Abstract
Inositol phosphates (IPs) comprise a family of ubiquitous eukaryotic signaling molecules. They have been linked to the regulation of a pleiotropy of important cellular activities, but low abundance and detection difficulties have hampered our understanding. Here we present a method to purify and enrich IPs or other phosphate-rich metabolites from mammalian cells or other sample types. Acid-extracted IPs from cells bind selectively via their phosphate groups to titanium dioxide beads. After washing, the IPs are easily eluted from the beads by increasing the pH. This technique, in combination with downstream analytical methods such as PAGE or SAX-HPLC, opens unprecedented investigative possibilities, allowing appropriate analysis of IPs from virtually any biological or non-biological source.
Keywords: Inositol polyphosphate, Inositol pyrophosphate, IP6, IP7, Metabolism, Metabolites, Signaling
Background
Inositol phosphates (IPs) are a family of conserved signaling molecules, ubiquitous in eukaryotes (Irvine and Schell, 2001; Tsui and York, 2010). They have been implicated in the regulation of a broad range of cellular activities, including calcium signaling, trafficking, and phosphate homeostasis ( Wilson et al., 2013 ; Thota and Bhandari, 2015; Azevedo and Saiardi, 2017). However, our understanding of IPs signaling has been hampered by the fact that they can be difficult to study.
Unlike other phosphate-rich molecules such as nucleotides, IPs do not absorb in the UV/Vis range, and are often present at relatively low abundance in cells. The traditional methodology for IP detection and analysis is to radioactively metabolically label cells with 3H-inositol that is taken up and incorporated into IPs over 1-5 days (Wilson and Saiardi, 2017). After labeling, IPs are extracted with perchloric acid; these extracts are neutralised before separation by strong anion exchange (SAX) HPLC and scintillation counting of each fraction (Azevedo and Saiardi, 2006). The use of 3H-labelled IPs and chromatography has also been required for in vitro biochemical studies. The requirement for radioactive IPs or metabolic labeling limits the possible lines of investigation. These are time-consuming, technically demanding and expensive experiments.
We previously developed a polyacrylamide gel electrophoresis (PAGE) method for resolving and visualizing IPs ( Losito et al., 2009 ; Loss et al., 2011 ). This technique was immediately useful in following in vitro reactions, as well as analyzing in vivo high abundance IPs such as IP6, IP7 and IP8 from Dictyostelium discoideum ( Pisani et al., 2014 ), or IP6 from plant seeds ( Desai et al., 2014 ; Kolozsvari et al., 2014 ). However, in the majority of cell types or model organisms, low IP concentrations make it impossible to run a PAGE gel of enough neutralized extract to visualize IPs while maintaining correct gel migration. In mammalian cells, the most abundant IP is IP6 at 40-100 µM (measured in cell lines such as HL60, C1866 and BAF3; French et al., 1991 ; Bunce et al., 1993 ), while the inositol pyrophosphate IP7 is thought to be present at sub-µM levels. We were therefore inspired to develop the present method that uses titanium dioxide beads to purify cold or radioactive IPs regardless of their abundance ( Wilson et al., 2015 ). Titanium dioxide binds the phosphate groups of the IPs. The concentrated IPs can be analyzed by PAGE, SAX-HPLC, or other techniques.
The use of titanium dioxide beads now enables analysis of total unlabeled IPs from any cell type ( Pavlovic et al., 2015 ; Wilson et al., 2015 ; Gu et al., 2016 ; Pavlovic et al., 2016 ). It also allows the study of IPs extracted from previously impossible sample types, including large volumes of liquid media, biofluids, or animal tissues. For biochemical work, the method can be used to remove salt and proteins from IPs preparations. Here we present the method as used for purifying IPs from cultured adherent mammalian cells.
Materials and Reagents
Pipette tips (Starlab, catalog number: S1112-1830)
1.5 ml Eppendorf-style microcentrifuge tubes (Starlab, catalog number: S1615-5500)
50 ml Falcon centrifuge tubes (Corning, catalog number: 352070)
15 cm tissue culture dishes (Thermo Fisher Scientific, catalog number: 168381)
pH test strips (Sigma-Aldrich, catalog number: P4536-100EA)
Titansphere TiO2 beads, 5 µm (Hichrom, catalog number: 5020-75000)
PBS (Thermo Fisher Scientific, catalog number: 20012019)
0.25% Trypsin-EDTA (Thermo Fisher Scientific, catalog number: 25200056)
Double distilled water (ddH2O) or Milli-Q water (Millipore)
Perchloric acid, 70% (Sigma-Aldrich, catalog number: 244252-1L)
Ammonium hydroxide, 28-30% (Sigma-Aldrich, catalog number: 221228-1L)
1 M perchloric acid (see Recipes)
~2.8% ammonium hydroxide (see Recipes)
Equipment
Pipettes (Gilson, models: P1000 and P200, catalog numbers: F123602, F123601)
Ice box
Balance (Acculab, model: ALC-80.4)
Humidified incubator (Eppendorf, model: Galaxy® 170 R, catalog number: CO17311002)
Benchtop centrifuge (Eppendorf, catalog number: 5702000365)
Benchtop centrifuge with cooling (LaboGene, model: ScanSpeed 1730R)
-
Rotator (Cole-Parmer, Stuart, model: SB3)
Note: This should be placed in a fridge or cold room.
Vortex mixer (Scientific Industries, model: Vortex Genie 2, catalog number: SI-0266)
Centrifugal evaporator (Martin Christ Gefriertrocknungsanlagen, catalog number: RVC 2-18)
Tilt table (Cole-Parmer, Stuart, catalog number: SSM4)
Cell scrapers (Greiner Bio One International, catalog number: 541070)
Procedure
-
Before starting the extraction
Switch on the cooled centrifuge and centrifugal evaporator.
-
Prepare 2.5% ammonium hydroxide and 1 M perchloric acid. Cool to 4 °C before use.
Note: The dilute ammonium hydroxide solution can be made in advance and stored at room temperature or 4 °C indefinitely. Check that the pH is > 10 and vortex mix before using.
-
Weigh titanium dioxide beads, 4 mg/sample, into an Eppendorf. Suspend all the beads together in 1 ml ddH2O by pipetting or vortexing, then centrifuge at 3,500 × g for 1 min at 4 °C. Remove supernatant and wash again using 1 ml 1 M perchloric acid. Remove supernatant and suspend beads in 50 µl 1 M perchloric acid per sample, then aliquot into the correct number of Eppendorfs.
Note: Beads can be mixed by pipetting or vortexing. They will not stay in suspension so mix frequently when aliquotting. The beads are not damaged by shear stress; standard pipette tips can be used for this protocol.
-
Preparing the cells
-
Culture the cell line of interest under standard conditions, for example in a 37 °C humidified incubator with 5% CO2.
Note: For PAGE analysis after titanium dioxide purification, prepare enough cells for 10 mg equivalent total protein per condition. Titanium dioxide purification with PAGE analysis has been validated in many cell types (Wilson et al., 2015). Depending on IPs of interest, cell type, or analysis method, more or fewer cells may be required.
Wash once with warm PBS then incubate in trypsin-EDTA until the cells have detached. Collect them into a Falcon tube and centrifuge at 200 × g for 3 min. Remove supernatant.
Resuspend cells in 1 ml cold PBS and transfer to Eppendorfs on ice. Adjust the volumes so all samples have approximately the same volume, then remove 40 µl into a separate Eppendorf. These cells are for normalization; they can be counted now or, for example, extracted to quantify protein concentration.
Centrifuge the cells at 200 × g for 3 min and remove supernatant. Samples can be processed immediately or frozen at -80 °C.
-
-
Perchloric acid extraction
-
Resuspend the pelleted cells in 1 ml cold 1 M perchloric acid. Mix by pipetting until fully suspended. The samples will immediately become white and cloudy as proteins precipitate.
Note: When using frozen cells, there is no need to defrost the samples before adding perchloric acid.
-
Incubate the samples on ice for 10-15 min, with frequent 2-5 sec vortex intervals.
Note: Many inositol phosphate species, especially inositol pyrophosphates, are labile under acidic conditions. Therefore all steps before addition of ammonium hydroxide should be performed at 4 °C to minimize degradation, and the acid incubation time should be kept to a minimum. Extraction from certain sample types may require longer incubation.
Centrifuge at 18,000 × g for 5 min at 4 °C. The pellet will contain membranes and proteins. Small polar molecules such as IPs will be in the supernatant.
-
-
Titanium dioxide beads purification
Transfer the supernatants from Step C3 to the Eppendorfs containing titanium dioxide beads prepared in Step A3. Vortex briefly to mix.
Rotate samples at 4 °C for 15-20 min.
-
Centrifuge samples at 3,500 × g for 1 min at 4 °C. Carefully discard supernatants as IPs will be bound to the titanium dioxide beads.
Note: Inositol pyrophosphates can be degraded by adsorption onto the beads as well as by acidic conditions. The protocol should take 1.5-2 h depending on sample number, excluding time taken for centrifugal evaporation.
Wash the beads by resuspending in 500 µl cold 1 M perchloric acid, centrifuging at 3,500 × g for 1 min at 4 °C, then removing supernatant. Repeat this step.
Resuspend the beads in 200 µl ~2.8% ammonium hydroxide to elute IPs. Vortex or pipette mix, then rotate samples for 5 min.
Centrifuge at 3,500 × g for 1 min. Transfer the supernatant to a new Eppendorf.
Elute again with another 200 µl ammonium hydroxide. After rotation and centrifugation, combine that supernatant with the first, for 400 µl sample volume.
-
Use the centrifugal evaporator to reduce the sample volume to 20-60 µl, or until the pH is 7-8. Do not store the samples until they reach neutral pH. The samples can be heated up to 60 °C during this process to speed up evaporation. If samples are accidentally dried, resuspend in ddH2O.
Note: Test pH of samples by spotting 1-2 µl onto pH test strips.
Store the neutralized samples at 4 °C. They are quite stable, and can be kept for a few weeks before analysis.
Notes
In this protocol, we have described harvesting cells by trypsinization, but other methods can be used depending on experimental goals. In metabolite extraction protocols it is desirable to quench the cellular metabolism as quickly as possible, to obtain the truest picture of intracellular concentrations. For optimum quenching of adherent cells: quickly remove the culture medium before washing twice in 5 ml cold PBS (for 15 cm dish). Add sufficient cold 1 M perchloric acid to cover the plate (3-5 ml). Incubate the dishes on a tilt table at 4 °C for 10-15 min. Carefully remove the perchloric acid into a Falcon tube and centrifuge to remove contaminating debris. Transfer the supernatant to a fresh tube and proceed with titanium dioxide purification. Almost complete recovery of IPs is possible from volumes up to 10-20 ml ( Wilson et al., 2015 ). Alternatively, after washing, scrape the cells in 1 ml cold PBS, transfer to Falcon or Eppendorf, centrifuge, discard supernatant, and resuspend in a more convenient volume of perchloric acid. The cellular yield is reduced when scraping compared to trypsinizing cells. The chief downside of harvesting by immediate extraction or scraping is that there is no potential for saving an aliquot of cells for normalization purposes. If the experiment will need normalization, e.g., by cell number, extra dishes must be prepared in parallel for this.
Elution of IPs from the titanium dioxide beads is very efficient, allowing reuse of the beads. Wash twice in ddH2O, and store in ddH2O at 4 °C. If carry-over is a concern, either elute again from the washed beads and perform the intended downstream assay to confirm lack of IPs, or use fresh beads each time. The beads we recommend are of defined diameter and intended for scientific use. Cheap cosmetic grade titanium dioxide powder is available to buy online; we found it to be poorer at purifying IPs from cell extracts, although perhaps using greater quantities would compensate.
Aside from IPs, other phosphate-containing molecules will co-purify from cells using acid extraction and titanium dioxide beads. These include nucleotides and other phosphate-containing metabolic intermediates. This protocol can therefore be used to enrich many different small molecules of interest for appropriate downstream analysis. Co-purifying nucleotides such as GTP and ATP resolve separately from IP6, IP7 and IP8 when analyzing by PAGE (Figure 1).
Figure 1. Higher IPs can be separated from co-purifying contaminant molecules by PAGE.
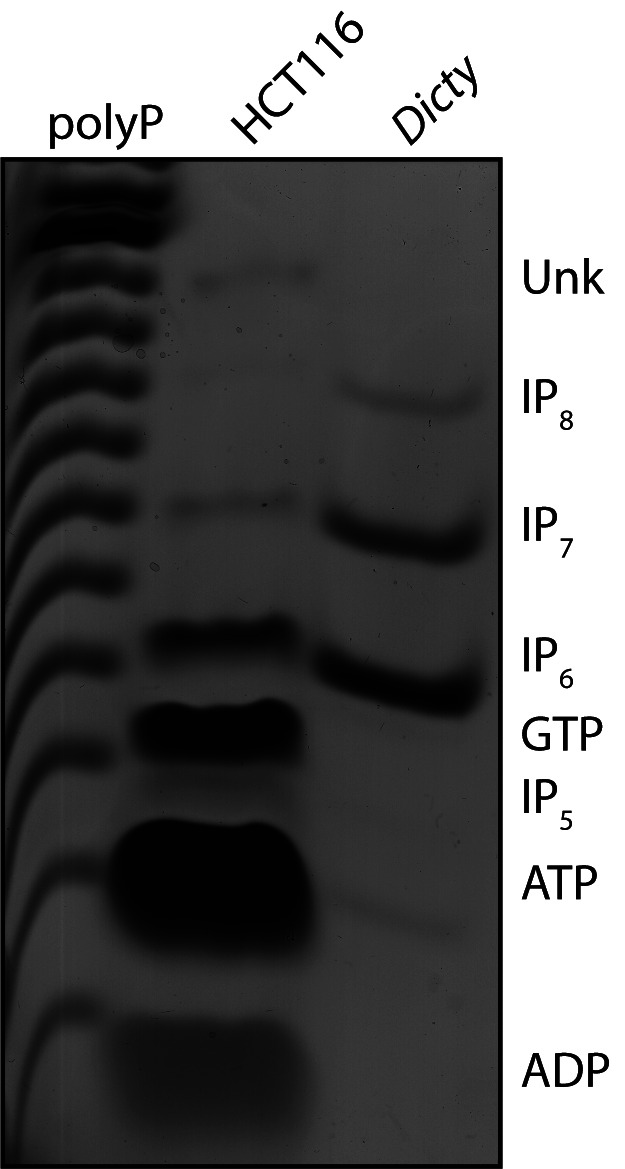
This representative PAGE gel, stained with toluidine blue, shows IPs purified from the human colon carcinoma cell line HCT116 using titanium dioxide beads. An amount of cells equivalent to 20 mg of total protein was used. Inorganic polyphosphate (polyP), and IP6, IP7 and IP8 derived from Dictyostelium discoideum are shown as markers. The identity of each band has been previously confirmed by co-migration with standards, and by enzymatic treatment (phytase to degrade IPs, apyrase to degrade nucleotides). The identity of the Unk band is unknown.
Recipes
-
1 M perchloric acid
For 200 ml, mix 17.2 ml stock (70%, 11.6 M) with 182.8 ml ddH2O
Store at room temperature or 4 °C
-
~2.8% ammonium hydroxide
For 30 ml, mix 3 ml stock (28-30%) with 27 ml ddH2O
Store at room temperature
Acknowledgments
This work was supported by the UK Medical Research Council (MRC) core support to the MRC/UCL Laboratory for Molecular Cell Biology University Unit (MC UU 1201814). This protocol is an expanded and slightly modified version of that originally described in ( Wilson et al., 2015 ). The authors declare that they have no conflicts of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Azevedo C. and Saiardi A.(2006). Extraction and analysis of soluble inositol polyphosphates from yeast. Nat Protoc 1(5): 2416-2422. [DOI] [PubMed] [Google Scholar]
- 2. Azevedo C. and Saiardi A.(2017). Eukaryotic phosphate homeostasis: The inositol pyrophosphate perspective. Trends Biochem Sci 42(3): 219-231. [DOI] [PubMed] [Google Scholar]
- 3. Bunce C. M., French P. J., Allen P., Mountford J. C., Moor B., Greaves M. F., Michell R. H., Brown G.(1993). Comparison of the levels of inositol metabolites in transformed haemopoietic cells and their normal counterparts. Biochem J 298(3): 667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desai M., Rangarajan P., Donahue J. L., Williams S. P., Land E. S., Mandal M. K., Phillippy B. Q., Perera I. Y., Raboy V. and Gillaspy G. E.(2014). Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J 80(4): 642-653. [DOI] [PubMed] [Google Scholar]
- 5. French P. J., Bunce C. M., Stephens L. R., Lord J. M., McConnell F. M., Brown G., Creba J. A., Michell R. H.(1991). Changes in the levels of inositol lipids and phosphates during the differentiation of HL60 promyelocytic cells towards neutrophils or monocytes. Proc Biol Sci 245(1314): 193-201. [DOI] [PubMed] [Google Scholar]
- 6. Gu C., Wilson M. S., Jessen H. J., Saiardi A. and Shears S. B.(2016). Inositol pyrophosphate profiling of two HCT116 cell lines uncovers variation in InsP8 levels. PLoS One 11(10): e0165286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Irvine R. F. and Schell M. J.(2001). Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol 2(5): 327-338. [DOI] [PubMed] [Google Scholar]
- 8. Kolozsvari B., Parisi F. and Saiardi A.(2014). Inositol phosphates induce DAPI fluorescence shift. Biochem J 460(3): 377-385. [DOI] [PubMed] [Google Scholar]
- 9. Losito O., Szijgyarto Z., Resnick A. C. and Saiardi A.(2009). Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS One 4(5): e5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loss O., Azevedo C., Szijgyarto Z., Bosch D. and Saiardi A.(2011). Preparation of quality inositol pyrophosphates. J Vis Exp(55): e3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pavlovic I., Thakor D. T., Bigler L., Wilson M. S., Laha D., Schaaf G., Saiardi A. and Jessen H. J.(2015). Prometabolites of 5-diphospho-myo-inositol pentakisphosphate. Angew Chem Int Ed Engl 54(33): 9622-9626. [DOI] [PubMed] [Google Scholar]
- 12. Pavlovic I., Thakor D. T., Vargas J. R., McKinlay C. J., Hauke S., Anstaett P., Camuna R. C., Bigler L., Gasser G., Schultz C., Wender P. A. and Jessen H. J.(2016). Cellular delivery and photochemical release of a caged inositol-pyrophosphate induces PH-domain translocation in cellulo. Nat Commun 7: 10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pisani F., Livermore T., Rose G., Chubb J. R., Gaspari M. and Saiardi A.(2014). Analysis of Dictyostelium discoideum inositol pyrophosphate metabolism by gel electrophoresis . PLoS One 9(1): e85533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thota S. G. and Bhandari R.(2015). The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J Biosci 40(3): 593-605. [DOI] [PubMed] [Google Scholar]
- 15. Tsui M. M. and York J. D.(2010). Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv Enzyme Regul 50(1): 324-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson M. S. C. and Saiardi A.(2017). Importance of radioactive labelling to elucidate inositol polyphosphate signalling. Top Curr Chem(Cham) 375(1): 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilson M. S., Bulley S. J., Pisani F., Irvine R. F. and Saiardi A.(2015). A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol 5(3): 150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson M. S., Livermore T. M. and Saiardi A.(2013). Inositol pyrophosphates: between signalling and metabolism. Biochem J 452(3): 369-379. [DOI] [PubMed] [Google Scholar]


