Abstract
A trace amount of coagulation factor VII (FVII) circulates in the blood in the activated form, FVIIa (EC 3.4.21.21), formed by internal proteolysis. To avoid disseminated thrombus formation, FVIIa remains in a conformation with zymogen-like properties. Association with tissue factor (TF), locally exposed upon vascular injury, is necessary to render FVIIa biologically active and initiate blood clotting. We have designed potent mutants of FVIIa by replacing residues believed to function as determinants for the inherent zymogenicity. The TF-independent rate of factor X activation was dramatically improved, up to about 100-fold faster than that obtained with the wild-type enzyme and close to that of the FVIIa-soluble TF complex. The mutants appear to retain the substrate specificity of the parent enzyme and can be further stimulated by TF. Insights into the mechanism behind the increased activity of the mutants, presumably also pertinent to the TF-induced, allosteric stimulation of FVIIa activity, were obtained by studying their calcium dependence and the accessibility of the N terminus of the protease domain to chemical modification. The FVIIa analogues promise to offer a more efficacious treatment of bleeding episodes especially in hemophiliacs with inhibitory antibodies precluding conventional replacement therapy.
In damaged blood vessels, subendothelial membrane-bound tissue factor (TF) contacts blood and forms a complex with factor VIIa (FVIIa, EC 3.4.21.21), which initiates the coagulation cascade (1). One role of TF is to enhance the activity of FVIIa, making it an efficient catalyst of factor IX and factor X (FX) activation. Alanine scanning and x-ray crystallography have identified a number of residues in FVIIa that are pivotal for its ability to bind and be stimulated by TF and to interact with and efficiently activate FX (2–4). In contrast, the features of the amino acid sequence of FVIIa that contribute to the stabilization of an enzymatically latent three-dimensional conformation of the free enzyme even after cleavage of the Arg-152–Ile-153 ({15–16}; hereafter, chymotrypsinogen numbering is given in curly brackets) peptide bond remain largely unknown. Complex formation with TF stabilizes the active conformation, but the crystal structures of free and TF-bound FVIIa have not been able to reveal much about the structural transformations associated with this event (3–7). One functionally important consequence of TF binding is the establishment of an intramolecular salt bridge between the N terminus of the protease domain of FVIIa and the side chain of Asp-343 {194} as illustrated by protection of the amino group from chemical modification (8). Active site occupancy also facilitates the N-terminal insertion (9), and presumably promotes other events that also can be induced by TF. This explains why the crystal structures, obtained with active-site inhibited FVIIa, do not shed light on the allosteric reorganizations occurring in FVIIa upon TF binding.
Without structural information about the latent, zymogen-like conformation of free FVIIa, the residues that should be mutated to convert it to the active conformation, or at least to a more active one, are not conspicuous. The considerable distance between the interface between TF and the protease domain of FVIIa and the active site allows for a multitude of possible allosteric pathways leading to the active state. Interestingly, conformational changes associated with locking FVIIa in the active state appear to spread as far as to the first epidermal growth factor-like domain (10). Nevertheless, two specific amino acid replacements in FVIIa have recently been shown to yield variants with increased intrinsic (TF-independent) activity. In one mutant, the hydrophobic residue Met-298 {156} was replaced by Gln (11) and in the other Val was substituted for Leu-305 {163} (12). In both cases, a residue more or less unique to FVIIa was replaced by one that occurs frequently in related enzymes. The activity of the mutants was significantly increased, but the enhancement is still moderate when compared with the activity of FVIIa bound to TF. A third FVIIa variant exhibiting increased activity has been reported in which the 311–322 {169–175} loop was replaced by the corresponding, shorter loop from trypsin (13). This molecule, however, displayed a changed substrate specificity. In this article, we have identified three residues that appear to contribute to the latency of FVIIa and used site-directed mutagenesis to create highly active FVIIa variants.
Materials and Methods
Reagents and Standard Methods.
The preparation of recombinant wild-type FVIIa, TF pathway inhibitor, and soluble TF (sTF) were carried out as described (14–16). FVIIa concentrations were determined by using a double mAb (both recognizing epitopes in the light chain of FVIIa) ELISA and sTF concentrations by absorbance measurements at 280 nm by using an absorption coefficient of 1.5 for a 1-mg/ml solution. FX and factor Xa (FXa) were from Enzyme Research Laboratories (South Bend, IN); thrombin was from Roche Molecular Biochemicals, factor V, prothrombin, FX (in the platelet experiments), and antithrombin (AT) were from Hematologic Technologies (Essex Junction, VT); potassium cyanate (KNCO) was from Merck, and the chromogenic substrates S-2288, S-2765, S-2238, and S-2366 were from Chromogenix (Mölndal, Sweden). SDS/PAGE was run on 8–25% gradient gels using the PhastSystem (Amersham Pharmacia) to verify the purity of the FVIIa variants.
Mutagenesis and Preparation of FVIIa Mutants.
The wild-type FVII expression plasmid pLN174 (17) was used as the starting template for site-directed mutagenesis, going through 1–4 rounds of PCR to construct the various mutant-encoding plasmids, depending on the number of mutations and their locations, using the QuikChange kit (Stratagene). The following primers (only sense primers given) with base substitutions in italics and the affected codons underlined were used to introduce the indicated mutations: V158D, GTG GGG GGC AAG GAC TGC CCC AAA GGG G; M298Q, GCC CTG GAG CTC CAG GTC CTC AAC GTG CCC; E296V/M298Q, GCC ACG GCC CTG GTG CTC CAG GTC CTC AAC GTG CCC; V158E, GTG GGG GGC AAG GAG TGC CCC AAA GGG G; E296R/M298K, GCC ACG GCC CTG AGG CTC AAG GTC CTC AAC GTG CCC; K337A, CGG ATG GCA GCG CGG ACT CCT GCA AGG G. L305V was introduced as described (12). Plasmids were prepared by using the QIAprep spin miniprep and QIAfilter plasmid midi kits (Qiagen, Valencia, CA). The entire cDNA encoding the mutants was verified by sequencing to exclude the presence of additional mutations. Baby hamster kidney cell transfection and selection, as well as expression, purification, concentration, and autoactivation of the FVII mutants, were carried out as described (18), with the exception that the protein was eluted from the anion-exchange column by stepping to equilibration buffer containing 25 mM CaCl2. The final FVIIa concentration was determined by ELISA and in the case of K337A-FVIIaIIa (V158D/E296V/M298Q-FVIIa) verified by densitometry analysis of the light chain SDS/PAGE band on a Gel Doc 2000 (Bio-Rad).
FVIIa Activity Assays.
All proteins were diluted in 50 mM Hepes, pH 7.4, containing 0.1 M NaCl, 5 mM CaCl2, and 0.1% (wt/vol) BSA (assay buffer), before analysis. All assays were run in a final volume of 200 μl. To measure the amidolytic activity in the absence of sTF, 100 nM wild-type or mutant FVIIa was mixed with 1 mM S-2288. This analysis was performed at ambient temperature and at 37°C and in the absence (CaCl2 replaced by 5 mM EDTA) and presence of Ca2+. To study substrate specificity, three additional chromogenic substrates (S-2765, S-2238, and S-2366), all at a concentration of 1 mM, were also used in this assay. The amidolytic activity in the presence of sTF and calcium was determined by measuring the hydrolysis of 1 mM S-2288 by a mixture of 10 nM wild-type or mutant FVIIa and 50 nM sTF. The amidolytic activity was monitored continuously at 405 nm in a SpectraMax 340 microplate spectrophotometer equipped with the software softmax pro, version 2.2 (Molecular Devices).
To measure the catalytic efficiency of the FVIIa variants in the absence of sTF, the kinetic parameters of FX activation were measured at a concentration of 50 nM wild-type FVIIa or 5 or 10 nM FVIIa mutant in a total volume of 100 μl. The effect of sTF on FVIIa-catalyzed FX activation was studied by mixing 5 nM FVIIa with 50 nM sTF. Both assays were started by adding FX (0.1–4.2 μM) and allowed to proceed for 20 min. The samples were then diluted with 50 μl assay buffer followed by the addition of 50 μl of 2 mM S-2765 (final concentration 0.5 mM) to measure the amount of FXa generated. The initial rate of FXa-catalyzed hydrolysis of S-2765 was measured for 2 min during which the absorbance development was linear. The FXa activity was corrected for the inherent activity of the FX and FVIIa preparations and of the FVIIa/sTF mixture, and the remaining absorbance generation was converted to a molar concentration of FXa by using a standard curve (0–5 nM).
The specific clotting activity was measured both in an assay containing relipidated human TF and a TF-free assay using phospholipid vesicles virtually as described (12).
Measurements of Thrombin Generation on Activated Platelets.
Prothrombin and FX were treated with a mixture of inhibitors to neutralize traces of active enzymes (19). FX (135 nM final concentration), prothrombin (1.2 μM), TF pathway inhibitor (0.1 μg/ml), and AT (2.5 μM) were mixed with CaCl2 (3 mM) and stored over night at 4°C. Peripheral blood from healthy volunteers was drawn into citrate. Platelets were isolated by density gradient centrifugation and gel filtration as described (20) and activated by incubation with 50 μg/ml thrombin receptor agonist peptide (SFLLRN) for 15 min at 37°C. An aliquot of the platelet suspension was transferred to TruCount tubes (Becton Dickinson), and the platelet density was determined by using fluorescent beads as the standard on a FACScan flow cytometer (Becton Dickinson) as described by the manufacturer. The final platelet density was ≈100,000/μl. Factor V (final concentration 7 μg/ml) was added to the protein/Ca2+ mixture. FVIIa variant (50 nM) was then added, followed by platelets, to give a final volume of 200 μl. At timed intervals, aliquots of 10 μl were transferred to 90 μl of 0.5 mM Chromozym TH (Roche Molecular Biochemicals) in 20 mM Hepes, pH 7.4, containing 150 mM NaCl, 1 mg/ml BSA, 1 mM EDTA, and 50 μM Pefabloc Xa (Pentapharm, Basel). Substrate hydrolysis was stopped after 20 min by adding 100 μl of 50% acetic acid, and the absorbance at 405 nm was measured. The thrombin concentration was calculated from a thrombin standard curve.
AT Inhibition.
The inhibition of 100 nM FVIIa variant alone, 10 nM FVIIa variant in the presence of 50 nM sTF, 2 nM FXa, or 5 nM thrombin, in the assay buffer described above, by AT (100 μg/ml) in the absence or presence of heparin (unfractionated, 1 unit/ml; Heparin Leo, Leo Pharmaceutical Products, Ballerup, Denmark) was measured by incubating the enzymes with the inhibitor for various time periods followed by the addition of S-2288 (1 mM) to measure the residual activity at 405 nm as described above.
Carbamylation of the N-Terminal Ile-153.
The FVIIa variants, alone at a concentration of 1 μM or at a concentration of 100 nM in the presence of 500 nM sTF, were incubated at room temperature with 0.2 M KNCO in 50 mM Hepes, pH 7.4, containing 0.1 M NaCl and 5 mM CaCl2. At different time points, an aliquot of the samples was diluted 10-fold in the same buffer containing 1 mg/ml BSA and 1 mM S-2288 followed by measurement of the residual amidolytic activity.
Surface Plasmon Resonance Measurements.
The conditions for sTF immobilization (≈500 resonance units) in the Biacore 1000 instrument (Biacore, Uppsala, Sweden), regeneration of the sTF-coated surface, and global evaluation of binding data were as described (21, 22). Wild-type or mutant FVIIa, in 50 mM Hepes, pH 7.5, containing 0.15 M NaCl, 5 mM CaCl2, and 0.02% Tween 80, was injected for 7 min at concentrations of 30 and 50 nM followed by a 10-min dissociation phase. The flow rate was 5 μl/min and the temperature 25°C.
Molecular Modeling.
The modeling package Quanta (Molecular Simulations, San Diego) was applied to explore differences between the serine proteases in the trypsin family. The inspected proteases, with Protein Data Bank entries in parentheses, include human FVIIa (1dan), human factor IXa (1pfx), human FXa (1hcg), activated human protein C (1aut), human α-thrombin (1ihs), bovine trypsinogen (1tgn), and bovine trypsin (1tgt). Models of FVIIa mutant structures were built by using the protein design facility in Quanta.
Results and Discussion
Mutagenesis Rationale.
A comparison of crystal structures of members of the trypsin family of serine proteases was undertaken with special focus on the region of the activation pocket. Two independent hypotheses then led to the design of the FVIIa mutants described. The first hypothesis is based on the observation that a Met residue sits in position 298 {156} in FVIIa, flanking the entrance to the pocket, whereas other enzymes in the family have a polar residue in the corresponding position (usually Gln or Lys). Stable insertion of the N terminus is facilitated by a water molecule present in virtually all inspected structures. The water molecule interacts with the main-chain amides of Gly residues 155 {18} and 156 {19} and is spatially close to residue 298 {156} (Fig. 1). When Gln (or Lys) resides in position 298 {156}, a hydrogen bond to the water molecule could strengthen the connection of the N terminus to the surface of the body of the protease domain (Fig. 2). We therefore constructed M298Q-FVIIa and V158D/E296V/M298Q-FVIIa (FVIIaIIa). In the latter mutant, Gln in position 298 {156} can also interact with the side chain of Asp in position 158 {21}. In addition, Val was introduced at position 296 {154} to avoid electrostatic repulsion between the native Glu residue and the introduced Asp at position 158 {21} (Fig. 2). This three-residue motif mimics thrombin, which in contrast to FVIIa possesses considerable constitutive activity. Analogously, we constructed another mutant containing the corresponding motif from FXa [V158E/E296R/M298K-FVIIa (FVIIaXa)]. The other approach was based on the idea that Lys-337 {188} in FVIIa, because of electrostatic repulsion, potentially prevents the N terminus of the protease domain from entering the activation pocket and approaching Asp-343 {194} to form a crucial ion pair. To test this hypothesis we generated K337A-FVIIa.
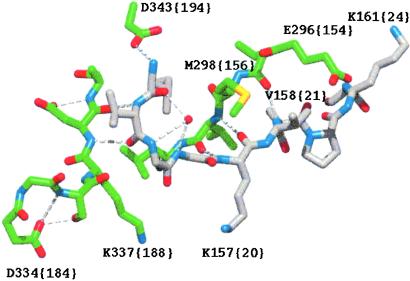
Figure 1.
Activation pocket region of FVIIa. The structure is from the complex between FVIIa and TF (3). The carbon atoms of N-terminal Ile-153 {16} to Lys-161 {24} are shown in gray and those of the amino acids constituting part of the activation pocket are in green. The water molecule (shown as a red sphere) interacting with main chain atoms of Gly-155 {18} and Gly-156 {19} lacks hydrogen bonds to the side chain of Met-298 {156}.
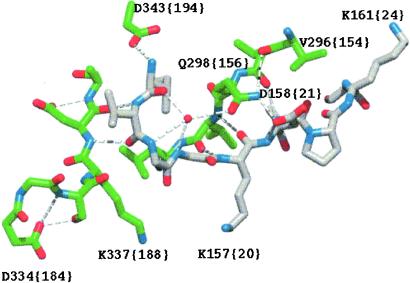
Figure 2.
Activation pocket region of FVIIa after mutating the residues in positions 158 {21}, 296 {154}, and 298 {156} to those occupying the corresponding positions in thrombin (i.e., Asp, Val and Gln, respectively). The backbone structure (3) and coloring scheme are the same as in Fig. 1. The introduced side chains are oriented as in the thrombin structure. Note that a hydrogen bond network between the water molecule, Gln-298 {156} and Asp-158 {21} is established.
Intrinsic Amidolytic Activity and Substrate Specificity of FVIIa Mutants.
The activity of the FVIIa mutants was initially measured in an amidolytic assay at room temperature in the absence of TF and presence of calcium ions (Table 1). The amidolytic activity of K337A-FVIIa was 4-fold that of wild-type FVIIa. Mutating Ser-336 {188A} to Gly, which based on the structure (Fig. 1) might facilitate ion pairing between the side chains of Lys-337 {188} and Asp-334 {186}, had no effect on FVIIa activity (not shown). We therefore propose that removal of the positive charge at position 337 {188}, or alternatively, reduction of the side-chain size, is required for enhanced intrinsic activity. The amidolytic activity of FVIIaIIa was increased about 8-fold as compared with that of wild-type FVIIa, whereas V158E/E296R/M298K-FVIIa (FVIIaXa) had 15-fold lower activity than wild-type FVIIa. The poor activity of FVIIaXa is probably caused by repulsion between the two inserted positive charges and K157 {20} and K161 {24}, and this variant was not examined further. FVIIaIIa was significantly more active than M298Q-FVIIa (11), which also in our hands displayed a 3-fold enhancement. The activity enhancement observed with the mutants as compared with wild-type FVIIa was the same at 37°C, indicating that the thermal stability was not affected by any of the mutations. K337A-FVIIa, like the wild-type enzyme, lost the majority (>85%) of activity upon the omission of Ca2+. Interestingly, FVIIaIIa behaved completely differently and retained ≈70% of its activity even in the absence of calcium. This resembles a property of FVIIa bound to sTF, namely that once the complex has formed, its amidolytic activity is independent of calcium ions (23, 24). This finding suggests that the mutations in FVIIaIIa mimic part of the allosteric effect of TF on FVIIa, establishing a structural epitope that normally requires Ca2+ binding to the 210–220 {70–80} loop in the protease domain. Because Glu-296 {154} contacts the Ca2+ binding loop, the E296V mutation is most likely responsible for the attenuated dependency on calcium ions (3, 5).
Table 1.
Amidolytic activity of FVIIa variants and kinetic parameters of FX activation
| FVIIa variant | Amidolytic activity
|
FX activation
|
||
|---|---|---|---|---|
| Mutant/wt | Km, μM | kcat, × 10−3 s−1 | kcat/Km, M−1⋅s−1 | |
| wt-FVIIa | 2.9 ± 0.5 | 0.094 ± 0.010 | 32 | |
| K337A-FVIIa | 3.9 ± 0.2 | 2.0 ± 0.6 | 0.28 ± 0.05 | 140 |
| L305V-FVIIa | 3.2 ± 0.2 | 2.9 ± 0.5 | 0.48 ± 0.06 | 170 |
| L305V/K337A-FVIIa | 7.2 ± 0.8 | 1.9 ± 0.4 | 0.37 ± 0.04 | 200 |
| FVIIaIIa | 7.8 ± 0.3 | 2.3 ± 0.7 | 2.6 ± 0.5 | 1,200 |
| K337A-FVIIaIIa | 11.0 ± 0.2 | 2.4 ± 0.1 | 4.4 ± 0.2 | 1,800 |
| L305V-FVIIaIIa | 6.7 ± 0.2 | 2.7 ± 0.1 | 4.2 ± 0.2 | 1,600 |
| L305V/K337A-FVIIaIIa | 11.5 ± 0.3 | 2.1 ± 0.2 | 6.8 ± 0.2 | 3,200 |
| M298Q-FVIIa | 3.4 ± 0.2 | 2.4 ± 0.2 | 0.52 ± 0.07 | 220 |
All values are means ± SD. The amidolytic activity is given as the ratio between the activity of the mutant and the activity of wild-type (wt) FVIIa in the presence of 1 mM S-2288 (n = 3). The kcat/Km values are calculated from the means (n = 2).
Combining the replacements in FVIIaIIa with K337A yielded a variant with an additively enhanced amidolytic activity (11-fold higher than wild-type FVIIa; Table 1). The introduction of the previously described L305V mutation (12) did not significantly affect the activity of this variant nor did it influence the activity of FVIIaIIa. Nonetheless, the double mutant L305V/K337A-FVIIa had 7-fold elevated activity, indicating an additive effect of these two mutations on FVIIa activity. This finding indicates that the L305V mutation and one or more of the substitutions in FVIIaIIa influence the activity of FVIIa by a common mechanism, and that the latter are more efficient in doing so because the activity of FVIIaIIa is higher than that of L305V-FVIIa. On the other hand, the K337A mutation appears to elicit a discrete activity-enhancing mechanism.
To assess whether any of the mutations influenced the substrate specificity of FVIIa, the rate of hydrolysis of a panel of four chromogenic substrates was measured. The substrates differ in their P3 and P2 positions and have revealed changes in the specificity of a previously described FVIIa mutant (13). The activity increase compared with FVIIa was 3.0- to 3.3-fold for K337A-FVIIa, 2.9- to 3.6-fold for L305V-FVIIa, 3.1- to 3.4-fold for M298Q-FVIIa, 8- to 10-fold for FVIIaIIa, and 10- to 17-fold for L305V/K337A-FVIIaIIa depending on which substrate was used. The rate of hydrolysis varied between substrates, but similar amidolytic activity enhancements of the individual variants as compared with wild-type FVIIa were observed with all substrates, indicating that the mutations had no, or a very small, impact on amidolytic substrate preference.
Cofactor Binding and Effect on Amidolytic Activity.
The amidolytic activities of M298Q-FVIIa and FVIIaIIa were enhanced about 7-fold and 3.5-fold, respectively, by sTF, resulting in an amidolytic activity of the complexes indistinguishable from that of the wild-type complex. The amidolytic activity of K337A-FVIIa was enhanced 10-fold by sTF, which makes the K337A-FVIIa–sTF complex about 1.5-fold more amidolytically active than the complex between wild-type FVIIa and sTF. K337A-FVIIa gained a bit more (about 25%) amidolytic activity than wild-type FVIIa and the other FVIIa variants upon sTF binding. The concentration of sTF used in this experiment (50 nM) gave about 95% saturation of all FVIIa variants according to the binding constants derived from surface plasmon resonance measurements (Table 2). Both FVIIaIIa and K337A-FVIIa appear to bind sTF with slightly higher affinity than wild-type FVIIa because of a slower dissociation from the cofactor. Thus the mutations change the protease domain of FVIIa into a conformational state that is more active, or shifts the equilibrium in the direction toward the active conformation, and enables the cofactor binding site to interact tighter with sTF. A similar observation has been made for M298Q-FVIIa (11). Modification of FVIIa with an active site inhibitor, assumed to lock the enzyme in the active conformation, has been shown to result in a qualitatively similar but more pronounced effect (25).
Table 2.
Kinetics of the interaction between the FVIIa variants and sTF
| FVIIa variant | kon, × 105 M−1⋅s−1 | koff, × 10−3 s−1 | Kd, nM |
|---|---|---|---|
| Wild-type FVIIa | 9.3 ± 0.3 | 2.1 ± 0.2 | 2.3 |
| K337A-FVIIa | 8.9 ± 1.1 | 1.7 ± 0.2 | 1.9 |
| FVIIaIIa | 9.6 ± 1.3 | 1.4 ± 0.2 | 1.5 |
Values are means ± SD (n = 3). Kd (koff/kon) values are calculated from the means.
Intrinsic Proteolytic Activity of FVIIa Mutants.
The proteolytic activity of the FVIIa variants in the absence of TF was characterized by using a two-stage assay. Because termination of FX activation by the addition of EDTA was not feasible in all cases, the mutants were allowed to act on FX for 20 min, followed by a 2-min quantification of the resulting FXa. K337A-FVIIa, L305V-FVIIa, and the variant containing both of these mutations displayed an increase in proteolytic activity similar to that seen for the amidolytic activity (Table 1). The proteolytic activity of M298Q-FVIIa was 7-fold that of wild-type FVIIa, which is in good agreement with a previous report (11) and twice the observed fold increase in amidolytic activity. In sharp contrast, all of the variants containing the mutations present in FVIIaIIa exhibited a dramatic enhancement of the proteolytic activity (between 37- and 100-fold that of wild-type FVIIa as compared with 8- to 11-fold increase in amidolytic activity). Even if one assumes that the Km for FX is the same for all variants, these mutants still have a 28- to 72-fold higher proteolytic activity than wild-type FVIIa. The maximal rate of FX activation catalyzed by the most active mutant, L305V/K337A-FVIIaIIa, was about one-third of that observed for wild-type FVIIa bound to sTF (not shown). The reason for the nonparallel increase in proteolytic and amidolytic activity is presently unknown. Because it appears not to be caused by a lower Km for FX (Table 1), it must be explained by a stabilization of structural elements that facilitate macromolecular substrate cleavage without affecting the hydrolysis of small peptidyl substrates. Alanine scanning of FVIIa has revealed that residues 158 {21}, 296 {154}, and 298 {156} are all important for optimal FX activation by FVIIa/TF (2). Multiple explanations could account for this finding. More interestingly in the context of the present study, Ala replacement of Glu-296 {154} has been shown to reduce the amidolytic activity of FVIIa–TF to a lesser extent than it reduces the proteolytic activity of the complex (26). We find that the proteolytic activity of free FVIIaIIa is enhanced to a larger extent than its amidolytic activity. In both cases, changing residue 296 {154} generates nonparallel effects on the activity measured with a macromolecular and a small substrate, respectively. Glu-296 {154} has been shown to be part of a macromolecular exosite and appears to be required for an allosteric linkage to the active site of FVIIa (26). It is possible that the ability of FX to promote its own conversion to FXa through the binding to this exosite has been enhanced by the E296V mutation even though the affinity of FX for FVIIaIIa, and other variants containing the same three replacements, appears to be unaltered. The M298Q mutation in FVIIaIIa possibly augments this ability because M298Q-FVIIa also displays a larger relative enhancement of the proteolytic activity as shown above and in a previous report (11).
The specific clotting activity of K337A-FVIIa, FVIIaIIa, and K337A-FVIIaIIa measured in plasma in the presence of TF were all similar to that of wild-type FVIIa (82–107%). When the procoagulant activity was analyzed in the absence of TF, using lipid vesicles, the specific activities of the three variants were increased 5-fold, 30-fold, and 52-fold, respectively, as compared with wild-type FVIIa. These results are in agreement with those obtained above in systems of purified components.
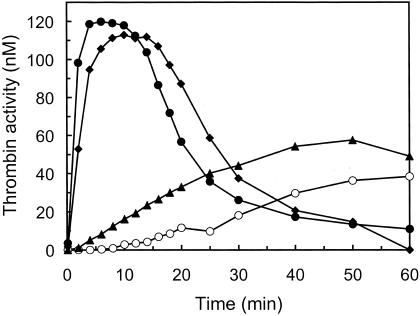
FVIIa has been shown to bind to activated platelets with low affinity and activate FX independently of TF, which subsequently leads to thrombin generation (27). At hemophilia-like conditions, i.e., in the absence of factor VIII or factor IX, 50 nM FVIIa generates enough FXa on activated platelets to give an amount of thrombin approaching the level obtained in samples with factor VIII, factor IX, TF, and a physiologically relevant concentration of FVIIa (0.2 nM). This finding demonstrates that FVIIa at superphysiological concentrations can compensate for the lack of factor VIII or IX, and it is currently used to treat bleedings in hemophiliacs with inhibitory antibodies to one of these factors. We measured the activity of some mutants on activated platelets, and Fig. 3 shows TF-independent thrombin generation by 50 nM wild-type FVIIa, K337A-FVIIa, FVIIaIIa, and K337A-FVIIaIIa. K337A-FVIIa had slightly higher activity than wild-type FVIIa, whereas FVIIaIIa and K337A-FVIIaIIa had substantially higher activity as seen by an increased rate of thrombin formation, shorter lag phase/earlier thrombin peak and higher maximal level of thrombin activity. M298Q-FVIIa had a thrombin generating activity between that of K337A-FVIIa and FVIIaIIa (not shown). Because this is a complex system, containing for instance AT that inhibits FVIIa (wild-type and mutants at different rates) as well as FXa and thrombin, separate experiments were performed to get an estimate of the relative potency of the different FVIIa variants. K337A-FVIIa at a concentration of 50 nM gave a thrombin generation curve between those obtained with 100 and 200 nM wild-type FVIIa, whereas 25 nM K337A-FVIIa gave a curve between those obtained using 50 and 100 nM wild-type FVIIa. This finding indicates that K337A-FVIIa was 2- to 4-fold more active than wild-type FVIIa in this system. FVIIaIIa at 2 nM and K337A-FVIIaIIa at 1 nM gave similar thrombin generation curves as 50 nM FVIIa, indicating 25- and 50-fold higher thrombin-generating activities as compared with wild-type FVIIa, respectively (not shown).
Figure 3.
Thrombin generation on activated platelets. Platelets were activated with the thrombin receptor agonist peptide SFLLRN, and 50 nM of wild-type FVIIa (○), K337A-FVIIa (▴), FVIIaIIa (⧫), or K337A-FVIIaIIa (●) was then added together with protein mixture and calcium (see Materials and Methods). Aliquots were removed and analyzed for thrombin amidolytic activity. The data shown are representative of three separate experiments.
Inhibition of FVIIa Mutants by AT.
The increased intrinsic activity of the FVIIa variants should be reflected in an increased rate of inhibition by AT as previously demonstrated for L305V-FVIIa (12). We found that the variants could be divided into three groups based on their rate of inhibition by AT in the presence of heparin (which was absolutely required for any significant inhibition) corresponding to the various levels of amidolytic activity (Table 3). The two variants, K337A-FVIIa and M298Q-FVIIa, with a 3- to 4-fold increase in amidolytic activity retained 40–50% of the activity after 15-min incubation with AT/heparin. Similar kinetics of inhibition were obtained with L305V-FVIIa, which also falls within this group (12). The variants with 6- to 8-fold increased amidolytic activity (L305V/K337A-FVIIa, FVIIaIIa, and L305V-FVIIaIIa) retained about 20% of the activity, and the two with 11 times higher activity (K337A-FVIIaIIa and L305V/K337A-FVIIaIIa) retained about 10% of the activity. Like FXa and thrombin, wild-type FVIIa and FVIIaIIa bound to sTF were both virtually completely inhibited after 15 min (not shown), and we assume that this must be true for all of the variants.
Table 3.
Rate of inhibition of FVIIa variants by AT in the presence of heparin
| FVIIa variant | Residual activity after 15 min, % |
|---|---|
| Wild-type FVIIa | 80 ± 7 |
| K337A-FVIIa | 41 ± 6 |
| L305V/K337A-FVIIa | 23 ± 5 |
| FVIIaIIa | 22 ± 5 |
| K337A-FVIIaIIa | 13 ± 4 |
| L305V-FVIIaIIa | 19 ± 4 |
| L305V/K337A-FVIIaIIa | 9 ± 3 |
| M298Q-FVIIa | 47 ± 6 |
Values are means ± SD (n = 3).
Accessibility of the Amino Group of Ile-153 {16}.
The accessibility of the N-terminal α-amino group of the protease domain of FVIIa has been shown to be influenced by TF binding (8), active site occupancy (9), and temperature (28). These observations suggest that the accessibility appears to be an inverse reflection of the activity state of wild-type FVIIa, with a less accessible N terminus in the active FVIIa conformation. However, neither an increased FVIIa activity obtained by mutagenesis (11, 13) nor a decreased FVIIa activity in the presence of zinc ions (29) appeared to correlate with an alteration of the susceptibility of the N terminus to carbamylation. After incubation with KNCO for 60 min, a similar loss of activity (50–70%) was observed for wild-type FVIIa, L305V-FVIIa, and K337A-FVIIa, whereas wild-type FVIIa bound to sTF lost only 20–25% of its activity. According to the result with K337A-FVIIa, removal of the positive charge/reduction of the size of the side chain does not facilitate insertion of the N terminus. The N terminus of FVIIaIIa and K337A-FVIIaIIa appeared to be significantly more buried as judged from the relatively small decrease in activity (14% and 9%, respectively). In addition, we found that the M298Q mutation protected the N terminus slightly (33% loss in activity). An explanation for the efficient protection of the N terminus in FVIIaIIa, superior to that induced by sTF binding, might be that the mutations are located in its immediate environment, creating new hydrogen bonds that tether the N terminus more firmly to the body of the protease domain. Thus the superactivity of the FVIIa mutants described in this article does occasionally correlate with a more buried N terminus, indicating that one or more of the M298Q, E296V, and V158D mutations somehow facilitate salt bridge formation with Asp-343 {194}.
Concluding Remarks.
To summarize, we describe a total of five positions in FVIIa that appear to regulate the activity of free FVIIa. The mutations in FVIIaIIa (V158{21}D, E296{154}V, and M298{156}Q), and to some extent the L305{163}V mutation (12), appear to mediate a structural transition also inducible by TF. This conclusion is based on the observations that FVIIaIIa has a diminished dependence on calcium ions, a protected N terminus, and the same activity in complex with sTF as wild-type FVIIa. On the contrary, the K337{188}A mutation appears to work by means of another mechanism, mainly because it confers some additional amidolytic activity to FVIIa not attainable by TF binding, but also because it leaves the Ca2+ dependence and the environment of the N terminus unaffected. The proximity to Asp-338 {189} and/or Asp-343 {194} and Ser-344 {195} might be a clue as to how the K337A mutation affects the activity of free FVIIa. Our most active mutant has an amidolytic and a proteolytic activity about one-third of those of the complex between wild-type FVIIa and sTF. This finding suggests that there are perhaps only a limited number of additional residues, apart from the five that have been replaced in this variant, that control the zymogenicity of free FVIIa. Superactive variants of recombinant FVIIa that provide more rapid FXa (and consequently thrombin) generation on activated platelets, independently of TF, could offer improved treatment of hemophiliacs with inhibitory antibodies by providing more effective hemostasis (27).
Acknowledgments
We thank Helle Bak, Anette Østergaard, Lone Odborg, and Berit Lassen for excellent technical assistance.
Abbreviations
- AT
antithrombin
- FVIIa
factor VIIa
- FVIIaIIa
V158D/E296V/M298Q-FVIIa
- FX
factor X
- FXa
factor Xa
- sTF
soluble tissue factor
- TF
tissue factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 2.Dickinson C D, Kelly C R, Ruf W. Proc Natl Acad Sci USA. 1996;96:14379–14384. doi: 10.1073/pnas.93.25.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banner D W, D'Arcy A, Chène C, Winkler F K, Guha A, Konigsberg W H, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhang E, St. Charles R, Tulinsky A. J Mol Biol. 1999;285:2089–2104. doi: 10.1006/jmbi.1998.2452. [DOI] [PubMed] [Google Scholar]
- 5.Pike A C W, Brzozowski A M, Roberts S M, Olsen O H, Persson E. Proc Natl Acad Sci USA. 1999;96:8925–8930. doi: 10.1073/pnas.96.16.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kemball-Cook G, Johnson D J D, Tuddenham E G D, Harlos K. J Struct Biol. 1999;127:213–223. doi: 10.1006/jsbi.1999.4158. [DOI] [PubMed] [Google Scholar]
- 7.Dennis M S, Eigenbrot C, Skelton N J, Ultsch M H, Santell L, Dwyer M A, O'Connell M P, Lazarus R A. Nature (London) 2000;404:465–470. doi: 10.1038/35006574. [DOI] [PubMed] [Google Scholar]
- 8.Higashi S, Nishimura H, Aita K, Iwanaga S. J Biol Chem. 1994;269:18891–18898. [PubMed] [Google Scholar]
- 9.Higashi S, Matsumoto N, Iwanaga S. J Biol Chem. 1996;271:26569–26574. doi: 10.1074/jbc.271.43.26569. [DOI] [PubMed] [Google Scholar]
- 10.Leonard B J N, Clarke B J, Sridhara S, Kelley R, Ofosu F A, Blajchman M A. J Biol Chem. 2000;275:34894–34900. doi: 10.1074/jbc.M001166200. [DOI] [PubMed] [Google Scholar]
- 11.Petrovan R, Ruf W. J Biol Chem. 2001;276:6616–6620. doi: 10.1074/jbc.M004726200. [DOI] [PubMed] [Google Scholar]
- 12.Persson E, Bak H, Olsen O H. J Biol Chem. 2001;276:29195–29199. doi: 10.1074/jbc.M102187200. [DOI] [PubMed] [Google Scholar]
- 13.Soejima K, Mizuguchi J, Yuguchi M, Nakagaki T, Higashi S, Iwanaga S. J Biol Chem. 2001;276:17229–17235. doi: 10.1074/jbc.M009206200. [DOI] [PubMed] [Google Scholar]
- 14.Thim L, Bjoern S, Christensen M, Nicolaisen E M, Lund-Hansen T, Pedersen A, Hedner U. Biochemistry. 1988;27:7785–7793. doi: 10.1021/bi00420a030. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen A H, Nordfang O, Norris F, Wiberg F C, Christensen P M, Moeller K B, Meidahl-Pedersen J, Beck T C, Norris K, Hedner U, Kisiel W. J Biol Chem. 1990;265:16786–16793. [PubMed] [Google Scholar]
- 16.Freskgård P-O, Olsen O H, Persson E. Protein Sci. 1996;5:1531–1540. doi: 10.1002/pro.5560050809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Persson E, Nielsen L S. FEBS Lett. 1996;385:241–243. doi: 10.1016/0014-5793(96)00400-0. [DOI] [PubMed] [Google Scholar]
- 18.Persson E, Nielsen L S, Olsen O H. Biochemistry. 2001;40:3251–3256. doi: 10.1021/bi001612z. [DOI] [PubMed] [Google Scholar]
- 19.Monroe D M, Roberts H R, Hoffman M. Br J Haematol. 1994;88:364–371. doi: 10.1111/j.1365-2141.1994.tb05032.x. [DOI] [PubMed] [Google Scholar]
- 20.Kjalke M, Oliver J A, Monroe D M, Hoffman M, Ezban M, Hedner U, Roberts H R. Thromb Haemostasis. 1997;78:1202–1208. [PubMed] [Google Scholar]
- 21.Persson E, Olsen O H, Østergaard A, Nielsen L S. J Biol Chem. 1997;272:19919–19924. doi: 10.1074/jbc.272.32.19919. [DOI] [PubMed] [Google Scholar]
- 22.Persson E. Haemostasis. 1996;26, Suppl. 1:31–34. doi: 10.1159/000217237. [DOI] [PubMed] [Google Scholar]
- 23.Neuenschwander P F, Morrissey J H. J Biol Chem. 1994;269:8007–8013. [PubMed] [Google Scholar]
- 24.Miyata T, Funatsu A, Kato H. J Biochem. 1995;117:836–844. doi: 10.1093/oxfordjournals.jbchem.a124784. [DOI] [PubMed] [Google Scholar]
- 25.Sørensen B B, Persson E, Freskgård P-O, Kjalke M, Ezban M, Williams T, Rao L V M. J Biol Chem. 1997;272:11863–11868. doi: 10.1074/jbc.272.18.11863. [DOI] [PubMed] [Google Scholar]
- 26.Shobe J, Dickinson C D, Ruf W. Biochemistry. 1999;38:2745–2751. doi: 10.1021/bi981951g. [DOI] [PubMed] [Google Scholar]
- 27.Monroe D M, Hoffman M, Oliver J A, Roberts H R. Br J Haematol. 1997;99:542–547. doi: 10.1046/j.1365-2141.1997.4463256.x. [DOI] [PubMed] [Google Scholar]
- 28.Petersen L C, Persson E, Freskgård P-O. Eur J Biochem. 1999;261:124–129. doi: 10.1046/j.1432-1327.1999.00258.x. [DOI] [PubMed] [Google Scholar]
- 29.Petersen L C, Olsen O H, Nielsen L S, Freskgård P-O, Persson E. Protein Sci. 2000;9:859–866. doi: 10.1110/ps.9.5.859. [DOI] [PMC free article] [PubMed] [Google Scholar]