2R38 is broadly tuned for bacterial compounds. PLoS One. 12(9):e0181302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Thomas SD, Zhang J. 2004. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 13:2671–2678. [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. 2004. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 74:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.