Abstract
The interaction between an origin sequence and the origin recognition complex (ORC), which is highly conserved in eukaryotes, is critical for the initiation of DNA replication. In this report, we have examined the interaction between the Schizosaccharomyces pombe (sp) autonomously replicating sequence 1 (ars1) and the spORC. For this purpose, we have purified the spORC containing all six subunits, a six-subunit complex containing the N-terminal-deleted spOrc4 subunit (spORCΔN-Orc4), and the spOrc4 subunit by using the baculovirus expression system. Wild-type spORC showed sequence-specific binding to ars1, and the spOrc4 protein alone showed the same DNA-binding properties as wild-type spORC. In contrast, the spORCΔN-Orc4 and the ΔN-spOrc4p alone did not bind significantly to ars1. These findings indicate that the N-terminal domain of the spOrc4 protein that contains multiple AT-hook motifs is essential for the ars1-binding activity. DNA-binding competition assays with fragments of ars1 and DNase I footprinting studies with full-length ars1 revealed that the spORC interacted with several AT-rich sequence regions of ars1. These DNA-binding properties of spORC correlate with the previously determined sequence requirements of the S. pombe ars1. These studies indicate that because of its unique Orc4 subunit, S. pombe uses a mechanism to recognize its origins different from that used by Saccharomyces cerevisiae.
DNA replication origin sequences in Saccharomyces cerevisiae are relatively short AT-rich sequences (100–150 bp) that include an essential autonomously replicating sequence (ARS) consensus sequence (ACS) (1–3). In contrast, replicators in higher eukaryotes are much longer and share no recognizable essential motifs (4, 5). The ARS elements of Schizosaccharomyces pombe are larger (longer than 700 bp) than those of Sac. cerevisiae, and contain several redundant AT-rich sequences that are important for ARS activity, but lack a conserved essential sequence motif such as an ACS (6–8).
The origin recognition complex (ORC) was initially purified as an ARS-binding protein from Sac. cerevisiae (9). ORC consists of six distinct subunits, Orc1p to Orc6p, which are all essential for DNA replication (10–12). Sac. cerevisiae ORC specifically recognizes and binds to the ACS containing an 11-bp AT-rich sequence in vivo and in vitro, and ATP is essential for this ARS-binding activity (9, 13, 14). ORC binds to replication origin DNA throughout the cell cycle and is required to recruit other essential initiation factors for the assembly of the prereplicative complex (15, 16). ORC is conserved in all eukaryotes and has been identified and purified from other species, including S. pombe, Drosophila melanogaster, Xenopus laevis, and humans (17–22). Although in vivo and in vitro studies suggest that ORC is essential for DNA replication in these species (15, 16), the interactions between these ORCs and their replication origin DNAs have not been characterized except for Sac. cerevisiae. D. melanogaster ORC interacts with multiple sites in ACE3, a sequence required for chorion gene amplification (23). However, the interaction of ORC with DNA replication origins in somatic Drosophila cells has not yet been characterized.
The S. pombe ORC (spORC) contains nine copies of a unique sequence motif in the N terminus of the spOrc4p, called “AT hooks,” which includes the core sequence, RGRP, flanked on each side by conserved positively charged amino acid residues (18). These AT-hook domains interact preferentially with the minor groove of stretches of AT-rich DNA recognizing such structures rather than a nucleotide sequence (24). The presence of these multiple AT-hook motifs suggests that the binding of spORC to origin DNA is likely to be influenced by this domain. Although all the spORC subunits have been identified and the complex has been reconstituted from in vitro transcription and translation products of each subunit (17), the origin DNA-binding properties of spORC have not been examined as yet.
In this study, we have reconstituted and purified the spORC by using the baculovirus expression system, and we have examined its ars1-binding properties by nitrocellulose filter binding, DNA-binding competition, and DNase I footprinting assays. The six-subunit complex containing the N-terminal truncated spOrc4p subunit (devoid of the AT-hook motifs) and the spOrc4p alone have also been expressed and purified by using the baculovirus system. The DNA-binding properties of these proteins and the wild-type spORC suggest that the complex interacts with multiple AT-rich sequence regions in ars1 specifically through the N-terminal domain of the spOrc4p.
Materials and Methods
Reagents.
Labeled dNTPs were obtained from NEN. Polynucleotides were from Amersham Pharmacia. Anti-FLAG M2 Ab-agarose and FLAG peptide were from Sigma. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Preparation of Baculoviruses Expressing ORC Protein.
For the preparation of the baculoviruses expressing individual spOrc proteins, cDNA fragments encoding full-length spOrc1p to spOrc6p were subcloned into the plasmid pFastBac1 (Life Technologies). Plasmid and recombinant baculoviruses were produced as described by the manufacturer (Bac-to-Bac baculovirus expression systems, Life Technologies). To produce the N-terminal truncated spOrc4p (ΔN-spOrc4p), the cDNA fragment encoding amino acids 516–972 of spORC4p was subcloned into the pFastBac1 plasmid. Specific tags were added to several proteins to facilitate the purification and detection of the recombinant proteins. These included the addition of two FLAG (2FLAG) epitope tags at the N terminus of spOrc2p, a six-histidine (6His) tag at the N terminus of spOrc4p, an influenza virus hemagglutinin (HA) epitope tag and 6His tag at the N terminus of spOrc5p, and a 2FLAG tag at the N terminus of ΔN-spOrc4p.
Purification of spORC, spORCΔN-Orc4, spOrc4p, and ΔN-spOrc4p.
SpORC containing all six subunits was purified from Sf9 cells (2 × 106 cells per ml, 1 liter) coinfected with baculoviruses expressing spOrc1, 2FLAG-spOrc2, spOrc3, 6His-spOrc4, HA-6His-spOrc5, and the spOrc6 proteins. After incubation at 27°C for 60 h, spORC was purified by the consecutive steps involving Ni2+-agarose (Invitrogen) and anti-FLAG M2 agarose chromatography as described (25) by using buffer A (20 mM Tris⋅HCl, pH 7.5/0.6 M NaCl/2 mM magnesium acetate/0.05% Nonidet P-40/15% glycerol, containing 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml aprotinin). Typically, this procedure yielded about 200 μg of spORC protein. For further purification, a portion of this fraction (30 μg of protein) was loaded onto a 0.5-ml hydroxyapatite column (Bio-Rad) equilibrated with buffer A containing 20 mM sodium phosphate, pH 7.4, and eluted with a 12-ml linear gradient of sodium phosphate buffer, pH 7.4, from 20 to 250 mM in buffer A. spORC eluted at 90 mM sodium phosphate (3 μg of protein recovered).
Isolation of the six-subunit spORC containing ΔN-spOrc4p was performed with baculoviruses expressing spOrc1, spOrc2, spOrc3, 2FLAG-ΔN-spOrc4, HA-6His-tagged spOrc5, and spOrc6 proteins that were coinfected into Sf9 cells. spORCΔN-Orc4 was purified as described above, yielding 100 μg of protein.
For the purification of full-length spOrc4p or the ΔN-spOrc4p, extracts of virus-infected Sf9 cells (500 ml) were prepared as described above, and the proteins were purified by using anti-FLAG M2 Ab-affinity column chromatography without the Ni-column purification step, yielding 1 mg of spOrc4p or ΔN-spOrc4p.
Nitrocellulose Filter Binding Assay.
For the preparation of labeled ars1 DNA used in the nitrocellulose filter binding assays, a 1.2-kb EcoRI fragment containing the ars1 was prepared from the pREP2 plasmid and labeled with [α-32P]dATP by using the Klenow fragment of DNA polymerase. Nitrocellulose filter binding assays were conducted in reaction mixtures (15 μl) containing 25 mM Hepes–NaOH (pH 7.5), 50 mM NaCl, 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 10 fmol of 32P-labeled DNA, 1 μg of poly(dA-dC)⋅(dG-dT) as competitor, and protein fractions as indicated. After incubation at 25°C for 30 min, the mixture was filtered through a nitrocellulose membrane (type HA, 0.45 μm) pretreated with alkali. The filter was then washed three times with 1 ml of washing buffer (25 mM Hepes–NaOH, pH 7.5/50 mM NaCl/5 mM magnesium acetate/1 mM DTT), and the radioactivity adsorbed to the filter was measured by liquid scintillation counting. In this study, the ars1 sequence contained within nucleotides 4360–3157 in GenBank accession number Z67961 has been renumbered as nucleotides 1–1204, respectively. Various ars1 DNA fragments used in the competition assays were generated by PCR with different combinations of the following forward and reverse primers: ars1F1, 5′-GACATAGTGACTTATCTGAC-3′; ars1F2, 5′-GGATAGAGGAGATTACATTG-3′; ars1F3, 5′-CTATCCTCTTTAACGCCATG-3′; ars1F4, 5′-CTGAGCTTATTTTGACATTTCG-3′; ars1F5, 5′-CATTTCGGACTCCCAAGGAC-3′; ars1R1, 5′-ATCGCTCCTCCATCAGATG-3′; ars1R2, TTGTACGTCCTTGGGAGTC-3′; ars1R3, 5′-GAGAAAACTTCAACGAATGTC-3′; ars1R4, 5′-GCTCATGGCGTTAAAGAGG-3′; and ars1R5, 5′-AGAAGATGCAATGTAATCTCC-3′. The resulting PCR-amplified DNA fragments included the following DNA regions: ars1-1, nucleotides 30–274; ars1-2, nucleotides 248–523; ars1-3, nucleotides 500–766; ars1-4, nucleotides 733–976; ars1-5, nucleotides 953-1179; ars1-6, nucleotides 30–976; ars1-7, nucleotides 248–976; ars1-8, nucleotides 500–976; ars1-9, nucleotides 30–766; and ars1-10, nucleotides 30–523.
DNase I Footprinting Assay.
DNaseI footprinting reactions were conducted in mixtures (15 μl) containing 25 mM Hepes–NaOH (pH 7.5), 50 mM NaCl, 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml BSA, 10 fmol of 32P-labeled ars1 fragments or pUR19N plasmid DNA containing ars1, 500 ng of λ DNA digested with HindIII as competitor, and indicated levels of the spORC. After incubation at 25°C for 30 min, 0.04 unit of DNase I was added, and the mixture was incubated at 25°C for 1 min. DNase I was inactivated by the addition of 4 μl of stop buffer (0.5% SDS/50 mM EDTA), and DNA was extracted with a phenol/chloroform mixture and precipitated with ethanol. When pUR19N DNA was used as the substrate, the DNase I-digested DNA was analyzed by the primer extension reaction with ars1F1, -F2, -F3, -F4, -R2, -R3, -R4, or -R5 primers as follows. The primers were labeled at their 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. The primer extension reactions were conducted in a standard PCR buffer (30 μl) containing 5 pmol of 32P-labeled primer and Taq DNA polymerase. Five reaction cycles were performed with 1-min denaturation at 94°C, 1-min annealing at 51°C, and 30-s extension at 72°C, and the synthesized DNA was analyzed by electrophoresis through a 6% polyacrylamide gel in 8 M urea.
Results
Reconstitution and Purification of spORC with the Baculovirus Expression System.
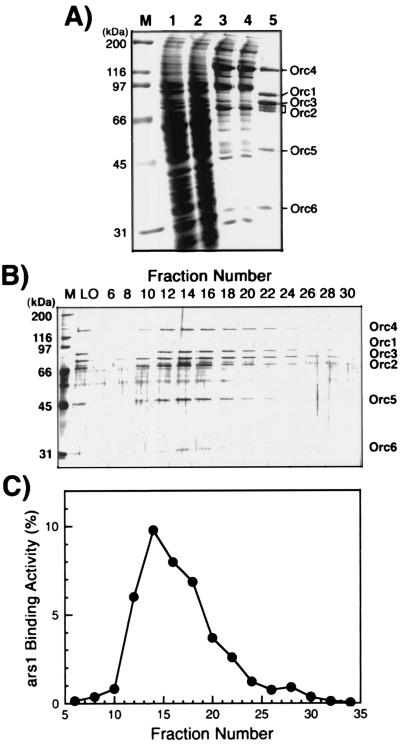
To examine the replication origin binding properties of spORC, the complex was reconstituted with use of the baculovirus expression system. The purification of the complex was facilitated by the insertion of a 6His tag and a 2FLAG tag onto the N termini of spOrc4p and spOrc2p subunits, respectively. After coinfection of baculoviruses expressing each spOrc subunit, a stable complex containing all six Orc polypeptides was purified by using Ni2+-agarose, anti-FLAG M2 Ab-agarose, and hydroxyapatite chromatography as described in Materials and Methods. The purified ORC was more than 90% homogeneous after the M2 Ab affinity chromatography step and contained near stoichiometric amounts of the six subunits (Fig. 1A). The ratio of Orc1p:Orc2p+Orc3p:Orc4p:Orc5:Orc6p was 1:1.8:1:1.3:1.1, as determined by densitometric scanning of a Coomassie blue-stained gel (Fig. 1A). Gel filtration and glycerol gradient sedimentation analyses suggest that these preparations were a mixture of monomers and dimers (data not presented). Multiple protein bands of Orc2p were detected and were converted to a single band after λ phosphatase treatment (data not presented).
Figure 1.
Purification of spORC from Sf9 cells. (A) Aliquots of each fraction obtained at each purification step were analyzed by electrophoresis on an SDS/10% polyacrylamide gel and stained with Coomassie blue. The lanes and the amount of protein analyzed were as follows: M, molecular mass markers; 1, 25 μg of crude extract; 2, 25 μg of the Ni-column flow-through fraction; 3, 6 μg of the Ni-column eluate; 4, 6 μg of the anti-FLAG Ab column flow-through fraction; and 5, 2 μg of the anti-FLAG Ab column eluate. (B) Hydroxyapatite column fractionation of the spORC. The material eluted from the anti-FLAG Ab column was subjected to chromatography. Aliquots (20 μl) of hydroxyapatite column fractions were analyzed by electrophoresis on an SDS/10% polyacrylamide gel and then stained with silver. M, molecular mass markers; LO, 50 ng of the anti-FLAG Ab column eluate. (C) ars1-binding activity of hydroxyapatite column fractions. Aliquots of fractions (1 μl) were assayed for their ability to bind full-length ars1 by using the nitrocellulose filter binding assay. DNA-binding activity was quantitated by measuring the percentage of the input radioactivity retained on the filter.
spORC Binds ars1 in a Sequence-Specific Manner.
The DNA-binding activity of the isolated spORC was examined by a nitrocellulose filter binding assay with full-length ars1 (1.2 kb) as substrate. The elution profile of the spORC protein complex (Fig. 1B) and DNA-binding activity (Fig. 1C) from the hydroxyapatite column coincided, indicating that this activity was intrinsic to spORC. As shown in Fig. 2A, the purified spORC complex interacted efficiently with ars1 DNA in the presence or absence of ATP. Low ATPase activity (about 0.25 pmol of ATP hydrolyzed per min per pmol of protein) was detected with the FLAG peptide-eluted spORC preparation (data not shown). However, the possibility that this low activity may be caused by trace contaminants present in the spORC fraction has not been ruled out. Thus, whether spORC resembles Sac. cerevisiae ORC in possessing low ATPase activity (14) remains to be determined.
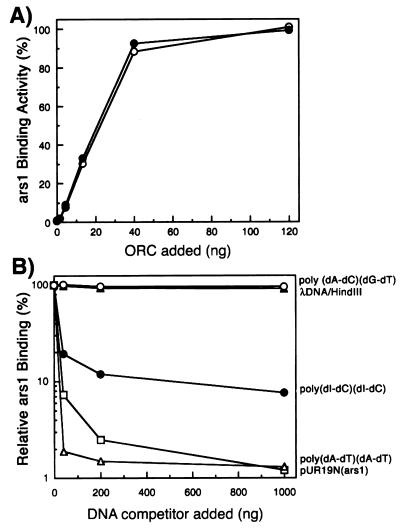
Figure 2.
Binding properties of spORC to ars1. (A) Effect of spORC on DNA binding. Increasing amounts of the FLAG peptide-eluted spORC preparation were incubated with 10 fmol of 32P-labeled ars1 DNA in the presence (○) or absence (●) of 1 mM ATP, and the extent of DNA binding was determined with the nitrocellulose filter binding assay. (B) Influence of various DNA competitors on spORC binding to ars1. The ars1-binding activity of the spORC protein (20 ng) was determined by the nitrocellulose filter binding assay in the presence of the various competitors as indicated. The relative ars1-binding activity (%) was calculated as the percentage of complex formed in the presence of competitor compared with the complex formed in the absence of competitor. The symbols indicated denote the competitor used. ○, poly(dA-dC)⋅(dG-dT); ●, poly(dI-dC)⋅(dI-dC); ▵, poly(dA-dT)⋅(dA-dT); ▴, λ DNA digested with HindIII; □, pUR19N plasmid containing ars1.
To examine whether the binding of spORC to ars1 is sequence specific, competition assays were performed by using levels of spORC capable of binding 50% of ars1 DNA. As shown in Fig. 2B, high levels of the copolymer poly(dA-dC)⋅(dG-dT) or phage λ DNA digested with HindIII did not compete the ars1-binding activity of spORC, whereas plasmid DNA containing ars1 DNA (pUR19N) efficiently blocked the reaction. These results suggest that spORC binds to ars1 in a sequence-specific manner. The copolymer poly(dA-dT)⋅(dA-dT) prevented spORC binding most efficiently and poly(dI-dC)⋅(dI-dC) also reduced ORC binding to ars1 DNA significantly (plateauing at ≈90% inhibition at high levels). Previous studies have shown that the DNA-binding activity of AT-rich sequence-binding proteins containing AT-hook motifs is significantly inhibited by poly(dI-dC)⋅(dI-dC), which has a minor groove similar in shape to that of AT sequences (26). Thus, spORC most likely interacted with ars1 at AT-rich sequences through the multiple AT-hook motifs at the N-terminal domain of spOrc4p.
spORC Interacts with Multiple Sites in ars1.
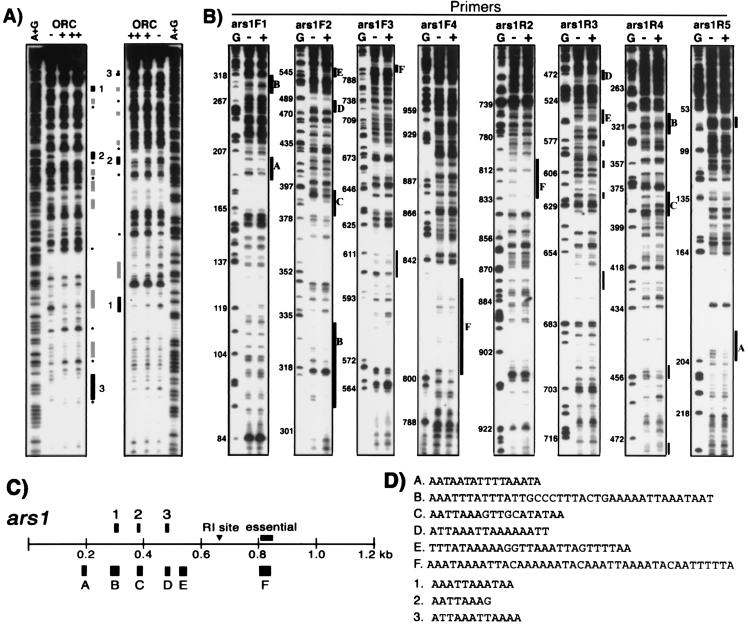
Fragments derived from ars1 were prepared and assayed for their ability to bind spORC to localize the spORC-binding sites within ars1. For this purpose, partial ars1 fragments of about 250 bp (ars1-1 to ars1-5, shown in Fig. 3A) were synthesized by PCR and used as competitors in the nitrocellulose filter binding assays. As shown in Fig. 3B, the addition of excess unlabeled DNA fragments, ars1-3 and ars1-5 barely reduced the binding of spORC to labeled ars1 DNA. In contrast, ars1-4 was the most effective competitor among the ars1 fragments examined (85% reduction). Fragments ars1-1 and ars1-2 also showed significant competition of spORC binding to ars1 (40–50% reduction). These results indicate that among these small fragments the ars1-4 region interacts most strongly with spORC, while ars1-1 and ars1-2 contact the complex as well. None of these small DNA fragments, however, was as effective as a competitor compared with full-length ars1. These observations suggest that ars1 DNA contains multiple spORC-interacting sites that are required for efficient binding of the complex. To support this notion, ars1 fragments of different lengths were generated, and their spORC-binding activity was evaluated in the competition assay. As shown in Fig. 3C, the larger DNA fragments containing more than two of the small spORC-interacting DNA regions, such as ars1-6, 1-7, 1-9, and 1-10, competed more effectively than the shorter ars1-4 fragment. The ars1-6 DNA fragment, which included all three DNA regions that interacted with the complex, showed the highest competition, similar to that observed with full-length ars1. These results suggest that the maximal binding of the spORC to ars1 DNA requires a relatively long DNA region containing multiple spORC-binding sites.
Figure 3.
Interaction of spORC with various regions of ars1. (A) Schematic diagram summarizing the regions in full-length ars1 from which the small ars1 fragments used were derived. The gray box indicates the region previously shown to be essential for ARS activity by deletion analysis of ars1 (6). The site labeled RI indicates the replication initiation site determined in vivo. (B and C) Influence of small ars1 fragments on spORC binding of labeled ars1 DNA. Nitrocellulose filter binding assays were performed with 20 ng of spORC and 10 fmol of full-length ars1 in the presence of various levels of the indicated small ars1 fragments.
spORC-Binding Sites in ars1 DNA Are AT-Rich Sequences.
DNase I footprinting assays were performed to determine the sequences of the multiple spORC-binding sites in ars1 DNA that were identified by the nitrocellulose filter binding assays. When the ars1-2 fragment was used as a substrate in the DNase I footprinting assay, several regions (as shown in Fig. 4 A, C, and D) were protected by spORC binding. These sites were virtually all AT sequences. Results with ars1-1 or ars1-4 DNA fragments as substrate also indicated that the complex interacted with AT-rich sequences (data not shown), suggesting that any AT-rich sequence can interact with the spORC. These findings are consistent with the observations that multiple AT-rich sequence regions are important for ARS activity in S. pombe cells and that the spOrc4p contains multiple motifs that interact with these sequences. However, the interaction between spORC and small ars1 fragments may not reflect the in vivo binding properties of spORC to ars1. To examine the interaction of spORC with full-length ars1, DNase I footprinting assays were performed with pUR19N DNA containing full-length ars1 as substrate. As shown in Fig. 4 B and C, six major spORC-binding sites in ars1 (indicated by black boxes, labeled A to F) were detected from the analysis of both strands. The sequences of these regions showed that the spORC interacted with sequences in ars1 that were almost exclusively AT-rich (Fig. 4D). A few additional DNA regions of limited protection, shown by gray boxes in Fig. 4B, were detected on only one strand. These regions also contained short AT-rich sequences. However, protection of many of these sites seemed to be caused by the excess spORC protein in the reaction, because they disappeared preferentially at lower concentrations of the complex.
Figure 4.
Determination of spORC-binding sites in ars1 by DNase I footprinting. (A) spORC binding sites in ars1-2 were identified by DNase I footprinting analysis using 15 ng (lane +) or 45 ng (lane ++) of spORC protein. Black boxes indicate the major protection sites, and gray boxes indicate weak protection sites. Asterisks (*) show hypersensitive sites. Lanes A+G represent the sequencing of ars1-2 fragment by using the Maxam and Gilbert method; lanes labeled − indicate reactions carried out without spORC protein. (B) spORC-binding sites in ars1 with pUR19N plasmid as substrate. DNase I-treated DNA was analyzed by primer extension with the primers indicated. The numbers on the left side of the autoradiogram indicate nucleotide positions in ars1. The black boxes labeled A to F denote sites protected in both DNA strands; gray boxes indicate minor sites protected only in one strand. Lanes G represent sequencing reactions that contained dideoxy-GTP; lanes labeled − and + denote reactions without the spORC protein, and reactions containing 60 ng of spORC, respectively. The locations and nucleotide sequences of the major protection sites are summarized in C and D, respectively.
N-Terminal AT-Hook Motifs of spOrc4 Are Essential for ars1 Binding.
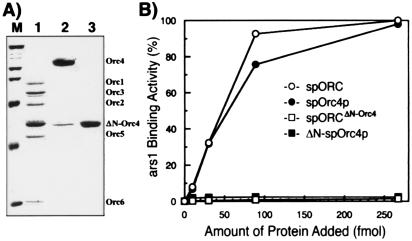
As reported previously, spOrc4p contains nine AT-hook motifs, which would be expected to interact with AT-rich sequences. To examine the role of this domain in the binding of the complex to ars1, we constructed and purified the spORCΔN-Orc4p containing the N-terminally truncated spOrc4 subunit (amino acids 516–972), the spOrc4 subunit alone, and the ΔN-spOrc4 subunit alone. When the ΔN-spOrc4 subunit, which is devoid of AT-hook domains, was expressed with the other spORC subunits, a complex containing all six subunits was isolated (Fig. 5A, lane 1), indicating that the N-terminal domain of spOrc4p is not required for the assembly of the spORC. As shown in Fig. 5B, ars1-binding activity was not observed with either the spORCΔN-Orc4 or the ΔN-spOrc4p in the nitrocellulose filter binding assay. In contrast, spOrc4p alone bound ars1 DNA as efficiently as the wild-type spORC (Fig. 5B), and this binding was competed by the copolymers poly(dA-dT)⋅(dA-dT) or poly(dI-dC)⋅(dI-dC) as observed with spORC (data not shown). Furthermore, the binding of spOrc4p to ars1 was reduced quantitatively by the addition of small ars1 DNA fragments (ars1-1 to ars1-10) in a manner identical with that observed with spORC (data not presented), suggesting that spOrc4p and the spORC interact with the same regions in ars1. These results indicate that the N-terminal AT-hook motifs of spORC4p mediate the interaction between spORC and ars1.
Figure 5.
DNA-binding properties of various spORC complexes and spORC4. (A) spORCΔN-Orc4, spOrc4, and ΔN-spOrc4 proteins were purified and analyzed by SDS/10% polyacrylamide gel electrophoresis followed by Coomassie blue staining. (B) Comparison of ars1-binding activities of spORC, the ORCΔN-Orc4, spOrc4, and ΔN-spOrc4 proteins. Indicated amounts of proteins were incubated with 10 fmol of 32P-labeled ars1, and the level of binding was examined by using the nitrocellulose filter binding assay. ○, spORC; ●, spOrc4p; □, ORCΔN-Orc4; ■, ΔN-spOrc4p.
Discussion
In this study, we examined the in vitro interaction of the spORC with ars1 to evaluate the cis-acting elements that are important for origin function in S. pombe. We have shown that the spORC, reconstituted and purified from Sf9 cells, binds to several AT-rich sequences in ars1. These results are in keeping with in vivo data that redundant clusters of AT-rich sequences within ARS elements are important for DNA replication in S. pombe. Our findings also indicate that the binding of spORC to ars1 is completely dependent on the N terminus of the spOrc4p subunit, which contains multiple AT-hook motifs. The DNA-binding properties of AT-hook motifs have been well characterized in the members of the high mobility group A (HMGA) family of proteins (24, 27). Mammalian HMGA proteins possess a set of three AT-hook motifs that are separated by unstructured linker peptides. A single AT-hook motif binds to the minor groove of an AT-rich sequence between four and six base pairs in length with the optimal binding site centered in the sequence AA(T/A)T. Because of its three AT-hook motifs, each HMGA protein is capable of interacting with multiple sites in a DNA substrate that are separated by various nucleotide lengths. Simultaneous binding of multiple AT-hooks increases the strength of the interaction between the HMGA protein and DNA and also affects the charge characteristics and conformation of the DNA. Such alterations in DNA structure, induced by HMGA binding, have been shown to play an important role in vivo for both enhanceosome formation in the human β-IFN promoter region and transcription of this gene. The results obtained from the analysis of spORC–ars1 interactions described in this study are in accord with the DNA-binding properties of AT-hook motifs. The most efficient binding of spORC to ars1 required about 1-kb DNA including all weak binding sites (Fig. 3), and all major protection sites revealed by the DNase I footprinting analysis were stretches of AT-rich sequences containing two or more potential AT-hook binding sites in close proximity (Fig. 4D). The N-terminal spOrc4p region, with its nine AT-hook motif repeats, may support prereplicative complex formation by acting in a manner analogous to the HMGA protein in enhanceosome formation. The binding of multiple AT-hooks to origin DNA could cause conformational changes in this DNA region that facilitate the formation of the prereplicative complex. In addition, the spacing between several AT-hook binding sites in ars1 might be important not only for spORC binding but also for the assembly of prereplicative complexes as well.
The ars1-3 DNA, which includes the reported in vivo replication initiation (RI) site (nucleotides 670–673) of ars1 (28), did not interact strongly with spORC. The absence of strong spORC-binding sites around the RI site was further confirmed by DNase I footprinting analysis with full-length ars1. As shown in Fig. 4C, five major protection sites found (labeled as A to E) were located between nucleotide 180 and nucleotide 550, and one relatively broad protection site (F) was located between nucleotide 801 and nucleotide 840. A gap of about 250 bp exists between these two protection sites, and the RI site is positioned in the middle of these two spORC-binding sites. This location, between spORC-binding sites, including the RI site, may be the region where the prereplicative complex assembles during the G1 phase. Consistent with this notion, recent studies indicated that spORC binds to two important AT-rich sequences, region I and region III of S. pombe ars2004 (29). The initiation site of DNA replication of ars2004 has been located between region Ι and region III. Furthermore, in vivo cross-linking and chromatin immunoprecipitation analysis showed that the MCM complex bound to DNA sequences near region II, located between the two spORC-binding sites during the G1 phase (T. Takahashi and H. Masukata, personal communication). Possibly, the binding of a monomeric or multimeric spORC to the two clusters of binding sites in ars1 (A to E and F), which are about 250 bp apart, may lead to the formation of a DNA loop that is required for the assembly of prereplicative complex. It may also cause conformational changes in this DNA region resulting in alterations in negative superhelicity, DNA bending, or DNA unwinding.
The binding studies described above indicated that spORC interacted with ars1 DNA only through the multiple AT-hook motifs in the N terminus of spOrc4p, and ATP was not required for this activity. In contrast, the origin DNA-binding activity of Sac. cerevisiae ORC and the binding of the Drosophila ORC to the ACE3 element were ATP-dependent (9, 23, 30). Considering the highly conserved nature of the mechanism of DNA replication, we cannot rule out the possibility that ATP affects the interaction of the spORC with origin DNA. Even if there were a weak ATP-dependent ars1-binding activity of spORC, as shown in the Drosophila ORC–ACE3 interaction, ATP may not affect the binding of spORC to ars1 DNA because of the strong ars1-binding activity of the spOrc4p N terminus. The initial binding of spORC to ars1 may be governed solely by the AT-hook motifs in spOrc4p, whereas the binding of ATP to other ORC subunits, such as spOrc1p, spOrc4p, or spOrc5p, leads to conformational changes that support the interactions of other ORC subunits with DNA. It is also possible that the phosphorylation of spORC affects its interaction with ars1. Our spORC preparations were heavily phosphorylated, judged by the presence of multiple bands of spOrc2p, which were reduced to a single protein band after λ phosphatase treatment. The phosphorylation of ORC by cyclin-dependent protein kinases is important for the regulation of the prereplicative complex in Sac. cerevisiae (31, 32).
Although the proteins involved in DNA replication and the fundamental aspects of this process seem to be conserved evolutionarily, the target sequences for origin activation in many eukaryotes are markedly different from those in Sac. cerevisiae. The results presented above suggest that the binding of spORC to its replication origins occurs through a mechanism that differs from that observed in Sac. cerevisiae.
Acknowledgments
We thank Dr. Z.-Q. Pan for the critical review of this paper. We are indebted to B. Phillips and I. Tappin for the preparation of Sf9 cells and the amplification of baculoviruses. This work was supported by Grant GM38559 from the National Institutes of Health.
Abbreviations
- ORC
origin recognition complex
- sp
Schizosaccharomyces pombe
- ARS
autonomously replicating sequence
- HMGA
high mobility group A
References
- 1.Marahrens Y, Stillman B. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 2.Marahrens Y, Stillman B. EMBO J. 1994;13:3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theis J F, Newlon C S. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DePamphilis M L. BioEssays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Spradling A C. Genes Dev. 1999;13:2619–2623. doi: 10.1101/gad.13.20.2619. [DOI] [PubMed] [Google Scholar]
- 6.Clyne R K, Kelly T J. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuno Y, Satoh H, Sekiguchi M, Masukata H. Mol Cell Biol. 1999;19:6699–6709. doi: 10.1128/mcb.19.10.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubey D D, Kim S M, Todorov I T, Huberman J A. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 9.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 10.Bell S P, Kobayashi R, Stillman B. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 11.Fox C A, Loo S, Dillin A, Rine J. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 12.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 13.Rao H, Stillman B. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 15.Kelly T J, Brown G W. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 16.Dutta A, Bell S P. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 17.Moon K Y, Kong D, Lee J K, Raychaudhuri S, Hurwitz J. Proc Natl Acad Sci USA. 1999;96:12367–12372. doi: 10.1073/pnas.96.22.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang R Y, Kelly T J. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossen M, Pak D T, Hansen S K, Acharya J K, Botchan M R. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 20.Rowles A, Chong J P, Brown L, Howell M, Evan G I, Blow J J. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 21.Tugal T, Zou-Yang X H, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- 22.Vashee S, Simancek P, Challberg M D, Kelly T J. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 23.Austin R J, Orr-Weaver T L, Bell S P. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves R, Beckerbauer L. Biochim Biophys Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee J K, Hurwitz J. J Biol Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 26.Solomon M J, Strauss F, Varshavsky A. Proc Natl Acad Sci USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bustin M. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez M, Antequera F. EMBO J. 1999;18:5683–5690. doi: 10.1093/emboj/18.20.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi, T. & Masukata, H. (2001) Genes Cells, in press. [DOI] [PubMed]
- 30.Klemm R D, Bell S P. Proc Natl Acad Sci USA. 2001;98:8361–8367. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen V Q, Co C, Li J J. Nature (London) 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 32.Vas A, Mok W, Leatherwood J. Mol Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]