Abstract
Introduction
Allosteric modulators of G-protein coupled receptors (GPCRs) hold the promise of improved pharmacology and safety over typical orthosteric GPCR ligands. These features are particularly relevant to the cannabinoid receptor 1 (CB1R) GPCR, since typical orthosteric CB1R ligands are associated with adverse events that limit their translational potential.
Areas covered
The contextual basis for applying allostery to CB1R is considered from pharmacological, drug-discovery, and medicinal standpoints. Rational design of small-molecule CB1R allosteric modulators as potential pharmacotherapeutics would be greatly facilitated by direct experimental characterization of structure-function correlates underlying the biological activity of chemically-diverse CB1R allosteric modulators, CB1R allosteric ligand-binding binding pockets, and amino acid contact residues critical to allosteric ligand engagement and activity. In these regards, designer covalent probes exhibiting well-characterized molecular pharmacology as CB1R allosteric modulators are emerging as valuable molecular reporters enabling experimental interrogation of CB1R allosteric site(s) and informing the design of new CB1R agents as drugs.
Expert opinion
Synthesis and pharmacological profiling of CB1R allosteric ligands will continue to provide valuable insights into CB1R structure-function correlates. The resulting data should expand the repertoire of novel agents capable of exerting therapeutic benefit by modulating CB1R-dependent signaling.
Keywords: Allosteric modulator, binding motif, cannabinoid GPCR, chemical probe, endocannabinoid system, isothiocyanate, ligand bias, molecular pharmacology, orthosteric modulator, signal transduction, transmembrane receptor
1. Exploiting allostery for G-protein-coupled receptor (GPCR) drug discovery
Approximately 800 annotated and 150 ‘orphan’ G-protein-coupled receptors (GPCRs) without known endogenous ligand/function constitute the largest superfamily of cell-surface integral-membrane proteins encoded by the human genome, within which GPCRs represent the third largest gene family [1,2]. Also known as seven-transmembrane receptors (7TMRs), GPCRs principally function as transducers of signals (light photons, ions, lipids, peptides, hormones, odorants, etc.) from the extracellular milieu across the plasma membrane and into the cytoplasmic compartment, where downstream intracellular signaling cascades modulate cell physiology [3]. The ubiquity of GPCRs and their importance to both human (patho)physiology and drug discovery are reflected in the fact that ~40–60% of known drugs interact with them, making GPCRs at present the largest class of therapeutic targets [4]. Most approved, GPCR-targeted drugs are synthetic small molecules that exert their therapeutic effects by engaging a GPCR at its primary ‘orthosteric’ site, classically defined as the intramolecular domain to which naturally occurring endogenous ligands bind [5].
Notwithstanding their undeniable contributions to clinical medicine and public health, GPCR orthosteric drugs share a propensity for adverse-event liabilities due to on-target (e.g. interference with constitutive physiological signaling; hyperefficacy) and/or off-target (e.g. lack of receptor selectivity) effects [6]. The off-target adverse effects are particularly noisome to therapeutics invention, since orthosteric binding sites within GPCR subfamilies are highly conserved evolutionarily and structurally, complicating the optimization and development of subtype-specific GPCR orthosteric ligands (particularly agonists) as drugs and inviting potential adverse interactions across functionally distinct GPCRs [1,7].
These limitations have stimulated exploration of new modes of molecular pharmacology in the search for candidate ligands that modulate GPCRs for therapeutic gain [8,9]. In this regard, increasing attention has been focused on the discovery implications of allostery, i.e. cooperative interactions among distinct sites within the conformational landscape of a multidomain molecular system (e.g. a protein drug target) [10]. Well recognized in multisite carrier proteins and enzymes, allosteric domain coupling has more recently been documented to occur in ‘druggable’ GPCRs across all subfamilies, especially among rhodopsinlike, class-A GPCRs [11]. GPCR binding sites that engage (drug-like) ligands with allosteric regulatory properties are structurally and topographically distinct from canonical orthosteric sites that bind endogenous ligands. The classic, ‘pure’ allosteric ligand has no intrinsic efficacy of its own, but once recognized and engaged by a GPCR allosteric binding site, modulates the binding and/or signaling character of an orthosteric agent. The (sub)molecular mechanism underlying this cooperative influence is believed to involve discrete changes in GPCR conformation, activation state, and signaling context that reflect coupled, cooperative structural and functional interactions between the receptor’s orthosteric site and its remote, allosteric ligand-binding site(s) [12–14]. Biological consequences of GPCR allosteric ligand engagement may include changes in the affinity and/or efficacy of the orthosteric ligand, temporal and spatial alterations in GPCR internalization dynamics and desensitization, and shifts in the directionality and magnitude of the GPCR’s information output and signaling characteristics (signal amplitude, frequency, intensity, and duration) [10,13]. Aside from this classic paradigm, allosteric effects may also arise through formation of GPCR heteromers/oligomers or complexes with accessory cell proteins [15]. Consequently, GPCR allosteric pharmacology is inherently complex and quite distinct from that of a typical orthosteric ligand.
Since the approval of the first allosteric medicine (cinacalcet, Sensipar®) by the United States Food and Drug Administration in 2004 [16], members of several prominent drug classes have been identified as allosteric receptor modulators [17]. Allosteric ligands offer attractive discovery features with potential therapeutic advantages over orthosteric GPCR ligands [17–19]. GPCR allosteric ligand-binding sites are not as highly conserved as orthosteric sites, inviting a higher degree of receptor-/target-level selectivity from allosteric ligands. As compared to orthosteric GPCR modulators, allosteric ligands offer greater potential and scope for therapeutically optimizing the signaling context of a target GPCR by stabilizing a receptor conformation preferentially directed along a particular (i.e. salutary) intracellular effector pathway and away from others (i.e. those eliciting adverse events) (so-called ‘biased agonism’ or ‘functional selectivity’) [20,21]. GPCR signaling character and consequent cell responses can also be biased or fine-tuned by an allosteric ligand whose effects are specific to a particular orthosteric effector (‘probe dependence’) [13]. Functioning cooperatively with endogenous orthosteric ligands to express activity, an allosteric modulator would preferentially exert its pharmacological effect only in those tissues with adequate orthosteric ligand tone. In this manner, an allosteric drug helps maintain the spatial and temporal fidelity of the endogenous effector and acts only when and where required, offering significant potential for tissue specificity with reduced adverse event risk [17–19]. The hallmark ‘ceiling effect’ of GPCR allosteric modulators reflects the saturable nature of their function and may provide significant clinical advantage, serving as an intrinsic safety feature that militates against the risks of a potentially harmful overpotentiation of efficacy and disruption of physiological signaling networks with dose escalation [22]. These salient properties of a GPCR allosteric ligand may lessen the need for additional pharmaceuticals, reduce the required dose of the orthosteric agent to achieve a therapeutic effect, lower any abuse risk associated with a central nervous system (CNS)-acting agent, increase the therapeutic index and enhance the therapeutic range [17,19,23].
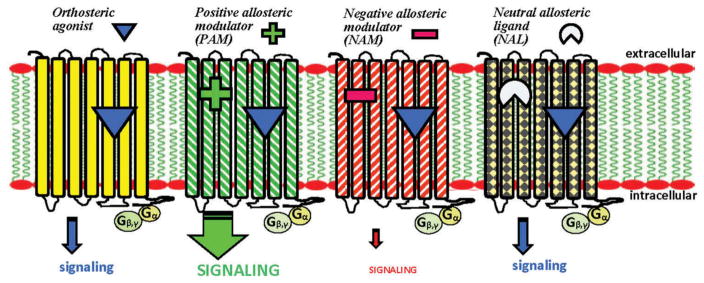
The phenotypes displayed by GPCR allosteric ligands are diverse and include ‘pure’ allosteric modulators with positive (potentiator) or negative (attenuator) cooperativity (PAMs or NAMs, respectively) as well as neutral allosteric ligands (NALs) that occupy a GPCR allosteric site while having no intrinsic cooperativity (Figure 1). Allosteric phenotypes may also encompass dual functionalities within a given ligand, e.g.: allosteric agonists coupled with PAM or NAM activity (ago-PAMs or ago-NAMs, respectively) or allosteric antagonists/inverse agonists coupled with PAM or NAM activity (IA-PAMs or IA-NAMs, respectively) [13,17,19]. Thus, allostery has great discovery potential to expand the GPCR-modulator chemical space, variegate the pharmacological modulation of a given GPCR target, offer a highly nuanced approach to therapeutic GPCR-dependent signal modulation by inducing unique and specific receptor conformational and functional states, and increase the number of druggable GPCRs. These perceived attractions have spurred recent medicinal chemistry efforts aimed at generating allosteric modulators as tool compounds and development candidates for drug targets including ligand-gated ion channels and, especially, GPCRs [24–26].
Figure 1.
Schematic representation of common phenotypes of GPCR allosteric modulators acting cooperatively with an orthosteric agonist. Diagramed are the potential effects of a positive allosteric modulator (PAM), negative allosteric modulator (NAM), and neutral allosteric ligand (NAL) on the signaling output of an orthosteric agonist. The comparison illustrates that a PAM can potentiate the signaling output of the orthosteric agonist by engaging the GPCR at a site distinct from the orthosteric site and enhancing the potency, affinity, and/or efficacy of the orthosteric agonist at the receptor, whereas a NAM has the opposite effect. In contrast, a NAL occupies the GPCR allosteric ligand-binding site without effect on the information transmission. Not shown are allosteric agonists with intrinsic activity in addition to their allosteric effects, endowing them, for example, with the capability of directly activating the receptor (e.g., ago-PAMs).
2. Cannabinoid receptor 1 (CB1R): functional and physiological characteristics
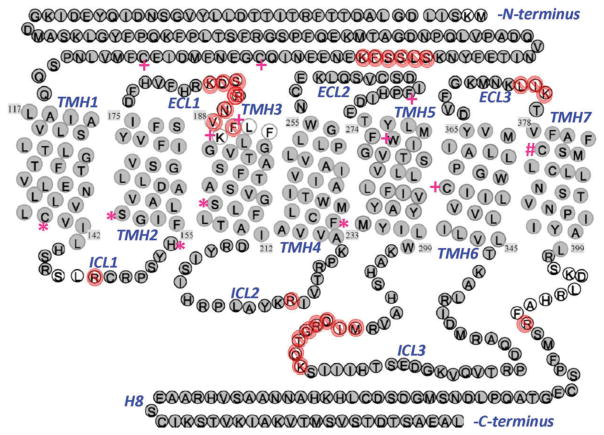
Perhaps best known as the GPCR engaged and activated by the phytocannabinoid Δ9-tetrahydrocannbinol (Δ9-THC), the main psychoactive component of Cannabis sativa (marijuana), the mammalian cannabinoid receptor 1 (CB1R) is a molecular constituent of the endogenous cannabinoid (‘endocannabinoid’) signaling system [27]. CB1R displays the general structural features of other integral-membrane, ‘rhodopsin-like,’ class-A GPCRs: an extracellular region consisting of the amino terminus and three extracellular loops (ECLs 1–3); a structural core of seven bundled transmembrane α-helices (TMHs 1–7); three intracellular loops (ICLs 1–3); a plasma-membrane proximal cytoplasmic region comprised of an amphipathic helix (H8) oriented parallel to the interior membrane face; and a carboxyl terminus longer than in most other class-A GPCRs (Figure 2) [28,29]. The extracellular region of CB1R modulates ligand access from the extracellular compartment; its TMH region engages orthosteric ligands and transduces information to the intracellular compartment through conformational change; and its intracellular, membrane-proximal region interfaces with cytoplasmic signaling proteins as a link to downstream information pathways. CB1R shares with most class-A GPCRs an activation mechanism involving disruption of a highly conserved salt bridge (‘ionic lock’) between an arginine residue of the TMH3 (D/E)RY motif and a negatively charged TMH6 aspartic or glutamic acid residue [30]. Human CB1R (hCB1R) activation involves disruption of the salt bridge between arginine residue R214(3.50) and aspartic acid residue D338(6.30), leading to a conformational change at rotameric tyrosyl Y397 (7.53) that also supports hCB1R ligand-independent (‘constitutive’) activity [31].
Figure 2.
Schematic representation of the human cannabinoid 1 receptor (hCB1R) structure. The serpentine plot identifies the hCB1R transmembrane helices (TMHs), intracellular loops (ICLs), extracellular loops (ECLs), and the membrane-juxtaposed loop (H8). The amino acid identities and sequence were determined experimentally by proteomic analysis of tryptic (grey circles) or chymotryptic (white circles) hCB1R digests. The 27 amino acid residues not identified with high confidence are designated by
 . Symbols (+, *, #) highlight specific CB1R amino acid residues implicated as constituents of the receptor’s allosteric binding sites for and/or be involved in the allosteric action of Org27569 (+) [32–34], Org27569alk3 (*) [35], or GAT100 (#) [36]. The figure has been adapted with permission from [37] Copyright 2010, the American Chemical Society.
. Symbols (+, *, #) highlight specific CB1R amino acid residues implicated as constituents of the receptor’s allosteric binding sites for and/or be involved in the allosteric action of Org27569 (+) [32–34], Org27569alk3 (*) [35], or GAT100 (#) [36]. The figure has been adapted with permission from [37] Copyright 2010, the American Chemical Society.
At the cellular level, the biological actions of CB1R depend upon its activation by endocannabinoid agonists, amide (e.g. anandamide) or ester (e.g. 2-arachidonoylglycerol) conjugates of arachidonic acid whose biosynthetic and inactivation pathways involve prominent representatives of the serine hydrolase enzyme class [27]. The lipophilic nature of cannabinergic ligands suggests that they access the orthosteric binding site in the CB1R TMH bundle through the lipid bilayer rather than directly from the aqueous extracellular milieu [38]. Canonical CB1R-dependent information transmission occurs through coupling with heteromeric G proteins of the Gi/o family to elicit multiple potential signaling events including inhibition of adenylyl cyclase and select voltage-dependent calcium channels and activation of inwardly rectifying potassium channels and mitogen-dependent protein kinases [39]. Under some conditions, CB1R can also interact with Gs or Gq and non-G-protein signaling partners such as GPCR-associated sorting protein 1 (GASP1) and auxiliary proteins including β-arrestins, factor associated with neutral sphingomyelinase (FAN), and adaptor protein-3 (AP-3) to control receptor information output, intracellular trafficking, and internalization/desensitization [40].
As the most abundant metabotropic GPCR in mammalian brain, CB1R is expressed presynaptically in many brain regions where it inhibits, in retrograde fashion, neurotransmitter release from postsynaptic neurons [41]. Compared to the levels of cannabinoid receptor 2 (CB2R) in the periphery, CB1R expression outside the CNS is more limited, being most prominent in tissues involved with substrate storage/mobilization and energy homeostasis, such as liver, adipose tissue, and skeletal muscle [27]. Within and outside the CNS, CB1R has been implicated in a wide range of biological processes. Diverse physiological actions ascribed to CB1R-medicated cannabinergic signaling contribute to prenatal and postnatal neurodevelopment; cardiovascular, renal, reproductive, and hepatic functions; metabolic control and energy balance; feeding behavior; nociception; memory; learning; salience; and emotional status [27,39,42–46]. Disease states associated with abnormal CB1R transmission include substance-use disorders, mood-related psychiatric disturbances (e.g. depression), motor disorders (e.g. multiple sclerosis and Huntington’s disease), diabetes, overweight/obesity, and cancer [47,48]. Polymorphisms of the hCB1R gene, CNR1, are implicated in most of these maladies and others, including irritable bowel syndrome, osteoporosis, inflammatory tissue injury, and coronary heart disease [49]. The chronic, complex nature of most of these disturbances as serious unsolved medical problems and global public health threats has firmly established CB1R as a prime GPCR discovery focus. Escalating interest in exploiting the health potential of medical marijuana has also increased attention on therapeutic CB1R modulation [50]. Albeit of variable quality and degree of association with adverse events, positive efficacy data with CB1R orthosteric agonists or antagonists/inverse agonists in laboratory disease models [27,39] and randomized cannabinoid clinical trials [51] offer additional validation of CB1R as a tractable therapeutic target.
3. CB1R as a prime discovery target for allosteric modulator drugs
Drug discovery efforts involving CB1R as therapeutic target have focused extensively on naturally occurring and synthetic small-molecule orthosteric ligands. However, a paucity of such agents has received regulatory approval over the past 26 years since CB1R was first cloned [52]. With reference to phytocannabinoids as medicines, the CB1R partial agonist Δ9-THC (Marinol® [dronabinol]) is approved for treating anorexic weight loss in acquired immune deficiency syndrome (AIDS) patients, and Sativex® (nabiximols), the combination of Δ9-THC and cannabidiol (CBD) – the latter a low-affinity CB1R ligand whose complex in vivo pharmacology includes indirect effects on CB1R signaling [53]–is approved for relieving multiple sclerosis spasticity and cancer pain [54]. A purified CBD preparation (Epidiolex®) was granted orphan drug designation for treating Dravet syndrome, a rare pediatric onset epilepsy [55]. Although some synthetic CB1R orthosteric ligands have proven quite useful as pharmacological tool compounds in the laboratory [27,56], only two have received regulatory approval. Cesamet® (nabilone) is a synthetic cannabinoid and potent CB1R and CB2R agonist approved to treat chemotherapy-induced nausea and emesis refractory to conventional antiemetics [57]. Rimonabant (Acomplia® [SR141716]), a CB1R antagonist/inverse agonist approved in 2006 the European Union as an adjunctive weight loss agent, was withdrawn by the manufacturer in 2009 due to its unacceptable risk:benefit ratio [58].
The very limited therapeutic actualization of both plant-derived and synthetic CB1R orthosteric ligands may be ascribed largely to their association with on-target, CNS-related liabilities that make definition of an effective therapeutic window challenging, complicate clinical development, and restrict – if not obviate – their medicinal utility. As with the phytocannabinoid Δ9-THC, conventional synthetic CB1R agonists carry abuse potential and the risk of psychological and mood-altering side effects (e.g. psychosis, panic, depression, and anxiety), particularly in predisposed individuals [39,54]. These liabilities are currently underscored by the proliferation of synthetic ‘street drugs’ designed to mimic cannabis, especially since they often act as orthosteric full CB1R agonists with greater potency than the partial agonist Δ9-THC [59]. In contrast to natural cannabis extracts, such synthetic CB1R agonist preparations do not contain other active phytochemicals such as CBD that can reduce the risk of psychosis and psychosis-like effects [60]. Adverse CNS effects of the CB1R antagonist/inverse agonist rimonabant in the clinic included anxiety, depression, and suicidal ideation [58] and may reflect, at least in part, its inverse agonist property, which can alter physiological CB1R constitutive signaling [8,61]. Peripherally disposed CB1R ligands having limited CNS penetration and so-called ‘silent’ or ‘neutral’ CB1R antagonists with no (or low) inverse agonist activity have been pursued as, respectively, pharmacodynamic and mechanistic attempts to circumvent the problems associated with therapeutic targeting of the CB1R orthosteric site [62,63]. Demonstrations of the preclinical therapeutic efficacy of CB1R neutral antagonists and peripheral antagonists/inverse agonists notwithstanding, none has yet reached advanced human trials, let alone the pharmacopoeia.
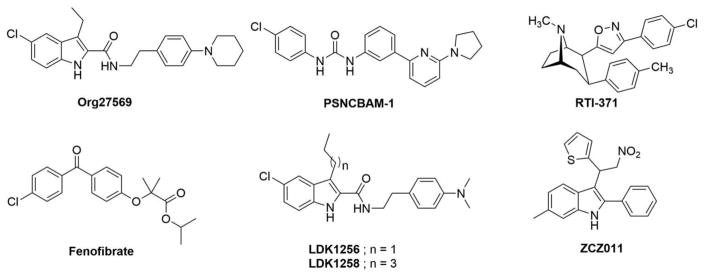
Consensus evidence demonstrates that most GPCRs are subject to allosteric regulation by chemically diverse natural and synthetic agents acting directly on the receptors with structure–activity relationships quite distinct from orthosteric ligands [11,13,17,64,65]. Initial evidence of a CB1R allosteric ligand-binding site was published in 2005 [64]. Accordingly, a tractable option for therapeutically modulating CB1R activity rests with ligands that target this GPCR’s allosteric site(s), given the potential benefits that this modality may bring for fine-tuning CB1R-dependent responses with greater control, precision, and selectivity than typical CB1R orthosteric agonists or antagonists (vide supra). The most intensively studied CB1R allosteric ligands are the synthetic indole Org27569 and the synthetic urea PSNCBAM-1 (Figure 3). These two prototypic CB1R allosteric ligands were initially characterized as having a paradoxical, contradictory pharmacological profile as PAMs of orthosteric agonist affinity, but NAMs of agonist efficacy [64,66]. Subsequent profiling of Org27659 demonstrated its ability to decrease orthosteric anatagonist/inverse agonist SR141716A binding, induce robust CB1R internalization, enhance agonist-induced β-arrestin-mediated extracellular-regulated MAP kinase 1/2 (ERK1/2) signaling in a G-protein-independent manner, act as a NAM of agonist-induced GTPγS binding and β-arrestin recruitment, and inhibit agonist-induced G-protein-mediated inhibition or stimulation of cAMP production [32,67]. These collective data indicate that Org27569 is a biased allosteric agonist in that it enhances agonist-induced ERK phosphorylation yet inhibits other agonist-induced signaling events [32]. Although both Org27569 and PSNCBAM-1 modulate signaling of the CB1R agonists CP55,940 and WIN55,212, they show probe dependence in that they are significantly more potent with CP55,940 than WIN55,212 [32]. Org27569 and PSNCBAM-1 also display (weak) inverse agonist activity in vitro, suggesting that these compounds may not act as ‘pure’ allosteric ligands under some conditions [32,68].
Figure 3.
Chemical structures of synthetic CB1R allosteric modulators discussed in the text.
Org27569, PSNCBAM-1, RTI-371, fenofibrate, LDK1256, LDK1258, ZCZ011
Direct analogs of these first-generation CB1R allosteric ligands have been synthesized, many of which apparently share the CB1R allosteric phenotype of the respective parent compound, albeit as characterized by less extensive experimental data than for the more well-studied Org27569 and PSNCBAM-1 [64,69–71]. For instance, LDK1256 and LDK1258 (Figure 3) are Org27569 analogs that share with the parent compound the ability to act as a PAM of orthosteric agonist (CP55,940) binding and a NAM of agonist-induced CB1R-G protein coupling while also capable of inducing β-arrestin-mediated ERK1/2 phosphorylation [69,72]. The dopamine transport inhibitor RTI-371 (Figure 3) has been identified as a synthetic CB1R PAM, and the peroxisome proliferator activated receptor-alpha (PPAR-α) agonist fenofibrate (Figure 3), as a synthetic CB1R NAM [73,74].
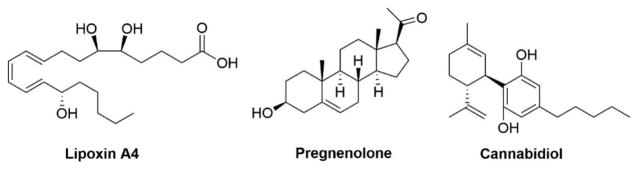
Natural substances identified as potential CB1R allosteric ligands include the nonclassical anti-inflammatory eicosanoid lipoxin A4 acting as a CB1R PAM of orthosteric ligand binding and adenylyl cyclase activity [75]; the steroid pregnenolone acting as a NAM on CB1R-mediated ERK1/2 phosphorylation without affecting cAMP-mediated signaling or orthosteric agonist binding affinity [76]; and CBD acting as a noncompetitive CB1R NAM [77] (Figure 4). Lipoxin A4 and pregnenolone allostery at CB1R, however, remains somewhat controversial [78]. Intriguingly, a family of hemopressin-related peptide endocannabinoids (‘pepcans’) present in rodent brain and plasma and human plasma act as CB1R NAMs on noradrenergic neurons in particular [79,80]. Identification of endogenous agents as putative CB1R allosteric ligands suggests the importance of allostery to physiological CB1R function.
Figure 4.
Chemical structures of small-molecule natural substances identified as CB1R allosteric modulators discussed in the text.
Lipoxin A4, pregnenolone, cannabidiol (CBD)
When examined in operational models of receptor allostery, binding parameters characterizing some of the above-referenced ligands as CB1R NAMs and PAMs are consistent with the allosteric ternary-complex model for 7TMRs and the proposition that CB1R has coupled, topographically and structurally distinct allosteric and orthosteric binding sites that engage modulatory ligands [64,78]. Reminiscent of allosteric modulators of other GPCRs [13,18–20,22], binding of at least some allosteric ligands to CB1R is considered to produce conformational ensembles and functional receptor states along the activity coordinate that are unique and unattainable by orthosteric ligands alone [65,81]. In silico modeling and experimental studies have begun to delineate and characterize putative discrete receptor sites involved in CB1R engagement of, especially, Org27569/PSNCBAM-1 and relate allosteric ligand engagement to subsequent CB1R functional modulation and intracellular trafficking (Section 4, vide infra).
As implied by the foregoing, most investigations of CB1R allostery have focused on defining CB1R NAM and PAM binding and signaling character in cellular (overexpression) systems using synthetic cannabinergic agonists such as WIN55,212–2 and, especially, CP55,940 as orthosteric ligands. Although definitive identification of a test compound as an allosteric GPCR modulator requires characterization of the compound’s effect on orthosteric ligand-binding kinetics by active receptor [13,19,24], the (patho)physiological significance of such biochemical data to biological systems is inherently limited, since allosteric pharmacology deduced from studies with synthetic agonists may be highly contextual (e.g. be probe dependent and/or test system specific) and of limited relevance to endogenous cannabinoid agonists. This view is supported by results from a recent electrophysiological study using cultured murine hippocampal neurons as a cell system with native CB1R signaling to characterize the molecular pharmacology of Org27569, PSNCBAM-1, and Pepcan-12 (RVDPVNFKLLSH) [82]. In this cell system, Org27569, PSNCBAM-1, and Pepcan-12 (RVDPVNFKLLSH) influenced 2-AG-mediated depolarization in a manner consistent with their predicted allosteric modulation as previously defined in cell-based studies using synthetic orthosteric ligands or the phytocannabinoid Δ9-THC [32,64,66,67,79]. In contrast, lipoxin A4 acted unexpectedly as a CB1R NAM against 2-AG-stimulated CB1R signaling and not as a PAM, as would have been predicted from a prior study [75]. Neither the purported PAM lipoxin A4 [75] nor the purported NAM pregnenolone [76] had any effect on 2-AG-mediated signaling in the neuronal cells. These data highlight the difficulty in reconciling data among test systems using synthetic orthosteric CB1R ligands versus endocannabinoids.
Emerging in vivo preclinical data from experimental disease models in laboratory animals indicate that CB1R allosteric modulation may have therapeutic utility. Lipoxin A4 reduced cognitive deficits in a murine model of β-amyloid-induced learning and memory impairment, suggesting that allosteric CB1R modulation may be a promising strategy for ameliorating the degenerative processes of Alzheimer’s and other neuroinflammatory diseases [75]. The significance of this finding to therapeutic CB1R allosteric modulation is made provisional by concerns that lipoxin A4 may not act as a true CB1R allosteric modulator [78]. In acute rat feeding models, PSNCBAM-1 and Org27569 reduced food intake and weight gain, indicating that CB1R allosteric modulation could represent a therapeutic modality for weight control and management of the cardiometabolic risks associated with overweight/obesity [66,83], although Org27659’s hypophagic effect may be CB1R independent [84]. Org27569 attenuated the cue-induced and drug-induced reinstatement of cocaine-seeking and methamphetamine-seeking behavior, inviting the possibility that CB1R allosteric modulators could represent effective pharmacotherapy for substance-use disorders [85]. The allosteric antagonist effect of PSNCBAM-1 on neuronal excitability suggests that CB1R allosteric modulation has therapeutic potential in diseases such as cerebellar ataxia [68]. In mice, pregnenolone inhibited several adverse behavioral and neurological effects of Δ9-THC intoxication and blunted the concordant potentiation of dopamine and glutamate release [76]. Arguably, the strongest experimental evidence in support of CB1R allostery as a therapeutic modality comes from recent in vivo demonstration in the conditioned place preference paradigm that a synthetic, brain-penetrant CB1R PAM, ZCZ011 (Figure 3), elicited CB1R-mediated antinociceptive effects in a mouse model of neuropathic pain without inducing the adverse psychoactivity or detrimental motivational effects (e.g. aversion) commonly associated with orthosteric cannabinoid agonists [86]. Caution should be exercised in ascribing any of these in vivo pharmacological effects solely, or even primarily, to on-target allosteric actions of the test ligands at CB1R, since the biological behaviors and molecular phenotypes of GPCR/CB1R allosteric ligands routinely delineated in cell overexpression systems employing synthetic orthosteric ligands may not be preserved in complex animal models in vivo.
4. Localization and architecture of CB1R allosteric ligand-binding sites and structure–function correlates
Optimal design of GPCR-targeted ligands as potential therapeutics depends upon detailed characterization of the molecular basis of ligand recognition, the receptor’s ligand-binding domains, and the conformational consequences of ligand engagement that generate specific signaling responses, all within the structural and energy dynamics of the resulting conformational ensemble. In these regards, orthosteric GPCR ligand-binding pockets continue to be studied most intensively with state-of-the-art structural biology techniques, given the role of natural ligands as templates for medicinal chemistry and the preponderance of GPCR-targeted orthosteric ligands among successful drugs [4,5]. For example, protein X-ray crystallography has provided atomic level structural information on druggable GPCRs in apo and liganded forms of value to drug discovery, notwithstanding inherent limitations (e.g. its static nature) and biases introduced by, for instance, protein engineering to facilitate crystallization [87,88]. A newer structural method, single-particle cryo-electron microscopy (cryo-EM), is also being used to map orthosteric drug-GPCR conformational states at near-atomic resolution [89].
In a more limited fashion as compared to GPCR orthosteric sites, computational and experimental approaches have been used to gain insight into the structural basis of GPCR allostery and characterize and quantify allosteric behavior. Recent reviews may be consulted for detailed exposition of these topics beyond this article’s scope [11,13,19,90–96]. Since most such investigations have focused on GPCRs other than CB1R [97], select examples among the most well-studied GPCRs are given to exemplify some of the approaches adopted to delineate GPCR allosteric structure–function correlates. Crystallographic structures with bound allosteric modulators of the class-A δ-opioid [98] and μ-opioid [99] and muscarinic acetylcholine (mAch) M1, M2, and M4 receptors [100,101]; the class-B glucagon-like peptide-1 (GLP-1) receptor [102]; and the class-C metabotropic glutamate (mGlu) receptors 1 [103] and 5 [104] have provided valuable insights into ligand binding-site topologies and docking poises involved in GPCR allosteric control. In silico and modeling approaches such as docking and molecular dynamics (MD) simulation have been utilized for probing ligand fit and calculating time-dependent GPCR conformational changes in the millisecond to femtosecond range following allosteric ligand engagement, in some cases adjunctive to crystallographic analyses [14,28,99,105–108]. Experimental techniques such as nuclear magnetic resonance spectroscopy (NMR) [109,110], site-directed mutagenesis coupled with quantitative pharmacological analyses [111–114] or long-timescale MD simulations [115] have also been applied to gain structural insight into GPCR allostery. Such approaches have also helped define structure–activity relationships among GPCR-targeted allosteric scaffolds [116,117].
Resulting data on the structural biology of GPCR allosteric ligand binding from the above-cited and other studies [118] have highlighted some generally shared characteristics of GPCR allosteric binding pockets, such as their usual, but not inevitable, location within the 7TMH helical bundle and not at the receptor surface. Specific amino acids serve as critical recognition and interaction points between particular allosteric ligands and target GPCRs critical to stabilizing receptor conformations in which the allosteric effector is cooperatively and functionally active within the given cellular context. Yet GPCR structural transitions consequent to allosteric ligand engagement that support allosteric communication and information transmission along discrete signaling circuits appear diverse and highly dynamic, ranging from local unfolding to rigid body motions, reflecting fundamental thermodynamic and structural properties of any given GPCR.
Given that characterization of allosteric binding sites is critical to rational, structure-based design of GPCR-targeted drugs, considerable attention is being paid to uncovering the properties of CB1R allosteric ligand-binding pockets and delineating the conformational sequelae of allosteric ligand engagement that determine CB1R signaling output.
4.1. CB1R Org27569 and PSNCBAM-1 binding regions
Efforts to elucidate the binding sites and structural consequences of allosteric ligand engagement by CB1R have involved mainly Org27569 and, to a lesser extent, PSNCBAM-1. The emphasis on Org27569 has been promulgated by its intriguing, contradictory pharmacological properties as a PAM of orthosteric agonist affinity, but a NAM of agonist efficacy [64,66].
The observation that mutating hCB1R tryptophan W5.43 abrogated Org27569’s ability to inhibit CP55,940 signaling implicated this residue in the Org27569 binding site [32]. W5.43 mutation also adversely affected binding of the orthosteric aminoalkylindole agonist WIN55,212 [119], suggesting the possibility that overlap between the binding pockets for WIN55,212 and Org27569 facilitates their allosteric cooperativity. A subsequent study identified a putative Org27569 allosteric binding site comprised of regions from CB1R’s TMH3, TMH6, and TMH7 [33]. Partial overlap between this binding pocket and that of rimonabant (SR141716A) presumably accounts for the ability of Org27569 to displace SR141716A; yet the Org27569 binding pocket features the more elaborated extracellular extension. Specific amino acid residues were identified as candidate Org27569-CB1R interaction points. In the absence of SR141716A orthosteric ligand, Org27569 interacts with CB1R phenylalanine residue F3.25 (189) to form a hydrogen bond between Org27569’s piperidine nitrogen and TMH3 lysine K3.28(192). Mutation and pharmacological data indicate that the Org27569-K3.28(192) hydrogen bond may be critical to CP55,940’s antagonist and inverse agonist actions. Extensive in silico modeling and MD simulation data for Org27569 docked into this putative CB1R binding pocket further suggest that Org27569 antagonizes the efficacy of CP55,940 by sterically hindering conformational adjustments important to CB1R activation in three complementary ways: blocking movements of the second extracellular loop (ECL2) by forming an aromatic interaction between Org27569’s indole ring and ECL2 phenylalanine Phe268; prohibiting a key electrostatic interaction between ECL3 lysine residue Lys373 and TMH2 aspartic acid residue D2.63(176); and hindering TMH6 movements important to CB1R activation and interaction with G proteins (Figure 2). The latter feature is congruent with the hypothesis [34] that Org27569 promotes establishment of a CB1R conformation intermediate between inactive and inactive states favoring receptors that preferentially bind orthosteric agonist, but are incapable of signaling through G-protein-mediated pathways.
Other studies have identified amino acids critical to CB1R allosteric ligand engagement and bioactivity through the use of pharmacologically active small molecules as covalent probes. Typical probe generation entails chemically modifying a noncovalent GPCR allosteric ligand with a warhead capable of reacting with defined amino acid residue(s) within (or very near) the target receptor’s ligand-binding domain. In this way, the probe serves as a chemical reporter of the target protein’s ligand-binding motif. This approach has met with conspicuous success for mapping orthosteric binding sites in (patho)physiologically important GPCR drug targets [120–123], including hCB1R [124]. In perhaps the most well-developed experimental paradigm along this line, designer orthosteric covalent probes have been integrated with mutational studies and peptide-level liquid chromatography-tandem mass spectrometry (LC/MS-MS) analysis of the covalently modified GPCR into a multidisciplinary, proteomics-based experimental paradigm termed ‘ligand-assisted protein structure’ (LAPS) to define interaction sites between CB1R or CB2R and orthosteric ligands [125,126].
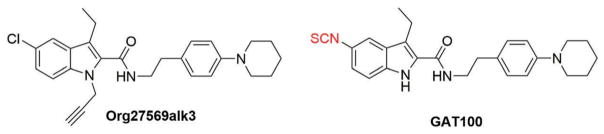
The first report to apply a probe for interrogating CB1R allosteric sites utilized an Org27569 derivative, Org27569alk3 (Figure 5), that features a reactive alkyne warhead capable of forming covalent adducts with amino acid nucleophiles containing either sulfhydryl (e.g. cysteine) or hydroxyl (e.g. serine, threonine, and tyrosine) groups [35]. Although Org27569alk3 was not characterized as to its CB1R binding affinity or (on-target) biological activity, the compound appeared sufficiently nontoxic to be tolerated by human embryonic kidney (HEK293) cells expressing recombinant green fluorescent protein (GFP)-labeled CB1R. Peptide-level LC-MS/MS analysis of CB1R-GFP from cells incubated with Org27569alk3 indicated that the probe covalently reacted with Ser2.45 and/or Ser3.42 in TMH2 and TMH3, respectively. Computational data along with mutational effects on Org27569 binding and activity analyzed with a fluorescent rimonabant (SR141716A) derivative indicated that the Org27569 binding pocket includes key Cys1.55, His2.41, and Phe4.46 residues of TMHs 1, 2, and 4, respectively (Figure 2). Intriguingly, homology analysis of CB1R with other class-A GPCRs suggested that the Org27569 binding site corresponds to a regulatory cholesterol consensus motif controlling CB1R’s regional disposition in the neuronal plasma membrane and ligand affinity: augmenting intracellular cholesterol shuttled CB1R to the axon while increasing orthosteric antagonist/inverse agonist binding, whereas reducing intracellular cholesterol enriched CB1R in neuron cell body (soma) membranes while decreasing Org27569 binding. In silico MD simulations and analyses of binding-mode stability analyses with rat brain membranes rich in CB1R suggested that Org27569 engagement elicits a displacement of TMH3, whose Leu3.29 and Ala3.34 residues are essential to Org27569’s allosteric effect on CP55,940 binding affinity, and a rearrangement of intracellular loop 1 (ICL1)-H8 that abrogates CB1R-G-protein coupling (Figure 2). This latter conformational response offers an alternative mechanistic rationalization to that proposed by Fay and Farrens [34,127] regarding the seemingly paradoxical effects of Org27569’s as both a CB1R-positive allosteric modulator and an antagonist of CB1R-G-protein coupling [32,64,67].
Figure 5.
Chemical probes for CB1 R allosteric sites discussed in the text.
Org27569alk3, GAT100
The first-reported library of small-molecule covalent probes purpose-designed to bind irreversibly to discrete amino acids within (or very near) the hCB1R allosteric site(s) for the prototypic CB1R NAMs, Org27569 and PSNCBAM-1, has recently been reported [36,128]. Probe design strategy involved substituting strategic positions on Org27569 and PSNCBAM-1 with either a photoactivatable azide (–N3) or benzophenone [–(C6H5)2CO] or an electrophilic isothiocyanate (–NCS) warhead. Among the resulting novel compounds, the Org27569 isothiocyanate analog, GAT100 (Figure 5), was identified as a PAM of CP55,940 binding to rat-brain CB1R and recombinant hCB1R and an exceptionally potent NAM across cell-based assays of CP55,940-, 2-AG-, and anandamide-dependent β-arrestin recruitment, GTP-γ-35S binding, and cAMP formation. Results from competitive kinetic and saturation-labeling experiments indicated that GAT100 irreversibly labeled hCB1R by virtue of its reactive isothiocyanate moiety. Extensive computational modeling and MD simulation studies identified TMH7 cysteine C7.38(382) as the hCB1R residue most likely to react with GAT100 (Figure 2) [36]. Identification of C7.38(382) as a key feature of GAT100’s binding motif is supported by the observation that, under the physiological incubation/reaction conditions (aqueous milieu, pH 7.4) used in the study, their thiol moiety renders protein cysteine residues the most reactive amino acid nucleophile toward isothiocyanate electrophiles [129–131]. From a therapeutic standpoint, the finding that GAT100 lacked the inverse agonism associated with Org27569 and PSNCBAM-1 is quite compelling [36,128], since the inverse agonist property of orthosteric CB1R antagonists/inverse agonists has been associated with undesirable on-target somatic and psychobehavioral adverse events that have severely undercut the clinical potential of this class of CB1R modulators [8,63]. Additional analogs of Org27569 bearing reactive photophore functionalities and that share the parent compound’s activity as CB1R NAMs of CP55,940-induced G-protein coupling may find utility as CB1R allosteric covalent probes, once validated and profiled for their biological activity [132].
Other, more mechanistic studies have focused on the conformational consequences of CB1R allosteric ligand engagement as related to the modulator’s functional pharmacology. By using wild-type CB1R and CB1R threonine mutants T210A and T210I that are, respectively, constitutively inactive and constituently active, Ahn et al. provided evidence suggesting that Org27569 promotes a conformational change in CB1R that generates a high-affinity agonist binding state biased toward downstream ERK phosphorylation [67]. In an attempt to link allosteric CB1R modulation to structural features of the {CP55,940-CB1 R-Org27569} conformational ensemble, Fay and Farrens constructed and purified two CB1R variants, a ‘minimal-cysteine’ mutant containing only the two cysteines required for CB1R function (Cys257 and Cys264) and another that introduced into that minimal-cysteine CB1R variant a cysteine residue at TMH6 site 6.34 [34]. The latter mutant enabled labeling of the cytoplasmic face of CB1R Cys6.34 with a site-directed fluorophore, a modification that did not alter the receptor’s functional affinity and efficacy for orthosteric (ant)agonists. Molecular modeling and fluorescence spectroscopy revealed that Org27569 is not a competitive inhibitor for the CB1R orthosteric site, but rather acts directly on CB1R by engaging a ‘nontraditional’ (allosteric) site to form a ternary complex with CP55,940 agonist and exert positive cooperativity, i.e. stimulate CP55,940 binding. Structurally, Org27569 blocked a key conformational change at the cytoplasmic end of TMH6 associated with agonist (but not antagonist) binding to CB1R (and to several other class-A GPCRs) so as to inhibit formation of a fully active CB1R structure. The authors suggest a structure–function correlate whereby Org27569 traps CB1R in a discrete conformational state that stimulates orthosteric agonist (i.e. CP55,940) binding to the receptor independent of the degree to which the receptor is coupled to G-protein for intracellular signaling, thus providing a structure-based rationale for Org27569’s paradoxical ability to enhance CP55,940 affinity and decrease its efficacy [32,64,66,67]. Suggestion was also made that this particular {CP55,940-CB1R-Org27569} ensemble constitutes a stabilized intermediate along a conformation and activity continuum from orthosteric agonist engagement to maximally active receptor [34].
A site-directed fluorescence study involving the minimal-cysteine (and other) CB1R mutants linked movements of CB1R TMH6 to G-protein activation for establishing a unique, inactive-like CB1R state stabilized by Org27569 and distinct from the orthosteric antagonist-bound, inactive state [127]. Org27569 allostery was further proposed to involve transitioning among CB1R activation states made possible by TMH5, TMH6, TMH7, and H8 rearrangements reflecting altered, water-mediated interhelical interactions. Conformational equilibrium in a TMH6-TMH7/H8 microdomain was suggested to be a decisive determinant of CB1R biased signaling between G-protein-independent (e.g. β-arrestin-mediated) and G-protein-dependent (e.g. adenylyl cyclase/cAMP-mediated) networks.
Predicated upon prior demonstration that most of CB1R’s extensive N-terminus is not necessary for orthosteric ligand binding, whereas mutations within the conserved N-terminal region proximal to the intracellular aspect of the plasma membrane impair that binding [133], the potential involvement of CB1R’s extensive N-terminal domain as contributor to the activity of CB1R allosteric ligands Org27569 and PSNCBAM-1 was investigated [134]. The findings indicate that a CB1R orthosteric binding pocket couples with the receptor’s N-terminal membrane-proximal region by virtue of the latter’s redox-sensitive, solvent-accessible, intramolecular disulfide bridge between Cys98 and Cys107 within the receptor’s extracellular N-terminus (Figure 2). Interplay among the binding domains of the orthosteric agonist CP55,940, the allosteric ligands Org27569 and PSNCBAM-1, and these two CB1R N-terminal cysteine residues was hypothesized to modulate allosterically the receptor’s affinity for (ant)agonists and regulate CB1R allosteric coupling by controlling ligand access to (an) allosteric binding pocket(s) at/near the receptor’s N-terminus.
4.2. CB1R CBD and pepcan-12 binding regions
As compared to Org27569 and PSNCBAM-1, information on CBD and pepcan binding sites in CB1R is at present very limited. In cell overexpression systems, an analysis of CB1R mutational variants [77] indicated that the negative, noncompetitive allosteric activity of CBD in the presence of either 2-AG or Δ9-THC is independent of the receptor’s first 89 N-terminal amino acids as well as the cysteine residues (Cys98 and Cys107) within the receptor’s extracellular N-terminus, the latter suggested [134] to play a role in Org27569’s and PSNCBAM-1’s allosterism by virtue of disulfide formation. In contrast, CBD’s allosteric activity at CB1R appeared to depend, at least in part, on the mere presence of polar amino acid residues (e.g. cysteine or serine) at N-terminal positions 98 and 107, independent of their ability to form a disulfide bridge [77].
Data from competitive displacement experiments with a fluorescent Pepcan-12 derivative as reporter demonstrated that N-terminal derivatization of Pepcan-12 abolished its binding to CB1R, whereas C-terminal derivatization had much less effect, inviting the postulate that allosteric pepcan binding occurs within the receptor’s N-terminal extracellular region and does not involve the CB1R TMH bundle [79].
5. Conclusion
Allostery invites new medicinal chemistry approaches and pharmacotherapeutic avenues for GPCR-targeted drugs with the promise of enhanced selectivity, increased signaling diversity, improved safety margins, and wider clinical application as compared to conventional orthosteric GPCR ligands. Therapeutic modulation of GPCR allosteric sites has particular importance to CB1R, a predominant CNS GPCR whose translational impact has been compromised by adverse effects associated with conventional orthosteric CB1R agonists and antagonists/inverse agonists. Accumulating data shed light on the relationship between the cell signaling effects of an increasing diversity of CB1R allosteric ligands and the receptor’s conformational and functional allosteric repertoires. While this information has helped inform the molecular pharmacology of CB1R allostery, further understanding of the structure–function correlates of CB1R allosteric modulation in living systems and how they relate to disease processes is required near-term to help extend CB1R’s potential as a druggable target. To this intent, purpose-designed covalent probes are emerging as important tools for interrogating CB1R allosteric binding sites and defining therapeutically relevant CB1R allosteric structural motifs.
6. Expert opinion
The dominance of GPCRs as the principal class of biomolecules targeted in drug discovery shows no sign of abating [135,136]. Most GPCR-based discovery pharmacology and drugs resulting therefrom have involved ligands whose medicinal effects reflect their activity at the orthosteric ligand-binding site of a therapeutically relevant GPCR [4,5]. Nonetheless, GPCR-related discovery initiatives are increasingly embracing the concept of allostery for designing and developing new chemical entities as proprietary drugs with the principal aim of leveraging the enhanced safety, selectivity, and pharmacological versatility that GPCR allosteric modulators may bring over orthosteric ligands [11,137,138]. The authors opine that discovery interest in GPCR allosteric ligands will increase, particularly in those instances where conventional orthosteric modulation of a GPCR disease target elicits preclinical liabilities that work against the target’s therapeutic exploitation and/or where clinically efficacious orthosteric modulators have elicited adverse events restricting or curtailing their medicinal use. Although CB1R has been implicated in a variety of disease processes as one of the most prominent CNS GPCRs [47–51], typical CB1R orthosteric ligands carry both these limitations [8,39,54,58,59,61]. Consequently, despite intensive medicinal chemistry efforts over the past two decades focused on CB1R orthosteric modulators and the public and private sector funds invested in the associated research, few new CB1R-modulator drugs have emerged [52,54,56]. Allosteric modulation may thus be considered a front-line molecular pharmacology approach for expanding the druggable CB1R landscape and generating new opportunities to modulate CB1R for health benefit with ‘smarter’ molecules. This proposition is particularly germane to the discovery and development of clinically useful CB1R PAMs, since most therapeutic applications of CB1R modulator cannabinergic agents involve this receptor’s pharmacological activation, which is associated with myriad unwanted psychobehavioral effects [39,54].
Given the association between direct CB1R activation by orthosteric agonists and on-target adverse events, the intrinsic agonist activity of ago-PAMs might appear a priori to obviate their candidacy as CB1R modulator drugs. Many factors, however, can alter the balance between the direct-agonist and PAM activities of an ago-PAM, including expression level of the target GPCR, distinctive signaling characteristics (bias, kinetics) between an orthosteric agonist alone and the ago-PAM, and differences among ‘active-state’ conformations stabilized by an orthosteric agonist versus an ago-PAM [26,139,140]. These and other factors are believed to contribute to contrasting observations that convulsions and seizures elicited in rodents by a metabotropic glutamate receptor subtype 5 (mGlu5) ago-PAM (and by a ‘pure’ mGlu5 PAM [141]) reflect pathological mGlu5 overstimulation by the ago-PAM [142]. In contrast, other agents characterized as ‘strong’ mGlu5 ago-PAMs in vitro evidenced similar preclinical therapeutic effects as antipsychotics in vivo to structurally related ‘pure’ PAMs, but induced no adverse events [140]. These considerations may also help account for the finding that an ago-PAM of the α7 nicotinic acetylcholine receptor (α7 nAchR) acts in vivo more effectively than ‘pure’ PAMs to reduce pain [26,143]. Thus, given the many contextual factors that could potentially influence the pharmacological profile of a CB1R ago-PAM, the molecular pharmacology of a CB1R ago-PAM as determined in recombinant cell lines or primary cells in vitro need not always predict its salutary and adverse activities in vivo, for at the systems level the relative proportion of direct-agonist versus PAM activity may vary.
Maturation of the field of CB1R allosteric modulators requires additional information on several fronts. At the (sub) molecular level, deeper exploration of the CB1R allosteric ligand chemical space, delineation of the precise location and structure of CB1R allosteric binding domains, and definition of preferred binding motifs for chemically and functionally distinct allosteric ligands are needed. For these purposes, pharmacologically well-characterized covalent probes should be of immense help in defining CB1R allosteric site location and topology and identifying specific amino acid residues involved in the recognition, engagement, and activity of CB1R allosteric ligands, particularly with respect to their structure– function correlates across distinct allosteric phenotypes (e.g. NAMs vs. PAMs) and/or pharmacological characteristics (probe dependence, binding-site selectivity, and signaling bias) and how these properties relate to CB1R’s conformational repertoire. Validated CB1R covalent allosteric probes could also be used to derive and refine mechanistic receptor homology models and help formulate novel (virtual) screening paradigms for new CB1R-targeted dugs that capture CB1R conformations in therapeutically attractive activation states. As exemplified by the LAPS paradigm for orthosteric CB2R (ant)agonists [125,126], purpose-designed covalent allosteric ligands with defined pharmacological and signaling activities are finding ready utility as probes for direct experimental interrogation of CB1R allosteric binding pockets and could be used as chemical reporters in peptide-level LC-MS/MS analysis of the covalent adducts formed. The resulting structural information is anticipated to help specify design criteria for drug-like CB1R allosteric modulators and identify privileged motifs for new drug templates and/or chemotype scaffolds toward hit and lead development in a rational discovery program to deliver allosteric modulators as CB1R-targeted drugs. To complement these conventional discovery approaches, the allosteric probes could themselves be used as leads for structure-guided drug design and as tools for identifying ‘orphan’ CB1R allosteric sites yet to be annotated. Covalent allosteric CB1R probes could prove therapeutically useful, especially if they combine the unique pharmacological merits of allosteric modulators (cooperative functionality, higher specificity, less adverse-event risk) at the potentially lower dosages afforded by a covalent drug, thereby helping ameliorate a perceived drawback of covalent drugs, off-target toxicity [144–146]. Pending further data regarding the utility of covalent GPCR modification with targeted allosteric ligands for eliciting salutary receptor responses, therapeutic application of CB1R covalent ligands remains theoretical.
An atomic level structure of CB1R in native form has remained elusive. Orthosteric ligands have aided the crystallization of several ‘druggable’ GPCRs for X-ray structural analysis [97]. However, the static nature of the resulting crystals obviates direct insight into the myriad functional distinctions among allosteric ligands that engage the same or similar CB1R site(s), but display distinct allosteric characters (e.g. with or without probe dependence). Although single-particle cryo-EM has been successfully used to identify conformational changes induced by the binding of a small-molecule allosteric inhibitor to a 93-kDa enzyme drug target [89], the molecular mass of hCB1R (~52.8 kDa) [37] is below the current limit of this technique. Despite the technical hurdles, the authors forecast that allosteric ligands will be used to facilitate CB1R crystallization and, as the technology develops, cryo-EM analysis of GPCR/CB1R conformations biased for a particular salutary allosteric phenotype.
Fundamental identification of a GPCR/CB1R ligand as an allosteric modulator is routinely made at the molecular level in vitro by establishing its cooperative effect on orthosteric ligand binding affinity and/or efficacy in a biochemical or cellular preparation enriched for the GPCR target of interest [82,147]. This principle notwithstanding, the influence of contextual factors on the phenotype and pharmacological effects of CB1R allosteric modulators and their cooperative nature (vide supra) demands multiparametric characterization of any such putative ligand in living systems, especially in those cases where the resulting data are being used to inform drug discovery. It is suggested that any CB1R ligand identified as an allosteric agent by the fundamental biochemical criterion of cooperative orthosteric ligand affinity/efficacy modulation be profiled across G-protein-dependent and G-protein-independent signaling pathways with both synthetic and endogenous CB1R orthosteric ligands in cell lines overexpressing recombinant hCB1R as well as in primary cells/tissue explants expressing native receptor. Use of such multiple in vitro cell-based models expressing both recombinant and native (h)CB1R helps build a reinforcing data set, allows unambiguous delineation of any influence of test system on probe or signaling pathway dependence and signaling bias, and could afford insight as to whether biased CB1R allosteric modulation and/or endogenous allosteric endocannabinoid tone affects the pharmacological profile/therapeutic potential of the test agent. For example, pathway-biased CB1R PAMs may offer a means to circumvent the undesired psychoactive effects associated with conventional CB1R agonists, especially in those diseases (e.g. substance use disorders, obesity, and nonalcoholic fatty liver) whose etiology has a component of CB1R (hyper)activation by endocannabinoids. This type of information could invite a more personalized approach for CB1R-targeted therapies, given that allosteric modulators must work in cooperation with endogenous orthosteric agonists to exert their pharmacological effects. To augment traditional ligand-binding and functional assays in GPCR pharmacology, agnostic, phenotypic cell-based assays involving label-free electrical impedance-based or optical-based biosensors to investigate the effects of CB1R allosteric modulators on cell shape, and structure [148], chemical proteomics approaches such as activity based protein profiling to interrogate the potential interaction of a CB1R allosteric ligand with other members of the cellular proteome [149], or surface plasmon resonance for direct measurement of CB1R-ligand interaction by [150] could offer a means to obtain a more comprehensive picture of CB1R allosteric agents in disease-relevant systems.
More information is also required regarding the salutary effects of exogenous allosteric CB1R modulators in laboratory disease models. As a potential therapeutic, any CB1R ligand characterized biochemically as an allosteric modulator would need to be tested for its preclinical therapeutic efficacy and mode of action in cell-based and laboratory animal models for its ability to redress associated disease markers/pathology in ways that would be attractive for further development as a candidate drug. This line of experimental inquiry should shed light on the relationship among a CB1R allosteric ligand, its therapeutic signaling, and the disease symptoms ameliorated. Furthermore, in vivo mechanistic information on CB1R allosteric modulators could open the door to developing drugs that selectively fine-tune CB1R receptors in different cellular subpopulations/tissue types [151] with distinct temporal (time-dependent) and spatial modulation of the effects of CB1R orthosteric ligands [152].
Future research will demonstrate whether exploitation of GPCR allostery could extend the accessible chemical space for drug-like CB1R ligands and thus revivify CB1R as a drug target with agents having greater selectivity, efficacy, and safety than conventional orthosteric CB1R (ant)agonists. It is hoped that this discourse stimulates further design and profiling of GPCR/CB1R allosteric modulators with an eye toward their eventual clinical application so as to broaden the impact of protein allostery on contemporary medicine.
Article highlights.
An allosteric ligand binds to a G-protein coupled receptor (GPCR) site structurally and topographically distinct from the classical orthosteric site that engages endogenous ligands. Cooperative conformational and functional coupling between allosteric and orthoseteric binding pockets enables an allosteric ligand to modulate the affinity/efficacy of an orthosteric ligand and the magnitude/direction of the GPCR’s signaling output.
Allosteric GPCR modulators display several unique properties that, if translatable into the clinic, could lead to improved, safer drugs.
The cannabinoid receptor 1 (CB1R) is a metabotropic class-A GPCR involved in diverse physiological activities and implicated in the pathology of several disease states with major unmet medical needs.
Despite the (pre)clinical therapeutic benefits associated with some novel orthosteric CB1R ligands, these agents have met with little translational success due to their on-target adverse events that might be circumvented with CB1R allosteric modulators.
Improved understanding of CB1R allosteric binding pocket(s) and the requirements for allosteric-ligand recognition, engagement, and activity is critical to inform the rational design of CB1R allosteric modulators as pharmacotherapeutics.
Covalent ligands pharmacologically active as allosteric modulators have recently been applied as designer probes to interrogate directly and experimentally the structure-function correlates of CB1R allosteric ligand-binding site(s).
Information on the interaction profiles and molecular pharmacology of CB1R allosteric ligands will continue to inform GPCR allostery as a therapeutic modality and guide the design, targeting, and application of new-generation CB1R ligands as potential drugs.
This box summarizes key points contained in the article.
Acknowledgments
Funding
GA Thakur was supported by National Institutes of Health grants DA027113 and EY024717.
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Roth BL, Kroeze WK. Integrated approaches for genome-wide interrogation of the druggable non-olfactory G protein-coupled receptor superfamily. J Biol Chem. 2015;290:19471–19477. doi: 10.1074/jbc.R115.654764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Eggert US. Non-traditional roles of G protein-coupled receptors in basic cell biology. Mol Biosyst. 2013;9:586–595. doi: 10.1039/c2mb25429h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 5.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 6.Bradley SJ, Tobin AB. Design of next-generation G protein-coupled receptor drugs: linking novel pharmacology and in vivo animal models. Annu Rev Pharmacol Toxicol. 2016;56:535–559. doi: 10.1146/annurev-pharmtox-011613-140012. [DOI] [PubMed] [Google Scholar]
- 7.Venkatakrishnan AJ, Deupi X, Lebon G, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 8.Janero DR, Lindsley L, Vemuri VK, et al. Cannabinoid 1 G protein-coupled receptor (periphero-)neutral antagonists: emerging therapeutics for treating obesity-driven metabolic disease and reducing cardiovascular risk. Expert Opin Drug Discov. 2011;6:995–1025. doi: 10.1517/17460441.2011.608063. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson KA. Structure-based approaches to ligands for G-protein-coupled adenosine and P2Y receptors, from small molecules to nanoconjugates. J Med Chem. 2013;56:3749–3767. doi: 10.1021/jm400422s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Guarnera E, Berezovsky IN. Allosteric sites: remote control in regulation of protein activity. Curr Opin Struct Biol. 2016;37:1–8. doi: 10.1016/j.sbi.2015.10.004. Excellent review of protein allostery and its functional consequences. [DOI] [PubMed] [Google Scholar]
- 11••.Gentry PR, Sexton PM, Christopoulos A. Novel allosteric modulators of G protein-coupled receptors. J Biol Chem. 2015;290:19478– 19488. doi: 10.1074/jbc.R115.662759. Recent chemistry overview of pharmacologically active allosteric ligands for therapeutic GPCRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelstein SJ, Le Novère N. Cooperativity of allosteric receptors. J Mol Biol. 2013;425:1424–1432. doi: 10.1016/j.jmb.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Mol Pharmacol. 2014;86:463–478. doi: 10.1124/mol.114.094342. [DOI] [PubMed] [Google Scholar]
- 14.Hertig S, Latorraca NR, Dror RO. Revealing atomic-level mechanisms of protein allostery with molecular dynamics simulations. PLoS Comput Biol. 2016;12:e1004746. doi: 10.1371/journal.pcbi.1004746. published online 10 June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidolin D, Agnati LF, Marcoli M, et al. G-protein-coupled receptor type A heteromers as an emerging therapeutic target. Expert Opin Ther Targets. 2015;19:265–283. doi: 10.1517/14728222.2014.981155. [DOI] [PubMed] [Google Scholar]
- 16.Cavanaugh A, Huang Y, Breitwieser GE. Behind the curtain: cellular mechanisms for allosteric modulation of calcium-sensing receptors. Br J Pharmacol. 2012;165:1670–1677. doi: 10.1111/j.1476-5381.2011.01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenthur CJ, Gentry PR, Mathews TP, et al. Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol. 2014;54:165–184. doi: 10.1146/annurev-pharmtox-010611-134525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wootten D, Christopoulos A, Sexton PM. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat Rev Drug Discov. 2013;12:630–644. doi: 10.1038/nrd4052. [DOI] [PubMed] [Google Scholar]
- 20.Rankovic Z, Brust TF, Bohn LM. Biased agonism: an emerging paradigm in GPCR drug discovery. Bioorg Med Chem Lett. 2016;26:241–250. doi: 10.1016/j.bmcl.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao ZG, Jacobson KA. Allosteric modulation and functional selectivity of G protein-coupled receptors. Drug Discov Today Technol. 2013;10:e237–243. doi: 10.1016/j.ddtec.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenakin T. Allosteric drugs and seven transmembrane receptors. Curr Top Med Chem. 2013;13:5–13. doi: 10.2174/1568026611313010003. [DOI] [PubMed] [Google Scholar]
- 23.Nickols HH, Conn PJ. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol Dis. 2014;61:55– 71. doi: 10.1016/j.nbd.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Lindsley CW, Emmitte KA, Hopkins CR, et al. Practical strategies and concepts in GPCR allosteric modulator discovery: recent advances with metabotropic glutamate receptors. Chem Rev. 2016;116:6707–6741. doi: 10.1021/acs.chemrev.5b00656. Review of discovery approaches for designing drug-like allosteric ligands targeted to a particular GPCR (i.e. glutamate) type. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao Y, Nichols SE, McCammon JA. Mapping of allosteric druggable sites in activation-associated conformers of the M2 muscarinic receptor. Chem Biol Drug Des. 2014;83:237–246. doi: 10.1111/cbdd.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horenstein NA, Papke RL, Kulkarni AR, et al. Critical molecular determinants of α7 nicotinic acetylcholine receptor allosteric activation: separation of direct allosteric activation and positive allosteric modulation. J Biol Chem. 2016;291:5049–5067. doi: 10.1074/jbc.M115.692392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu HC, Mackie K. An introduction to the endogenous cannabinoid system. Biol Psychiatry. 2016;79:516–525. doi: 10.1016/j.biopsych.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst DP, Schmeisser M, Reggio PH. Endogenous lipid activated G protein-coupled receptors: emerging structural features from crystallography and molecular dynamics simulations. Chem Phys Lipids. 2013;169:46–56. doi: 10.1016/j.chemphyslip.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadel R, Ahn KH, Kendall DA. The cannabinoid type-1 receptor carboxyl-terminus, more than just a tail. J Neurochem. 2011;117:1– 18. doi: 10.1111/j.1471-4159.2011.07186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nebane NM, Kellie B, Song ZH. The effects of charge-neutralizing mutation D6.30N on the functions of CB1 and CB2 cannabinoid receptors. FEBS Lett. 2006;580:5392–5398. doi: 10.1016/j.febslet.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Ahn KH, Scott CE, Abrol R, et al. Computationally-predicted CB1 cannabinoid receptor mutants show distinct patterns of salt-bridges that correlate with their level of constitutive activity reflected in G protein coupling levels, thermal stability, and ligand binding. Proteins. 2013;81:1304–1317. doi: 10.1002/prot.24264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baillie GL, Horswill JG, Anavi-Goffer S, et al. CB1 receptor allosteric modulators display both agonist and signaling pathway specificity. Mol Pharmacol. 2013;83:322–338. doi: 10.1124/mol.112.080879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shore DM, Baillie GL, Hurst DH, et al. Allosteric modulation of a cannabinoid G protein-coupled receptor: binding site elucidation and relationship to G protein signaling. J Biol Chem. 2014;289:5828–5845. doi: 10.1074/jbc.M113.478495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fay JF, Farrens DL. A key agonist-induced conformational change in the cannabinoid receptor CB1 is blocked by the allosteric ligand Org 27569. J Biol Chem. 2012;287:33873–33882. doi: 10.1074/jbc.M112.352328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stornaiuolo M, Bruno A, Botta L, et al. Endogenous vs exogenous allosteric modulators in GPCRs: a dispute for shuttling CB1 among different membrane microenvironments. Sci Rep. 2015;5:15453. doi: 10.1038/srep15453. published online 20 October 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Laprairie RB, Kulkarni AR, Kulkarni PM, et al. Mapping cannabinoid 1 receptor allosteric site(s): critical molecular determinant and signaling profile of GAT100, a novel, potent, and irreversibly binding probe. ACS Chem Neurosci. 2016;7:776–798. doi: 10.1021/acschemneuro.6b00041. Pharmacological profiling of the first covalent allosteric CB1R ligand and its validation as a molecular probe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zvonok N, Xu W, Williams J, et al. Mass spectrometry-based GPCR proteomics: comprehensive characterization of the human cannabinoid 1 receptor. J Proteome Res. 2010;9:1746–1753. doi: 10.1021/pr900870p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurst DP, Grossfield A, Lynch DL, et al. A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J Biol Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Pertwee RG. Endocannabinoids and their pharmacological actions. Handb Exp Pharmacol. 2015;231:1–37. doi: 10.1007/978-3-319-20825-1_1. Comprehensive introduction to the endocannabinoid signaling system and its endogenous ligands. [DOI] [PubMed] [Google Scholar]
- 40.Howlett AC, Blume LC, Dalton GD. CB(1) cannabinoid receptors and their associated proteins. Curr Med Chem. 2010;17:1382–13893. doi: 10.2174/092986710790980023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohno-Shosaku T, Kano M. Endocannabinoid-mediated retrograde modulation of synaptic transmission. Curr Opin Neurobiol. 2014;29:1–8. doi: 10.1016/j.conb.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 42.Ligresti A, De Petrocellis L, Di Marzo V. From phytocannabinoids to cannabinoid receptors and endocannabinoids: pleiotropic physiological and pathological roles through complex pharmacology. Physiol Rev. 2016;96:1593–1659. doi: 10.1152/physrev.00002.2016. [DOI] [PubMed] [Google Scholar]
- 43.Benyó Z, Ruisanchez É, Leszl-Ishiguro M, et al. Endocannabinoids in cerebrovascular regulation. Am J Physiol Heart Circ Physiol. 2016;310:H785–H801. doi: 10.1152/ajpheart.00571.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper ME, Regnell SE. The hepatic cannabinoid 1 receptor as a modulator of hepatic energy state and food intake. Br J Clin Pharmacol. 2014;77:21–30. doi: 10.1111/bcp.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battista N, Bari M, Maccarrone M. Endocannabinoids and reproductive events in health and disease. Handb Exp Pharmacol. 2015;231:341–365. doi: 10.1007/978-3-319-20825-1_12. [DOI] [PubMed] [Google Scholar]
- 46.Panagis G, Mackey B, Vlachou S. Cannabinoid regulation of brain reward processing with an emphasis on the role of CB1 receptors: a step back into the future. Front Psychiatry. 2014;5:92. doi: 10.3389/fpsyt.2014.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iannotti FA, Di Marzo V, Petrosino S. Endocannabinoids and endocannabinoid-related mediators: targets, metabolism and role in neurological disorders. Prog Lipid Res. 2016;62:107–128. doi: 10.1016/j.plipres.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Maccarrone M, Bab I, Bíró T, et al. Endocannabinoid signaling at the periphery: 50 years after THC. Trends Pharmacol Sci. 2015;36:277– 296. doi: 10.1016/j.tips.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasileiou I, Fotopoulou G, Matzourani M, et al. Evidence for the involvement of cannabinoid receptors’ polymorphisms in the pathophysiology of human diseases. Expert Opin Ther Targets. 2013;17:363–377. doi: 10.1517/14728222.2013.754426. [DOI] [PubMed] [Google Scholar]
- 50.Hill KP. Medical marijuana for treatment of chronic pain and other medical and psychiatric problems: a clinical review. JAMA. 2015;313:2474–2483. doi: 10.1001/jama.2015.6199. [DOI] [PubMed] [Google Scholar]
- 51.Whiting PF, Wolff RF, Deshpande S, et al. Cannabinoids for medical use: a systematic review and meta-analysis. JAMA. 2015;313:2456– 2473. doi: 10.1001/jama.2015.6358. [DOI] [PubMed] [Google Scholar]
- 52.Vemuri VK, Makriyannis A. Medicinal chemistry of cannabinoids. Clin Pharmacol Ther. 2015;97:553–558. doi: 10.1002/cpt.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McPartland JM, Duncan M, Di Marzo V, et al. Are cannabidiol and Δ (9)-tetrahydrocannabivarin negative modulators of the endocannabinoid system? A systematic review. Br J Pharmacol. 2015;172:737–753. doi: 10.1111/bph.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pertwee RG. Targeting the endocannabinoid system with cannabinoid receptor agonists: pharmacological strategies and therapeutic possibilities. Philos Trans R Soc Lond B Biol Sci. 2012;367:3353– 3363. doi: 10.1098/rstb.2011.0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GW Pharmaceuticals provides update on orphan program in childhood epilepsy for Epidiolex®. [cited 2016 Oct 1]. Available from: http://www.gwpharm.com/GW%20Pharmaceuticals%20Provides%20Update%20on%20Orphan%20Program%20in%20Childhood%20Epilepsy%20for%20Epidiolex.aspx.
- 56.Janero DR, Makriyannis A. Cannabinoid receptor antagonists: pharmacological opportunities, clinical experience, and translational prognosis. Expert Opin Emerg Drugs. 2009;14:43–65. doi: 10.1517/14728210902736568. [DOI] [PubMed] [Google Scholar]
- 57.Davis MP. Oral nabilone capsules in the treatment of chemotherapy-induced nausea and vomiting and pain. Expert Opin Investig Drugs. 2008;17:85–95. doi: 10.1517/13543784.17.1.85. [DOI] [PubMed] [Google Scholar]
- 58.Krentz AJ, Fujioka K, Hompesch M. Evolution of pharmacological obesity treatments: focus on adverse side-effect profiles. Diabetes Obes Metab. 2016;18:558–570. doi: 10.1111/dom.12657. [DOI] [PubMed] [Google Scholar]
- 59.Tait RJ, Caldicott D, Mountain D, et al. A systematic review of adverse events arising from the use of synthetic cannabinoids and their associated treatment. Clin Toxicol. 2016;54:1–13. doi: 10.3109/15563650.2015.1110590. [DOI] [PubMed] [Google Scholar]
- 60.van Amsterdam J, Brunt T, van den Brink W. The adverse health effects of synthetic cannabinoids with emphasis on psychosis-like effects. J Psychopharmacol. 2015;29:254–263. doi: 10.1177/0269881114565142. [DOI] [PubMed] [Google Scholar]
- 61.Meye FJ, Ramakers GM, Adan RA. The vital role of constitutive GPCR activity in the mesolimbic dopamine system. Transl Psychiatry. 2014;4:e361. doi: 10.1038/tp.2013.130. Published online 11 February 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunos G, Osei-Hyiaman D, Bátkai S, et al. Should peripheral CB(1) cannabinoid receptors be selectively targeted for therapeutic gain? Trends Pharmacol Sci. 2009;30:1–7. doi: 10.1016/j.tips.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janero DR. Cannabinoid-1 receptor (CB1R) blockers as medicines: beyond obesity and cardiometabolic disorders to substance abuse/drug addiction with CB1R neutral antagonists. Expert Opin Emerg Drugs. 2012;17:17–29. doi: 10.1517/14728214.2012.660916. [DOI] [PubMed] [Google Scholar]
- 64•.Price MR, Baillie GL, Thomas A, et al. Allosteric modulation of the cannabinoid CB1 receptor. Mol Pharmacol. 2005;68:1484–1495. doi: 10.1124/mol.105.016162. Initial report of CB1R allosteric modulation. [DOI] [PubMed] [Google Scholar]
- 65.Ross RA. Allosterism and cannabinoid CB(1) receptors: the shape of things to come. Trends Pharmacol Sci. 2007;28:567–572. doi: 10.1016/j.tips.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Horswill JG, Bali U, Shaaban S, et al. PSNCBAM-1, a novel allosteric antagonist at cannabinoid CB1 receptors with hypophagic effects in rats. Br J Pharmacol. 2007;152:805–814. doi: 10.1038/sj.bjp.0707347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahn KH, Mahmoud MM, Kendall DA. Allosteric modulator ORG27569 induces CB1 cannabinoid receptor high affinity agonist binding state, receptor internalization, and Gi protein-independent ERK1/2 kinase activation. J Biol Chem. 2012;287:12070–12082. doi: 10.1074/jbc.M111.316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Horswill JG, Whalley BJ, et al. Effects of the allosteric antagonist 1-(4-chlorophenyl)-3-[3-(6-pyrrolidin-1-ylpyridin-2-yl) phenyl]urea (PSNCBAM-1) on CB1 receptor modulation in the cerebellum. Mol Pharmacol. 2011;79:758–767. doi: 10.1124/mol.110.068197. [DOI] [PubMed] [Google Scholar]
- 69.Khurana L, Ali HI, Olszewska T, et al. Optimization of chemical functionalities of indole-2-carboxamides to improve allosteric parameters for the cannabinoid receptor 1 (CB1) J Med Chem. 2014;57:3040–3052. doi: 10.1021/jm5000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.German N, Decker AM, Gilmour BP, et al. Diarylureas as allosteric modulators of the cannabinoid CB1 receptor: structure-activity relationship studies on 1-(4-chlorophenyl)-3-{3-[6-(pyrrolidin-1-yl) pyridin-2-yl]phenyl}urea (PSNCBAM-1) J Med Chem. 2014;57:7758–7769. doi: 10.1021/jm501042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen T, German N, Decker AM, et al. Structure-activity relationships of substituted 1H-indole-2-carboxamides as CB1 receptor allosteric modulators. Bioorg Med Chem. 2015;23:2195–2203. doi: 10.1016/j.bmc.2015.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Picone RP, Kendall DA. From the bench, toward the clinic: therapeutic opportunities for cannabinoid receptor modulation. Mol Endocrinol. 2015;29:801–813. doi: 10.1210/me.2015-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Navarro HA, Howard JL, Pollard GT, et al. Positive allosteric modulation of the human cannabinoid (CB) receptor by RTI-371, a selective inhibitor of the dopamine transporter. Br J Pharmacol. 2009;156:1178–1184. doi: 10.1111/j.1476-5381.2009.00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Priestley RS, Nickolls SA, Alexander SP, et al. A potential role for cannabinoid receptors in the therapeutic action of fenofibrate. FASEB J. 2015;29:1446–1455. doi: 10.1096/fj.14-263053. [DOI] [PubMed] [Google Scholar]
- 75.Pamplona FA, Ferreira J, Menezes de Lima O, Jr, et al. Anti-inflammatory lipoxin A4 is an endogenous allosteric enhancer of CB1 cannabinoid receptor. Proc Natl Acad Sci USA. 2012;109:21134– 21139. doi: 10.1073/pnas.1202906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vallée M, Vitiello S, Bellocchio L, et al. Pregnenolone can protect the brain from cannabis intoxication. Science. 2014;343:94–98. doi: 10.1126/science.1243985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laprairie RB, Bagher AM, Kelly ME, et al. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br J Pharmacol. 2015;172:4790–4805. doi: 10.1111/bph.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khajehali E, Malone DT, Glass M, et al. Biased agonism and biased allosteric modulation at the CB1 cannabinoid receptor. Mol Pharmacol. 2015;88:368–379. doi: 10.1124/mol.115.099192. [DOI] [PubMed] [Google Scholar]
- 79.Bauer M, Chicca A, Tamborrini M, et al. Identification and quantification of a new family of peptide endocannabinoids (pepcans) showing negative allosteric modulation at CB1 receptors. J Biol Chem. 2012;287:36944–36967. doi: 10.1074/jbc.M112.382481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hofer SC, Ralvenius WT, Gachet MS, et al. Localization and production of peptide endocannabinoids in the rodent CNS and adrenal medulla. Neuropharmacology. 2015;98:78–89. doi: 10.1016/j.neuropharm.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 81.Iliff HA, Lynch DL, Kotsikorou E, et al. Parameterization of Org27569: an allosteric modulator of the cannabinoid CB1 G protein-coupled receptor. J Comput Chem. 2011;32:2119–2126. doi: 10.1002/jcc.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Straiker A, Mitjavila J, Yin D, et al. Aiming for allosterism: evaluation of allosteric modulators of CB1 in a neuronal model. Pharmacol Res. 2015;99:370–376. doi: 10.1016/j.phrs.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ding Y, Qiu Y, Jing L, et al. Behavioral effects of the cannabinoid CB1 receptor allosteric modulator ORG27569 in rats. Pharmacol Res Perspect. 2014;2:e00069. doi: 10.1002/prp2.69. published online 24 August 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gamage TF, Ignatowska-Jankowska BM, Wiley JL, et al. In-vivo pharmacological evaluation of the CB1-receptor allosteric modulator Org-27569. Behav Pharmacol. 2014;25:182–185. doi: 10.1097/FBP.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jing L, Qiu Y, Zhang Y, et al. Effects of the cannabinoid CB1 receptor allosteric modulator ORG 27569 on reinstatement of cocaine- and methamphetamine-seeking behavior in rats. Drug Alcohol Depend. 2014;143:251–256. doi: 10.1016/j.drugalcdep.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ignatowska-Jankowska BM, Baillie GL, Kinsey S, et al. A cannabinoid CB1 receptor-positive allosteric modulator reduces neuropathic pain in the mouse with no psychoactive effects. Neuropsychopharmacology. 2015;40:2948–2959. doi: 10.1038/npp.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87•.Jazayeri A, Dias JM, Marshall FH. From G protein-coupled receptor structure resolution to rational drug design. J Biol Chem. 2015;290:19489–19495. doi: 10.1074/jbc.R115.668251. Overview of GPCR structural biology and its relevance to drug design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Topiol S. X-ray structural information of GPCRs in drug design: what are the limitations and where do we go? Expert Opin Drug Discov. 2013;8:607–620. doi: 10.1517/17460441.2013.783815. [DOI] [PubMed] [Google Scholar]
- 89.Merk A, Bartesaghi A, Banerjee S, et al. Breaking Cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kenakin T. Analytical pharmacology and allosterism: the importance of quantifying drug parameters in drug discovery. Drug Discov Today Technol. 2013;10:e229–235. doi: 10.1016/j.ddtec.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 91.Bennett KA, Doré AS, Christopher JA, et al. Structures of mGluRs shed light on the challenges of drug development of allosteric modulators. Curr Opin Pharmacol. 2015;20:1–7. doi: 10.1016/j.coph.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 92.Zhu S, Paoletti P. Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Curr Opin Pharmacol. 2015;20:14–23. doi: 10.1016/j.coph.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 93.Munk C, Harpsøe K, Hauser AS, et al. Integrating structural and mutagenesis data to elucidate GPCR ligand binding. Curr Opin Pharmacol. 2016;30:51–58. doi: 10.1016/j.coph.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ehlert FJ. Functional studies cast light on receptor states. Trends Pharmacol Sci. 2015;36:596–604. doi: 10.1016/j.tips.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Changeux JP, Christopoulos A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell. 2016;166:1084–1102. doi: 10.1016/j.cell.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 96.Martí-Solano M, Schmidt D, Kolb P, et al. Drugging specific conformational states of GPCRs: challenges and opportunities for computational chemistry. Drug Discov Today. 2016;21:625–631. doi: 10.1016/j.drudis.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 97.Shonberg J, Kling RC, Gmeiner P, et al. GPCR crystal structures: medicinal chemistry in the pocket. Bioorg Med Chem. 2015;23:3880–3906. doi: 10.1016/j.bmc.2014.12.034. [DOI] [PubMed] [Google Scholar]
- 98.Fenalti G, Giguere PM, Katritch V, et al. Molecular control of delta-opioid receptor signalling. Nature. 2014;506:191–196. doi: 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang W, Manglik A, Venkatakrishnan AJ, et al. Structural insights into μ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kruse AC, Ring AM, Manglik A, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thal DM, Sun B, Feng D, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wootten D, Reynolds CA, Koole C, et al. A hydrogen-bonded polar network in the core of the glucagon-like peptide-1 receptor is a fulcrum for biased agonism: lessons from class B crystal structures. Mol Pharmacol. 2016;89(3):335–347. doi: 10.1124/mol.115.101246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu H, Wang C, Gregory KJ, et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Doré AS, Okrasa K, Patel JC, et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- 105.Bartuzi D, Kaczor AA, Matosiuk D. Activation and allosteric modulation of human μ opioid receptor in molecular dynamics. J Chem Inf Model. 2015;55:2421–2434. doi: 10.1021/acs.jcim.5b00280. [DOI] [PubMed] [Google Scholar]
- 106.Feng Z, Hu G, Ma S, et al. Computational advances for the sevelopment of allosteric modulators and bitopic ligands in G protein-coupled receptors. Aaps J. 2015;17:1080–1095. doi: 10.1208/s12248-015-9776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shang Y, Yeatman HR, Provasi D, et al. Proposed mode of binding and action of positive allosteric modulators at opioid receptors. ACS Chem Biol. 2016;11:1220–1229. doi: 10.1021/acschembio.5b00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deganutti G, Cuzzolin A, Ciancetta A, et al. Understanding allosteric interactions in G protein-coupled receptors using supervised molecular dynamics: A prototype study analysing the human A3 adenosine receptor positive allosteric modulator LUF6000. Bioorg Med Chem. 2015;23:4065–4071. doi: 10.1016/j.bmc.2015.03.039. [DOI] [PubMed] [Google Scholar]
- 109.Isogai S, Deupi X, Opitz C, et al. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature. 2016;530:237–241. doi: 10.1038/nature16577. [DOI] [PubMed] [Google Scholar]
- 110.Sounier R, Mas C, Steyaert J, et al. Propagation of conformational changes during μ-opioid receptor activation. Nature. 2015;524:375– 378. doi: 10.1038/nature14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chan WY, McKinzie DL, Bose S, et al. Allosteric modulation of the muscarinic M4 receptor as an approach to treating schizophrenia. Proc Natl Acad Sci USA. 2008;105:10978–10983. doi: 10.1073/pnas.0800567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valant C, Felder CC, Sexton PM, et al. Probe dependence in the allosteric modulation of a G protein-coupled receptor: implications for detection and validation of allosteric ligand effects. Mol Pharmacol. 2012;81:41–52. doi: 10.1124/mol.111.074872. [DOI] [PubMed] [Google Scholar]
- 113.Khatri A, Burger PB, Swanger SA, et al. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Mol Pharmacol. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Farinha A, Lavreysen H, Peeters L, et al. Molecular determinants of positive allosteric modulation of the human metabotropic glutamate receptor 2. Br J Pharmacol. 2015;172:2383–2396. doi: 10.1111/bph.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dror RO, Green HF, Valant C, et al. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 116.Rovira X, Malhaire F, Scholler P, et al. Overlapping binding sites drive allosteric agonism and positive cooperativity in type 4 metabotropic glutamate receptors. FASEB J. 2015;29:116–130. doi: 10.1096/fj.14-257287. [DOI] [PubMed] [Google Scholar]
- 117.Jang JW, Cho NC, Min SJ, et al. Novel scaffold identification of mGlu1 receptor negative allosteric modulators using a hierarchical virtual screening approach. Chem Biol Drug Des. 2016;87:239–256. doi: 10.1111/cbdd.12654. [DOI] [PubMed] [Google Scholar]
- 118.Zhang D, Zhao Q, Wu B. Structural studies of G protein-coupled receptors. Mol Cells. 2015;38:836–842. doi: 10.14348/molcells.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McAllister SD, Rizvi G, Anavi-Goffer S, et al. An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. J Med Chem. 2003;46:5139–5152. doi: 10.1021/jm0302647. [DOI] [PubMed] [Google Scholar]
- 120.Gregory KJ, Velagaleti R, Thal DM, et al. Clickable photoaffinity ligands for metabotropic glutamate receptor 5 based on select acetylenic negative allosteric modulators. ACS Chem Biol. 2016;11:1870–1879. doi: 10.1021/acschembio.6b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bueno AB, Showalter AD, Wainscott DB, et al. Positive allosteric modulation of the glucagon-like peptide-1 receptor by diverse electrophiles. J Biol Chem. 2016;291:10700–10715. doi: 10.1074/jbc.M115.696039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huber T, Sakmar TP. Chemical biology methods for investigating G protein-coupled receptor signaling. Chem Biol. 2014;21:1224–1237. doi: 10.1016/j.chembiol.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 123.Weichert D, Gmeiner P. Covalent molecular probes for class A G protein-coupled receptors: advances and applications. ACS Chem Biol. 2015;10:1376–1386. doi: 10.1021/acschembio.5b00070. [DOI] [PubMed] [Google Scholar]
- 124.Janero DR, Yaddanapudi S, Zvonok N, et al. Molecular-interaction and signaling profiles of AM3677, a novel covalent agonist selective for the cannabinoid 1 receptor. ACS Chem Neurosci. 2015;6:1400–1410. doi: 10.1021/acschemneuro.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Szymanski DW, Papanastasiou M, Melchior K, et al. Mass spectrometry-based proteomics of human cannabinoid receptor 2: covalent cysteine 6.47(257)-ligand interaction affording megagonist receptor activation. J Proteome Res. 2011;10:4789–4798. doi: 10.1021/pr2005583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pei Y, Mercier RW, Anday JK, et al. Ligand-binding architecture of human CB2 cannabinoid receptor: evidence for receptor subtype-specific binding motif and modeling GPCR activation. Chem Biol. 2008;15:1207–1219. doi: 10.1016/j.chembiol.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fay JF, Farrens DL. Structural dynamics and energetics underlying allosteric inactivation of the cannabinoid receptor CB1. Proc Natl Acad Sci USA. 2015;112:8469–8474. doi: 10.1073/pnas.1500895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kulkarni PM, Kulkarni AR, Korde A, et al. Novel electrophilic and photoaffinity covalent probes for mapping the cannabinoid 1 receptor allosteric site(s) J Med Chem. 2016;59:44–60. doi: 10.1021/acs.jmedchem.5b01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 130.Tahtaoui C, Balestre MN, Klotz P, et al. Identification of the binding sites of the SR49059 nonpeptide antagonist into the V1a vasopressin receptor using sulfydryl-reactive ligands and cysteine mutants as chemical sensors. J Biol Chem. 2003;278:40010–40019. doi: 10.1074/jbc.M301128200. [DOI] [PubMed] [Google Scholar]
- 131.Wu X, Zhou QH, Xu K. Are isothiocyanates potential anti-cancer drugs? Acta Pharmacol Sin. 2009;30:501–512. doi: 10.1038/aps.2009.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qiao CJ, Ali HI, Ahn KH, et al. Synthesis and biological evaluation of indole-2-carboxamides bearing photoactivatable functionalities as novel allosteric modulators for the cannabinoid CB1 receptor. Eur J Med Chem. 2016;121:517–529. doi: 10.1016/j.ejmech.2016.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Andersson H, D’Antona AM, Kendall DA, et al. Membrane assembly of the cannabinoid receptor 1: impact of a long N-terminal tail. Mol Pharmacol. 2003;64:570–577. doi: 10.1124/mol.64.3.570. [DOI] [PubMed] [Google Scholar]
- 134.Fay JF, Farrens DL. The membrane proximal region of the cannabinoid receptor CB1 N-terminus can allosterically modulate ligand affinity. Biochemistry. 2013;52:8286–8294. doi: 10.1021/bi400842k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Garland SL. Are GPCRs still a source of new targets? J Biomol Screen. 2013;18:947–966. doi: 10.1177/1087057113498418. [DOI] [PubMed] [Google Scholar]
- 136.Kinch MS. 2015 in review: FDA approval of new drugs. Drug Discov Today. 2016;21:1046–1050. doi: 10.1016/j.drudis.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 137.Jacobson KA. New paradigms in GPCR drug discovery. Biochem Pharmacol. 2015;98:541–555. doi: 10.1016/j.bcp.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miao Y, McCammon JA. G-protein coupled receptors: advances in simulation and drug discovery. Curr Opin Struct Biol. 2016;41:83–89. doi: 10.1016/j.sbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Desai AJ, Henke BR, Miller LJ. Elimination of a cholecystokinin receptor agonist ‘trigger’ in an effort to develop positive allosteric modulators without intrinsic agonist activity. Bioorg Med Chem Lett. 2015;25:1849–1855. doi: 10.1016/j.bmcl.2015.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Noetzel MJ, Rook JM, Vinson PN, et al. Functional impact of allosteric agonist activity of selective positive allosteric modulators of metabotropic glutamate receptor subtype 5 in regulating central nervous system function. Mol Pharmacol. 2012;81:120–133. doi: 10.1124/mol.111.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Parmentier-Batteur S, Hutson PH, Menzel K, et al. Mechanism based neurotoxicity of mGlu5 positive allosteric modulators–development challenges for a promising novel antipsychotic target. Neuropharmacology. 2014;82:161–173. doi: 10.1016/j.neuropharm.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 142.Bridges TM, Rook JM, Noetzel MJ, et al. Biotransformation of a novel positive allosteric modulator of metabotropic glutamate receptor subtype 5 contributes to seizure-like adverse events in rats involving a receptor agonism-dependent mechanism. Drug Metab Dispos. 2013;41:1703–1714. doi: 10.1124/dmd.113.052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bagdas D, Wilkerson JL, Kulkarni A, et al. The α7 nicotinic receptor dual allosteric agonist and positive allosteric modulator GAT107 reverses nociception in mouse models of inflammatory and neuropathic pain. Br J Pharmacol. 2016;173:2506–2520. doi: 10.1111/bph.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mah R, Thomas JR, Shafer CM. Drug discovery considerations in the development of covalent drugs. Bioorg Med Chem Lett. 2014;24:33–39. doi: 10.1016/j.bmcl.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 145.Nussinov R, Tsai CJ. The design of covalent allosteric drugs. Annu Rev Pharmacol Toxicol. 2015;55:249–267. doi: 10.1146/annurevpharmtox-010814-124401. [DOI] [PubMed] [Google Scholar]
- 146.Schreiber SL, Kotz JD, Li M, et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell. 2015;161:1252–1265. doi: 10.1016/j.cell.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Langmead CJ. Determining allosteric modulator mechanism of action: integration of radioligand binding and functional assay data. Methods Mol Biol. 2011;746:195–209. doi: 10.1007/978-1-61779-126-0_10. [DOI] [PubMed] [Google Scholar]
- 148.Rocheville M, Martin J, Jerman J, et al. Mining the potential of label-free biosensors for seven-transmembrane receptor drug discovery. Prog Mol Biol Transl Sci. 2013;115:123–142. doi: 10.1016/B978-0-12-394587-7.00003-8. [DOI] [PubMed] [Google Scholar]
- 149.Janero DR. The future of drug discovery: enabling technologies for enhancing lead characterization and profiling therapeutic potential. Expert Opin Drug Discov. 2014;9:847–858. doi: 10.1517/17460441.2014.925876. [DOI] [PubMed] [Google Scholar]
- 150.Aristotelous T, Hopkins AL, Navratilova I. Surface plasmon resonance analysis of seven-transmembrane receptors. Methods Enzymol. 2015;556:499–525. doi: 10.1016/bs.mie.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 151.García C, Palomo-Garo C, Gómez-Gálvez Y, et al. Cannabinoid-dopamine interactions in the physiology and physiopathology of the basal ganglia. Br J Pharmacol. 2016;173:2069–2079. doi: 10.1111/bph.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cawston EE, Connor M, Di Marzo V, et al. Distinct temporal fingerprint for cyclic adenosine monophosphate (cAMP) signaling of indole-2-carboxamides as allosteric modulators of the cannabinoid receptors. J Med Chem. 2015;58:5979–5988. doi: 10.1021/acs.jmedchem.5b00579. [DOI] [PubMed] [Google Scholar]