Abstract
The natural ability of stem cells to self-organize into functional tissue has been harnessed for the production of functional human intestinal organoids. Although dynamic mechanical forces play a central role in intestinal development and morphogenesis, conventional methods for the generation of intestinal organoids have relied solely on biological factors. Here, we show that the incorporation of uniaxial strain, by using compressed nitinol springs, in human intestinal organoids transplanted into the mesentery of mice induces growth and maturation of the organoids. Assessment of morphometric parameters, transcriptome profiling, and functional assays of the strain-exposed tissue revealed higher similarities to native human intestine, with regards to tissue size and complexity, and muscle tone. Our findings suggest that the incorporation of physiologically relevant mechanical cues during the development of human intestinal tissue enhances its maturation and enterogenesis.
Keywords: Uniaxial Strain, Tissue Engineering, Transplantation, Human Pluripotent Stem Cells, Small Intestine
INTRODUCTION
The establishment of a three-dimensional in vitro model of the human intestine has required a deep understanding of embryonic intestinal development1. These complex structures are created from human embryonic stem cells and/or induced pluripotent stem cells (PSCs) by the perturbation of signaling pathways through a temporal series of growth factor manipulations2, 3. This exclusively biological and mechanically static methodology to intestinal tissue generation has been successful in creating both functional intestinal lineages (e.g. Paneth, Goblet, enteroendocrine, and enterocyte) and architecture similar to that of native intestine (e.g. crypts, villi, and smooth muscle layers)2, 4. These tissues, termed human intestinal organoids (HIOs) have been shown to be functional and possess the ability to engraft in vivo4, 5. However, the maturation status of transplanted HIOs (tHIOs) best approximates that of human fetal intestinal tissue, which ultimately limits their utility6. Current methods have yielded a powerful tool for use in both basic science and clinical applications. HIOs are an avenue to study intricate physiological interactions and personalize medicine for patients across the globe7, 8. However, in order to build upon these uses it is imperative that we begin to scale and generate tissues with increased size, maturity and function, because creating tissue for transplantation remains an unmet clinical need.
Recently, there has been a renewed interest in not only the biological cues impacting development and morphogenesis, but the mechanics of development as well9. In particular, Shyer and Savin have elegantly demonstrated that strain plays a role in intestinal development. Gut looping, villification, and the localization of intestinal stem cells to the crypt have all been associated with mechanical strain10–13. As strain is a common contributing factor between several architectural features of the intestine, we thought to incorporate this pulling or tension force within the generation of HIOs. Perhaps both mechanical and biological cues will prove necessary in the generation of larger scale tissues with a maturation status beyond that of human fetal intestine.
In order to incorporate strain into the current protocol of HIO generation, we combined in vivo transplantation with the repurposing of a lengthening device designed for the treatment of short bowel syndrome: the spring. Much like other endoluminal lengthening devices, the spring has been shown to stimulate an adaptive morphometric response in the setting of mature tissues, though additional effects have not been thoroughly characterized14–18. Of the various lengthening devices previously described, we chose to utilize the spring because its geometry and applied force could be scaled for our purposes and it allows for the accumulation of mucous as the tHIO is a closed system14–17. In combining these transplantation strategies, we hypothesized that the application of strain in a fetal setting would elicit tissue maturation and overall growth of the tHIO.
In this study, we have combined a common mechanic of development, uniaxial strain, with the generation of HIOs. Grafts that had undergone strain (transplanted HIO with strain; tHIO+S) were found to have increased intestinal and maturation features compared to those that did not experience applied strain, including a transcriptional, morphological, and functional shifts toward postnatal human intestine. Herein, we report the first description of mechanically manipulating tHIOs in vivo to result in the successful induction of maturation and enterogenesis.
RESULTS
Nitinol Spring as an Endoluminal Lengthening Device for tHIOs
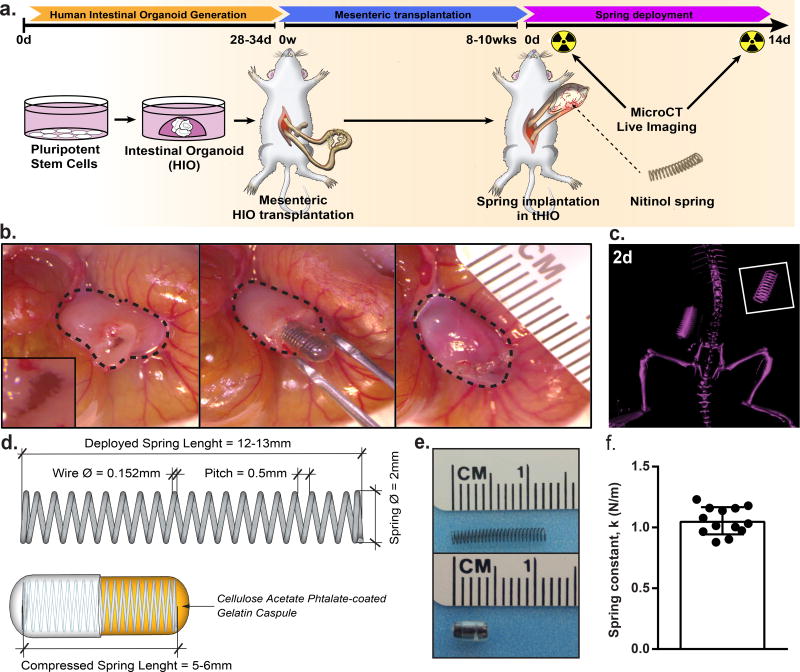
Our groups previously developed a method of stepwise growth factor manipulations to differentiate human pluripotent stem cells into intestinal organoids2–5. Upon transplantation into the mesentery of NOD-SCID IL-2Rγnull (NSG) mice these organoids indeed engraft and go on to closely resemble native intestine with well-defined crypt regions, villi, and smooth muscle layers4. After ten weeks, the tHIO has drastically grown in size. At this time, a secondary procedure was performed wherein a compressed spring was implanted inside the tHIO (Figure 1a). 14 days (14d) post implantation of the springs, the grafts were harvested and their human origins confirmed (Supplementary Figure 1a,b)19. The spring implantation first involves opening the tHIO, briefly flushing any accumulated mucous and inserting the encapsulated compressed spring into the luminal space of the tHIO before it is closed (Figure 1b). Sham experiments of implanting empty capsules were also performed (Supplementary Figure 1c). The survival rates between sham and spring implanted tHIOs were similar (Supplementary Figure 1d). The spring’s deployment could be monitored in vivo through micro computed-tomography (microCT) and was observed to be linear (Figure 1c).
Figure 1. Transplantation of springs into tHIOs.
(a) 28–34 day old HIOs were transplanted into the mesentery of NSG mice and allowed to grow for 8–10 weeks. Then, a second procedure was performed wherein a compressed NiTi spring was implanted inside the tHIO. Harvest occurred 14 days post spring implantation. (b) Procedural images of the spring insertion into a tHIO. Dashed line indicates perimeter of tHIO. (c) MicroCT of a linearly deployed spring in vivo 2 days after implantation. (d) Schematic of springs used in experiments. Springs utilized for transplantation had a relaxed length of 12–13 mm, compressed length of 5–6 mm and an outside diameter of 2 mm. Compression of springs was maintained through use of a gelatin capsule subsequently coated with a polymer to delay deployment. (e) Photographs of springs used in relaxed (top) and compressed/encapsulated (bottom) forms. (f) The spring constant of those used was 1.05 ± 0.11 N/m (n=13 independent samples). Data are represented as mean ± SD.
Our use of the spring was based upon previous studies describing endoluminal lengthening devices14–17, 20. We found the nickel-titanium (nitinol, NiTi) spring as an ideal device, because it had been successfully used in rats and did not require manipulation post implantation as hydraulic devices do14, 17. Another advantage in utilizing the NiTi spring is that a large amount of negative space was retained within the tHIO after insertion, which permitted mucous accumulation. The spring’s geometry was based upon the constraint of a commercially available gelatin capsule used to maintain the spring in a compressed state during transplantation. An enteric coating of cellulose acetate phthalate (C-A-P) was used in order to delay the degradation of the gelatin capsule during implantation. The springs had a relaxed length of 12–13 mm, outer diameter of 2 mm and when encapsulated were 5–6 mm in length (Figure 1d,e). The pitch of the spring was designed to achieve a spring constant of 1.05 ± 0.11 N/m (Figure 1f). Springs with higher constants deployed in a nonlinear fashion causing bowel obstructions and total mortality (Supplementary Figure 1d).
Morphological Maturation as a Result of Strain in the tHIO
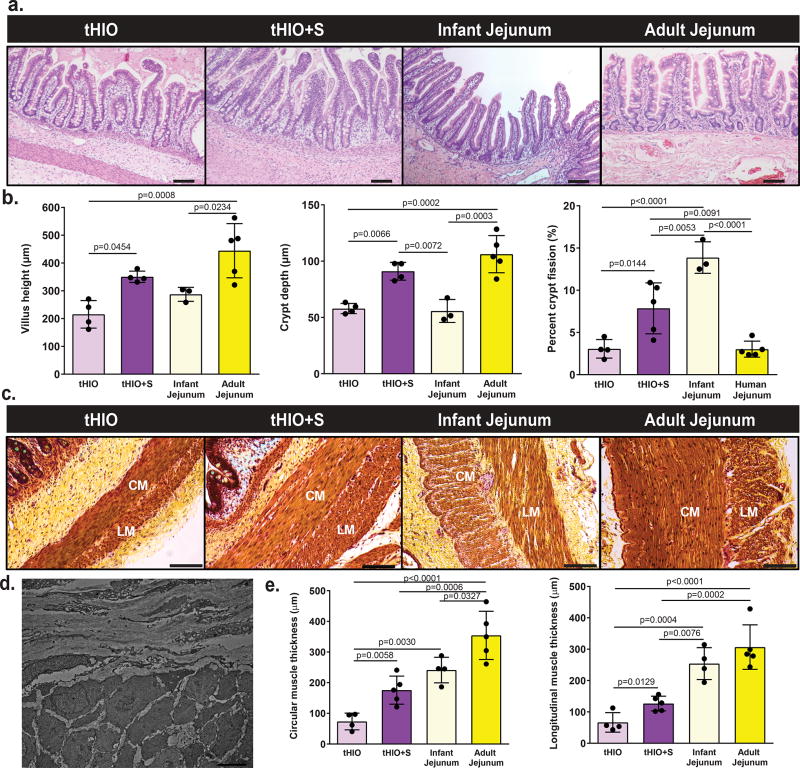
At the time of harvest, tHIO+S had a more tubular appearance verses the saccular shape of sham or tHIO and grew to about 10–14 mm (Supplementary Figure 1a). To examine if the application of strain produced an adaptive response, architectural features were quantified in tHIO, tHIO+S, and human surgical samples of infant and adult jejunum from hematoxylin and eosin stained sections (Figure 2a). In tHIO+S, villus height, crypt depth and crypt fission were observed to be increased significantly over that of tHIO (Figure 2b). The increase in both villus height and crypt depth approximated that of adult jejunum, while the level of crypt fission was higher in tHIO+S (Figure 2b). Upon quantifying these features in sham operated grafts, no statistically significant differences were found when compared to tHIO indicating that the changes observed in tHIO+S were in association with strain and not surgically induced (Supplementary Figure 1d). All subsequent assays used the tHIO as a developmental control, as its attributes represent the current state of the field. Smooth muscle layers were also quantified across sample types using pentachrome stained sections (Figure 2c). Perpendicular orientation of muscle fibers was observed in micrographs obtained using a transmission electron microscope (TEM) in both tHIO and tHIO+S (Figure 2d, only tHIO+S shown). This observation confirmed the presence of circular and longitudinal muscle layers. The thicknesses of which were found to be increased significantly in tHIO+S over tHIO (Figure 2e). While increased muscle thickness was observed in the tHIO+S, it did not yet approximate that of human adult jejunum. These data all suggest that a regulated tensile force improves the overall structure and geometry of tHIOs.
Figure 2. tHIO+S samples exhibit increased morphological characteristics.
(a) Representative H&E sections of tHIO, tHIO+S, infant jejunum and adult jejunum. Scale bar = 50 µm. This experiment was repeated three times independently and findings were similar. (b) Morphometric quantification of tissue sections was plotted. Villus height, crypt depth and crypt fission were increased in tHIO+S compared to tHIO and better approximate human tissue in the cases of villus height and crypt depth. Sample sizes are the following: tHIO n=4, tHIO+S n=4, infant jejunum n=3, and adult jejunum n=5. All samples are biologically independent. (c) Representative pentachrome sections of tHIO, tHIO+S, infant jejunum and adult jejunum. Scale bar = 100 µm. This experiment was repeated three times independently and findings were similar. (d) Transmission electron micrograph displaying perpendicular orientation of muscle fibers in tHIO+S, similar orientation was observed in tHIO. This experiment was repeated three times independently and findings were similar. (e) Quantification of muscle in pentachrome sections was plotted. Layers of circular and longitudinal muscle (CM; LM respectively) were thicker in tHIO+S compared to tHIO and trend toward that of human jejunum. Sample sizes are the following: tHIO n=4, tHIO+S n=5, infant jejunum n=3, and adult jejunum n=5. All samples are biologically independent. For (b,e) data are represented as the mean ± SD. An ANOVA followed by Tukey’s post hoc tests were performed and the statistical significance cutoff was p < 0.05.
Strain’s Impact on Transcriptional Changes in tHIOs
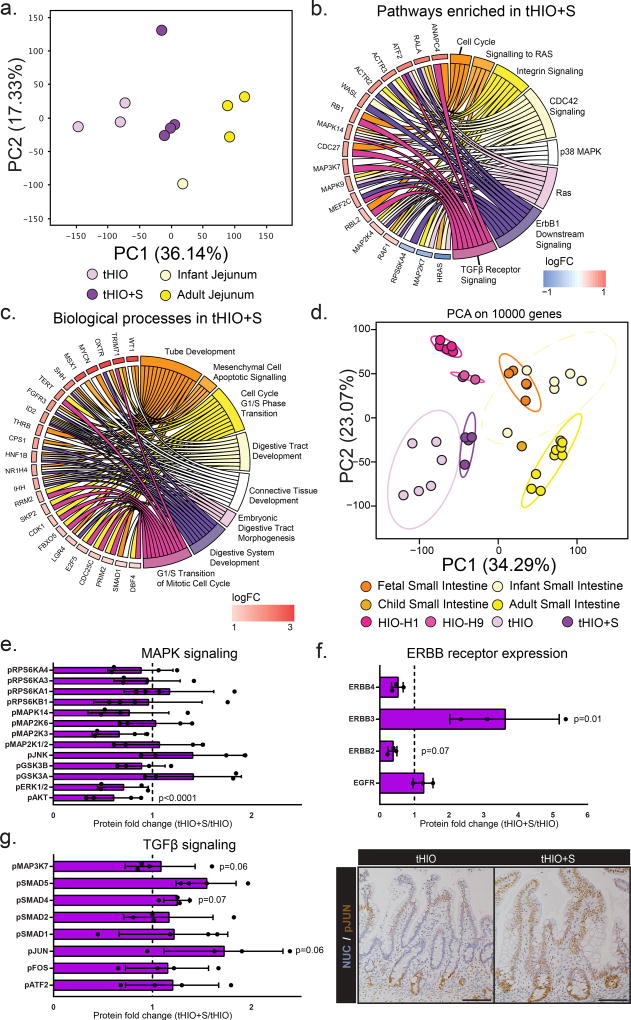
We examined the differential gene expression in tHIO+S compared to tHIO and human jejunum (all full thickness samples) using RNA sequencing. Out of 23,366 genes annotated in the genome, 4,537 genes were significantly differentially regulated amongst the samples. Samples were analyzed using a scaled centered principle component analysis (PCA) to visualize the multi-dimensional variation between samples. Principal component 1 (36.14%), discriminated the samples among their types and suggested a higher degree of similarity between tHIO+S and human infant tissue (Figure 3a). Unsupervised hierarchical clustering based on the Cosine distance and Spearman rank correlation, revealed similarities between tHIO+S and human infant tissues suggesting a pro-maturation effect of the applied strain (r2>0.75) (Supplemental Figure 2a). We then performed a functional enrichment analysis on the PC loadings. We found that PC1 segregated PSCs-derived tissues from Patient-derived tissues encompassing biological processes involving development (negative) and an immune signature (positive). PC2 segregated an overall intestinal signature from immune-competent intestinal tissues, encompassing immune and defense processes (negative) and digestion and metabolic processes (positive) (Supplementary Figure 2b–e).
Figure 3. Transcriptionally tHIO+S are matured beyond tHIO.
(a) Scaled Centered Principal Component Analysis of the tHIOs, tHIO+S, human infant and adult jejunum was performed. tHIO+S clustered closer to human jejunum tissues. Sample sizes are the following: tHIO n=3, tHIO+S n=4, infant jejunum n=1, and adult jejunum n=3. All samples are biologically independent. (b) Functional enrichment of the pathways upregulated in tHIO+S compared to tHIOs. Sample sizes are the following: tHIO n=3, tHIO+S n=4. All samples are biologically independent. (c) Differential expression gene analysis between tHIO+S and human tissues demonstrated upregulated biological processes in the GO categories concerning intestinal development. Sample sizes are the following: tHIO+S n=4 and adult jejunum n=3. All samples are biologically independent. (d) Scaled Centered Principal Component Analysis of samples retrieved from our study and several publicly available databases. Sample sizes are the following: fetal small intestine n=5, infant small intestine n=6, child small intestine n=1, adult small intestine n=9, HIO-H1 n=5, HIO-H9 n=3, tHIO n=6, and tHIO+S n=4. All samples are biologically independent. (e) Fold-change in MAPK signaling protein array. n=4 biologically independent samples for all groups. (f) Fold-change in ERBB receptor protein expression. n=3 biologically independent samples for all groups. (g) (Left panel) Fold-change in TGFβ signaling protein array. n=4 biologically independent samples for all groups. (Right panel) Immunostaining of phosphorylated JUN in tHIO and tHIO+S. Data are representative of n=4 for all groups. For (e–g) data are represented as the mean ± SD. The unpaired student t-test statistical significance cutoff was p < 0.05.
In order to examine the biological processes induced by strain in tHIO, we compared the differentially expressed genes (DEGs) between the tHIO+S and tHIO groups. In tHIO+S compared to tHIOs, 454 and 243 genes were respectively up- and down-regulated. Next, a functional enrichment analysis was performed to highlight biological processes within these gene sets. 14d after the initial application of strain implantation, cell cycle and overall catabolic and protein assembly processes were captured, suggesting an impact of strain upon cell proliferation and tissue formation (Supplementary Figure 3a). When further examining the gene ontology analysis of pathways enriched in tHIO+S compared to tHIO, significant pathways concerning the cell cycle, MAPK, ERBB and TGFβ signaling were observed (Figure 3b). Since the tHIO+S displayed a higher similarity with human jejunum tissues, we compared their profiles. 254 genes were uniquely up-regulated in tHIO+S when compared to human jejunum tissues (Supplementary Figure 3b). Interestingly, the functional enrichment highlighted biological processes involving tube and digestive tract development, mesenchymal cell functions and cell cycle (Figure 3c) that were not seen in tHIO when compared to human tissues (Supplementary Figure 3c). We hypothesized that strain would enhance maturation of the tHIO. To test our hypothesis, we re-processed a collection of public RNAseq databases including fetal, newborn, human small intestinal tissues and HIO, tHIO and tHIO+S. A scaled centered PCA was performed to dimensionally cluster the samples. As previously demonstrated, the HIO clustered with fetal tissues. Interestingly, the tHIO+S were clustering toward human infant tissues thereby confirming our hypothesis (Figure 3d; Supplementary Figure 4a). To gain additional insight from the principal components (PCs), we performed a functional enrichment analysis on the PC loadings. We found that PC1 segregated PSCs-derived tissues from Patient-derived tissues encompassing biological processes involving developmental processes and morphogenesis (negative) and an immune signature (positive). PC2 segregated fetal intestinal tissues from intestinal matured tissues, encompassing biological processes involving system development processes (positive) and digestion and metabolic processes (negative) (Supplementary Figure 4b–e).
To broadly investigate some of the pathways enriched with exposure to strain beyond the level of transcription, protein analysis was performed. A protein phosphorylation array for MAPK signaling was executed and quantified (Figure 3e). The fold changes in protein phosphorylation levels related to MAPK signaling were not consistently up or down across the panel, making it difficult to draw a conclusion as to its potential role. (For this and all subsequent protein analysis, array maps, developed membranes and normalized protein expression levels can be found in Supplementary Figure 5a–f). Pan ERBB receptor expression was also measured and quantified (Figure 3f). ERBB3 was found to be significantly increased in tHIO+S over tHIO. Finally, TGFβ signaling was investigated; a protein phosphorylation array was performed and quantified (Figure 3g, left panel). While, statistical significance was not found due to high variability, there did appear to be an overall increasing trend in TGFβ pathway activation as measured by phosphorylation. The most striking difference was found to be in phosphorylated Jun Proto-Oncogene, AP-1 Transcription Factor Subunit (pJUN), which was further validated through immunohistochemistry (Figure 3g, right panel). In staining for pJUN, an obvious visual increase was observed. With strain, pJUN expression expanded along the entirety of the villus rather than being predominantly expressed in the crypt with low level expression towards the tips of the villi without strain exposure. This expansion of pJUN provides support for the earlier pathway analysis.
Strain’s Impact on Proliferation and the Stem Compartment in tHIOs
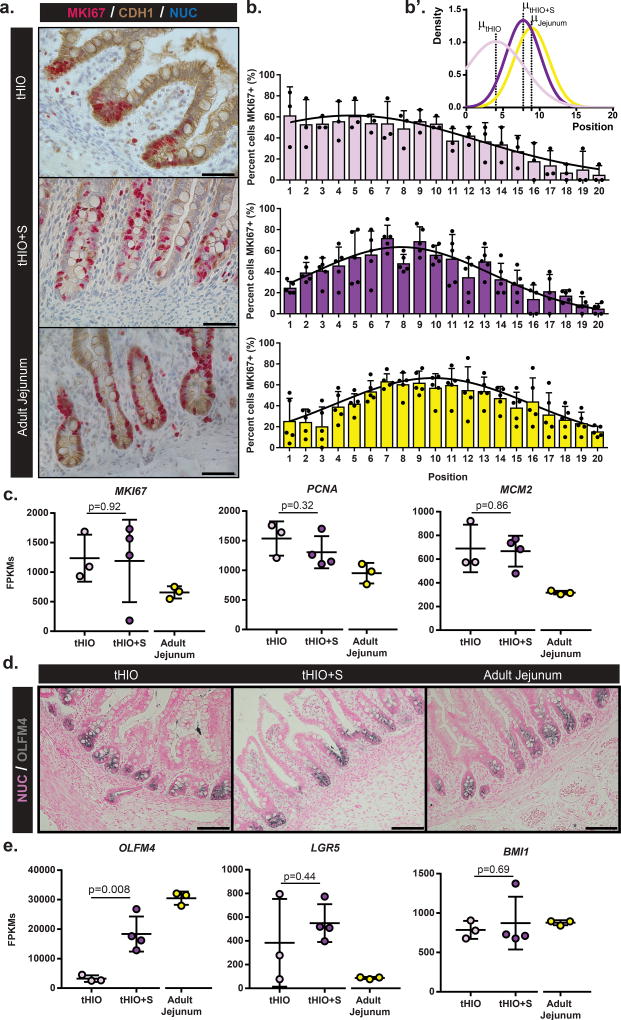
To examine whether strain could impact the intestinal stem cell/transit-amplifying compartment, we analyzed proliferation in the crypts of tHIOs and tHIO+S and human adult jejunum. Dual chromogenic staining for Marker of Proliferation KI67 (MKI67) and Cadherin-1 (CDH1) was performed for the purpose of quantification (Figure 4a). Total epithelial proliferation within the crypt was similar between groups (Supplemental Figure 6a). However, when comparing the means of Gaussian curve fits of the proliferation profiles by position, the tHIO+S exhibited an upward shift within the crypt compartment, which followed a pattern that was similar to that observed in adult jejunum (Figure 4a–b’). This suggests the development of a transient amplifying zone within tHIO+S. The amount of proliferation in sham operated tissues was also quantified and found to follow a similar pattern to that of the tHIO (Supplemental Figure 6b–c’). In total tissue, transcripts for the cell cycle related genes MKI67, Proliferating Cell Nuclear Antigen (PCNA) and Minichromosome Maintenance Complex Component 2 (MCM2) were observed as similar between tHIO+S and tHIO, while both were elevated compared to adult jejunum (Figure 4c). To further examine the intestinal stem cell compartment, staining for Olfactomedin 4 (OLFM4) was performed and all sample types displayed strong positivity exclusive to the crypt (Figure 4d). Transcripts for OLFM4 were found to be significantly increased in tHIO+S compared to tHIO, while additional intestinal stem cell markers Leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5) and BMI1 Proto-Oncogene, Polycomb Ring Finger (BMI1) were observed to be similar between the two groups (Figure 4e).
Figure 4. tHIO+S samples display a shift in proliferation and expansion of the stem compartment.
(a) Double chromogenic staining for Marker of Proliferation KI67 (MKI67), red, and Cadherin 1 (CDH1), brown, on sections of tHIO, tHIO+S and adult jejunum. Scale bar = 50µm. This experiment was repeated three times independently and findings were similar. (b) Proliferation quantified by MKI67 and CDH1 positivity and cell position in tHIO, tHIO+S, and adult jejunum was plotted. An upward shift in proliferation is observed in tHIO+S versus tHIO. Sample sizes are the following: tHIO n=3, tHIO+S n=5, infant jejunum n=3, and adult jejunum n=5. All samples are biologically independent. Data are represented as the mean + SD. (b’) Gaussian curve fits were plotted for each group. (c) Normalized FPKMs were plotted for tHIO, tHIO+S and adult jejunum for cell cycle related genes MKI67, Proliferating Cell Nuclear Antigen (PCNA) and Minichromosome Maintenance Complex Component 2 (MCM2). In all cases, HIO’s levels were elevated above adult jejunum. Sample sizes are the following: tHIO n=3, tHIO+S n=4, and adult jejunum n=3. All samples are biologically independent. Data are represented as the mean ± SD. The unpaired student t-test statistical significance cutoff was p < 0.05. (d) Positive expression of Olfactomedin 4 (OLFM4, gray) in immunohistochemistry sections of tHIO, tHIO+S, and adult jejunum. Scale bar = 100 µm. This experiment was repeated three times independently and findings were similar. (e) Normalized FPKMs were plotted for tHIO, tHIO+S and adult jejunum for stem cell compartment related genes OLFM4, Leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5), and BMI1 Proto-Oncogene, Polycomb Ring Finger (BMI1). OLFM4 was significantly elevated in tHIO+S compared to tHIO, while LGR5 and BMI remained similar between the engineered tissues. Sample sizes are the following: tHIO n=3, tHIO+S n=4, and adult jejunum n=3. All samples are biologically independent. Data are represented as the mean ± SD. The unpaired student t-test statistical significance cutoff was p < 0.05.
Epithelial Response of tHIOs to Strain
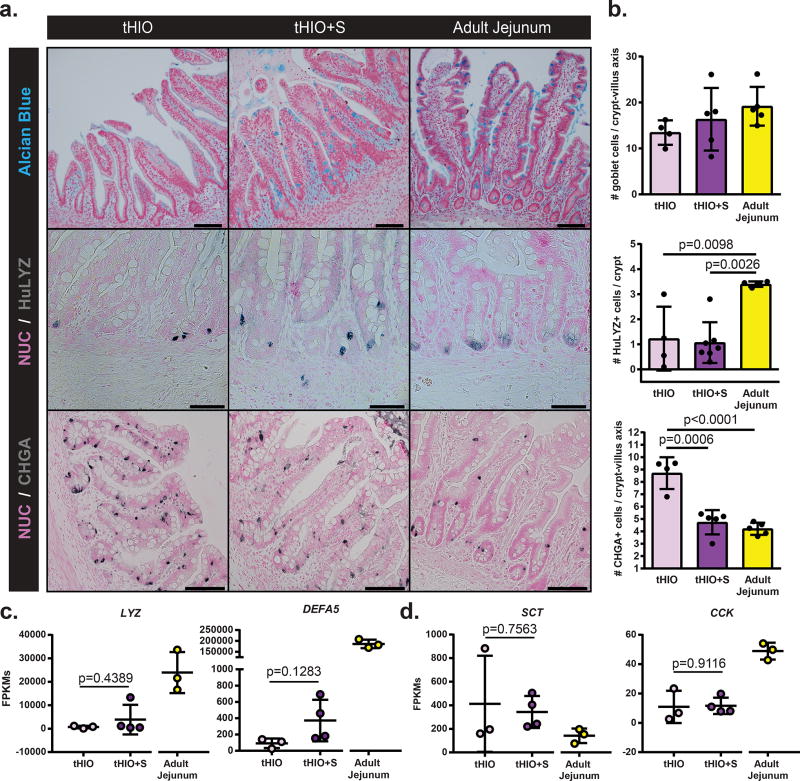
To evaluate if strain impacted cell fate, secretory epithelial cell types were quantified. Alcian blue staining was performed for the identification of Goblet cells, human specific Lysozyme (LYZ) for Paneth cells and Chromogranin-a (CHGA) for enteroendocrine cells (EECs) which were all subsequently quantified (Figure 5a,b). No difference in goblet cell counts were observed across sample types. The number of Paneth cells per crypt was also found similar between tHIO+S and tHIO, yet significantly lower than that of adult jejunum. However, when looking at antimicrobial peptide alpha-Defensin 5 (DEFA5) transcription levels, an increasing trend was observed while those for LYZ remained unchanged (Figure 5c). The number of CHGA+ EECs was significantly reduced in tHIO+S compared to tHIO and approximated that of adult jejunum. Interestingly, the transcripts for EEC secreted hormones Serotonin (SCT) and Cholecystokinin (CCK) were not reduced (Figure 5d). Therefore, this data may suggest strain can negatively regulate EEC specification or that they serve as a progenitor pool. Taken together, the overall impact of strain on the developing graft’s epithelium appears to be one of functional promotion and cell type normalization.
Figure 5. Strain’s impact on secretory lineages.
(a) Sections with staining for Goblet cells (alcian blue), Paneth cells (HuLYZ), and enteroendocrine cells (CHGA) in tHIO, tHIO+S and adult jejunum. Scale bar = 50 µm. These experiments were repeated three times independently and findings were similar. (b) Quantification of cell types in (a). No significant differences are observed in goblet cells, though the intensity of staining visually increases toward that of adult jejunum. The number of Paneth cells is reduced in HIOs compared to adult jejunum. The number of enteroendocrine cells followed a decreasing trend and was significantly less in tHIO+S than tHIO. Sample sizes are the following: tHIO n=4, tHIO+S n=5, and adult jejunum n=5. All samples are biologically independent. Data are represented as the mean ± SD. An ANOVA followed by Tukey’s post hoc tests were performed and the statistical significance cutoff was p < 0.05. (c) Normalized FPKMs were plotted for tHIO, tHIO+S and adult jejunum for the Paneth cell produced antimicrobial peptides Lysozyme (LYZ) and Alpha-Defensin 5 (DEFA5). For both, expression follows an increasing trend in tHIO+S over tHIO, while much lower than that of adult jejunum, though not significant. (d) Normalized FPKMs were plotted for tHIO, tHIO+S and adult jejunum for enteroendocrine cell produced Serotonin (SCT) and Cholecystokinin (CCK). For (c,d) sample sizes are the following: tHIO n=3, tHIO+S n=4, and adult jejunum n=3. All samples are biologically independent. Data are represented as the mean ± SD. The unpaired student t test statistical significance cutoff was p < 0.05.
Epithelial topography of tHIO, tHIO+S, and adult jejunum samples was observed to see if strain impacted features of the surface ultrastructure (Figure 6a). These scanning electron micrographs depicted ridge-like protrusions in the tHIO, while long finger-like villus projections were found in tHIO+S. This increase in structuration suggests that strain indeed promoted the villification process, as previously described11. Deep corrugations in PSC generated tissues were quite rare, but found to be a prominent feature of the adult jejunum’s large finger-like villi. This additional epithelial folding drastically increases the tissue’s surface area increasing the gut’s efficiency in digestive processes21. These observations suggest strain improved the architectural features of the tHIO, while further complexities, like corrugations, remain largely underdeveloped.
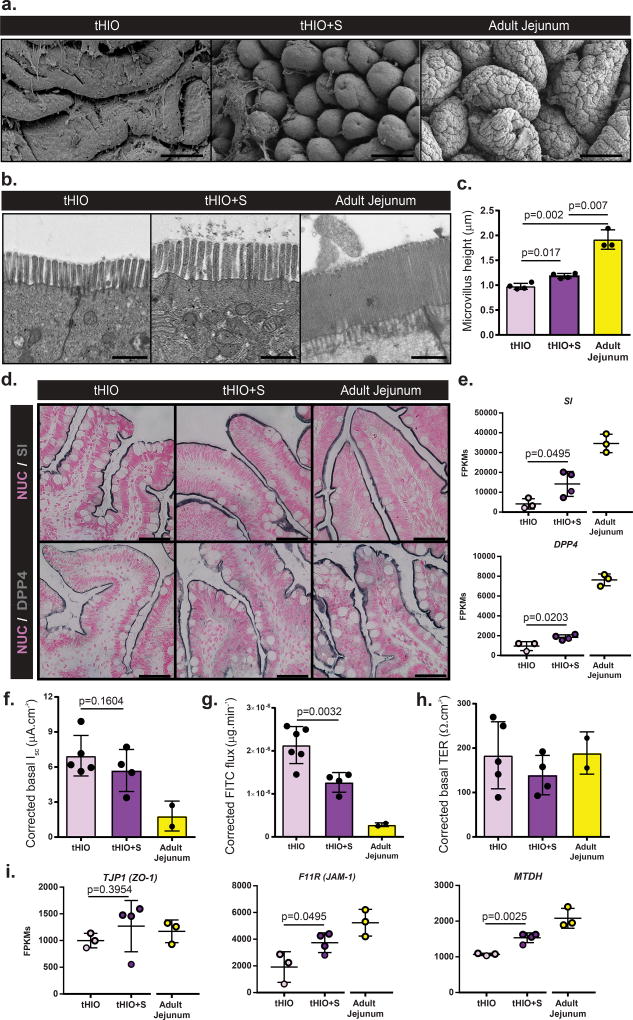
Figure 6. Epithelial integrity is retained and function improved in tHIO+S.
(a) Scanning electron micrographs of tHIO, tHIO+S, and adult jejunum epithelial surfaces. Scale bar = 100 µm. This experiment was repeated three times independently and findings were similar. (b) Transmission electron micrographs of tHIO, tHIO+S and adult jejunum microvilli. Scale bar = 1 µm. This experiment was repeated three times independently and findings were similar. (c) Quantification of microvilli in (b). While both are much lower than adult jejunum, microvilli in tHIO+S are longer than tHIO. Sample sizes are the following: tHIO n=4, tHIO+S n=4, and adult jejunum n=3. All samples are biologically independent. Data are represented as the mean ± SD. An ANOVA followed by Tukey’s post hoc tests were performed and the statistical significance cutoff was p < 0.05. (d) Sections with immunohistochemistry for Sucrase-Isomaltase (SI) and Dipeptidyl Peptidase 4 (DPP4) in in tHIO, tHIO+S and adult jejunum. All samples displayed positivity for the brush border markers. Scale = 25 µm. These experiments were repeated three times independently and findings were similar. (e) Normalized FPKMs were plotted for tHIO, tHIO+S and adult jejunum for SI and DPP4. Significant increases in transcripts were found in tHIO+S when compared to tHIO. (f) Corrected short circuit current of tHIO, tHIO+S and adult jejunum was plotted. A decreasing trend is observed, but changes are not statistically significant.. (g) Corrected calculated FITC dextran flux for tHIO, tHIO+S and adult jejunum was plotted. Flux was significantly decreased in tHIO+S compared to tHIO and trended toward the level of adult jejunum. (h) Corrected transepithelial resistance of tHIO, tHIO+S and adult jejunum was plotted. Observations across groups were similar. For (e–h) sample sizes are the following: tHIO n=5, tHIO+S n=4, and adult jejunum n=2. All samples are biologically independent. Data are represented as the mean ± SD. An ANOVA followed by Tukey’s post hoc tests were performed and the statistical significance cutoff was p < 0.05. (i) Normalized FPKMs were plotted for tHIO, tHIO+S and adult jejunum for tight junction components Tight Junction Protein 1 (TJP1), Junctional Adhesion Molecule 1 (F11R) and Metadherin (MTDH). For F11R and MTDH, the expression level in tHIO+S was significantly increased above that in the tHIO. Sample sizes are the following: tHIO n=4, tHIO+S n=4, and adult jejunum n=3. All samples are biologically independent. Data are represented as the mean ± SD. The unpaired student t test statistical significance cutoff was p < 0.05.
In addition to larger scale structural features, we also wanted to examine aspects related to epithelial function to see if strain had modulated them. On top of overall villus length, the brush border’s size impacts the available surface area for nutrient exchange within the gut22. Transmission electron micrographs depict the brush border in tHIO, tHIO+S and adult jejunum (Figure 6b). Individual microvilli were quantified and a significant increase was seen in their length in tHIO+S verses tHIO (Figure 6c). While an increase was observed, the microvilli in adult jejunum were much longer than those in tHIO+S (Figure 6c). When quantifying the density of microvilli per unit length, an increasing trend was observed; however differences were not significant between groups (Supplementary Figure 7a). When performing immunohistochemistry for the brush border enzymes Sucrase-isomaltase (SI) and Dipeptidyl Peptidase-4 (DPP4), a strong presence was observed across all sample types (Figure 6d). Transcriptionally, a significant increase in expression levels of SI and DPP4 was observed in tHIO+S compared to tHIO indicating the capacity for carbohydrate digestion of the tHIO may be increased with exposure to strain (Figure 6e).
An Ussing chamber was used to measure the epithelial characteristics of short circuit current (Isc), fluorescein isothiocyanate (FITC)-dextran flux and transepithelial resistance (TER) to observe any functional changes related to strain. All values were corrected using a correction factor calculated from morphometric observations (Supplementary Figure 7b). Due to the differences in villus height between sample types a correction factor was applied to further normalize the available surface area of the samples while in the Ussing chamber. High variability was observed in Isc measurements and there was not a significant difference between tHIO+S and tHIO (Figure 6f; uncorrected in Supplementary Figure 7c). However, a trend toward adult jejunum was observed. This measure indicated that the level of active ion transport may not be heavily influenced by strain23. FITC-dextran flux or permeability was also observed and found to be significantly decreased in tHIO+S compared to tHIO, again moving toward the levels observed in adult jejunum (Figure 6g; uncorrected in Supplementary Figure 7d). This indicates less paracellular permeability and increased barrier function. Levels of TER, an indicator of tight junction establishment23, displayed moderate variability and was similar between groups (Figure 6h; uncorrected in Supplementary Figure 7e). Transcripts for tight junction components Tight Junction Protein 1 (TJP1/ZO-1), F11 Receptor (F11R/JAM-1), and Metadherin (MTDH) were all measured (Figure 6i). Significant increases in transcripts for F11R and MTDH were observed in tHIO+S compared to tHIO, while there was no difference in TJP1 expression levels between groups. Together these data suggest a positive impact of strain on barrier function. While in some cases the effect of strain on the epithelium was slight, some improvements were seen and there was no evidence of epithelial disruption in association with strain.
Ex Vivo Muscle Function in Response to Strain in tHIO
We next tested ex vivo muscle properties to see if the increased muscle thickness further correlated to improved muscle function. In a previous study, the presence of intestinal cells of Cajal (ICCs) was confirmed in tHIOs5. Here, with Anoctamin1 (ANO1), a marker more specific for ICCs than CD117 which also stains mast cells, we observe their presence in all sample types24. In both tHIO and tHIO+S, ICCs were localized primarily within the smooth muscle layers, though not always at the interface of the longitudinal and circular muscle layers as was typically observed in adult jejunum (Figure 7a). Spontaneous isometric contractions related to ICC activity in tHIO and tHIO+S were recorded and generally found to have a higher amplitude and duration in tHIO+S compared to tHIO (Figure 7a, lower right). When observing isometric contractions in human jejunum this same association holds true, however due to the presence of the enteric nervous system they are not solely relatable to ICC activity. Thus it was not surprising that they were observed to have a higher amplitude and frequency in comparison to PSC generated samples (Figure 7a, lower right panel). A larger sampling of isometric force contractions were plotted to describe variability between biological repeats (Supplementary Figure 8a).
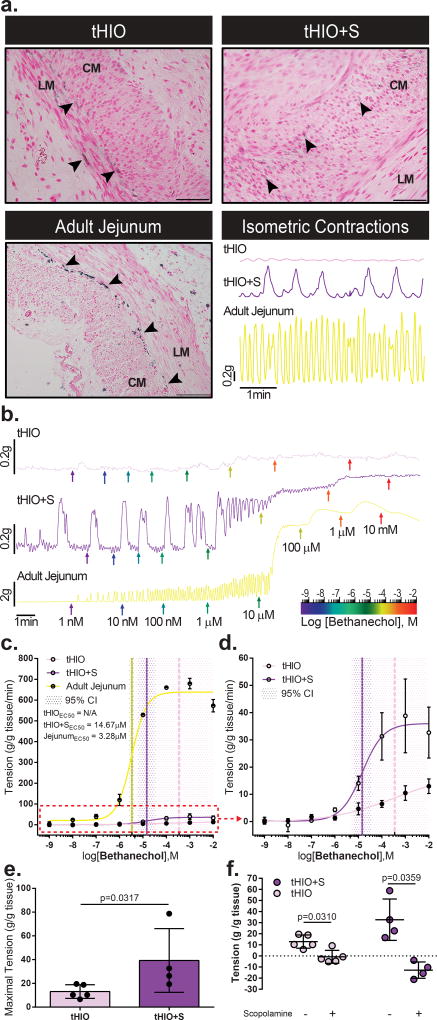
Figure 7. Muscle function is improved in tHIO+S.
(a) Immunohistochemistry for ICCs using Anoctamin 1 (ANO1) in tHIO, tHIO+S and adult jejunum. ICCs are observed in all sample types. Arrow heads indicate ANO1+ cells. LM and CM denote longitudinal and circular muscle layers respectively. The lower right panel depicts representative recordings of spontaneous (not stimulated) muscle contractions in tHIO, tHIO+S and adult jejunum after an equilibration period. Phasic contractions related to ICC presence are measurable in all tissue types. These experiments were repeated three times independently and findings were similar. (b) Plotted dose response curves of tHIO, tHIO+S and adult jejunum to bethanechol. Colored arrows indicate logarithmic dose administration. These experiments were repeated three times independently and findings were similar. (c) The effective concentration for half the maximal response (EC50) to bethanechol was calculated for tHIO, tHIO+S and adult jejunum. For the tHIO, the EC50 could not be accurately calculated with a 95% confidence interval. For tHIO+S and adult jejunum the EC50 dosages were found to be 14.67 µM and 3.28 µM respectively. Sample sizes are the following: tHIO n=5, tHIO+S n=4, and adult jejunum n=2. All samples are biologically independent. Data are represented as the mean ± SD. (d) Upon removal of the adult jejunum data in (c), the difference between the tHIO and tHIO+S EC50 curves is better observed. (e) The maximal tension normalized per tissue mass was plotted and observed to be significantly increased in tHIO+S compared to tHIO. Sample sizes are the following: tHIO n=5 and tHIO+S n=4. All samples are biologically independent. Data are represented as the mean ± SD. The Mann-Whitney statistical significance cutoff was p < 0.05. (f) Plotted tension before and after administration of scopolamine in tHIO and tHIO+S. Relaxation was successfully induced in both sample types. Sample sizes are the following: tHIO n=5 and tHIO+S n=4. All samples are biologically independent. Data are represented as the mean ± SD. The paired student t test statistical significance cutoff was p < 0.05.
After an equilibration period, muscle force contraction was assayed using a logarithmic series of bethanechol doses ranging in concentration from 1 nM to 10 mM. Representative tensile response recordings are plotted for tHIO, tHIO+S and adult jejunum (Figure 7b). From these data the effective concentration to achieve half of the maximal response (EC50) per tissue type was calculated (Figure 7c). To better visualize the differences between tHIO and tHIO+S the data was also plotted sans adult jejunum (Figure 7d). Due to such a low overall tensile response of tHIO to bethanechol, an EC50 with 95% confidence could not be determined. In tHIO+S, the response was more robust and an EC50 of 14.67 µM was calculated, which was much higher than that of adult human jejunum at 3.28 µM. Muscarinic receptor expression was confirmed transcriptionally in tHIO and tHIO+S, indicating that the decrease in reactivity was functional and not a reflection of poor receptor expression in PSC derived tissues (Supplementary Figure 8b). In calculating the maximal tension produced in tHIO and tHIO+S, we observed the tHIO+S to have a significantly higher tensile capability (Figure 7d). With the application of scopolamine, a nonspecific antimuscarinic, a significant amount of relaxation is observed in both tissues (Figure 7f). These values were also measured in adult jejunum, which had a much greater tensile capacity than the PSC derived samples. Interestingly, when assaying infant jejunum, the responses were within the same order of magnitude as PSC derived samples (Supplementary Figure 8c,d). This suggests that significant muscle development occurs postnatally and supports that the maturation status of the tHIO+S is most similar to infant jejunum.
DISCUSSION
We have combined the principles of embryonic intestinal development with the mechanics of development to successfully engineer human pluripotent stem cell-derived intestinal tissue with maturity and function, which exceeds that of those produced with a strictly biological approach. We have shown the application of strain induced gross, microscopic, and ultrastructural changes in the morphology of transplanted organoids making them more similar to native human samples. These structural changes were reflected transcriptionally, where a closer correlation to human tissue was observed with the application of strain. When testing function, we also saw a positive effect of strain in the tHIO. Increasing trends in TER coupled with increases in tight junction protein transcripts suggest that barrier function increased as a result of strain. We also observed an increase in overall muscle activity and tone indicating that strain promoted not only muscle growth, but strength as well. This dual approach to HIO generation seemed an improvement over the singular approach that is common in the field.
While the present study has not clearly elucidated the specific mechanism of mechanotransduction in the tHIO+S system that results in maturation, there is evidence that the TGFβ and MAPK pathways contribute as they were both enriched for transcriptionally. Protein analysis revealed an increase in TGFβ signaling activity as marked by an overall elevation in protein phosphorylation across the board, while the data was less convincing for MAPK signaling. However, with further investigation of pJUN, which is a transcription factor downstream in both MAPK and TGFβ signaling, expression was observed to be visually increased with strain by immunohistochemistry. Furthermore, pJUN has been described as one of the transcription factors controlling responses to mechanical strain in mechanotransduction pathways25, making these pathways worthy candidates for investigation. In subsequent studies we hope to better describe the mechanism by which the maturation observed is initiated, because it may also provide new developmental insights, where currently lacking.
Vertebrate gut development and organogenesis involves not only specification and growth, which have been well described, but also mechanical processes26–30. Briefly, the gastrointestinal tract and its adjacent organs initially develop from an endodermal sheet, which folds to form a midline tube along the anterior-posterior axis of the embryo which can be divided into the foregut, midgut and hindgut regions28. Following tube formation in humans is a significant elongation process which results in a hairpin fold, subsequent rotations and looping of the gut in the human embryo31. Evidence in animal models suggests that strain between the gut tube and mesentery, caused by differential growth rates, influences loop formation13. When this interaction is disrupted by detaching a portion of the mesentery from the gut tube looping fails to occur in that area of the gut13. Furthermore, the development and growth of the gut’s smooth muscle layers have been shown to influence the epithelial architecture within the gut tube. Prior to the presence of smooth muscle, the luminal surface of the gut is smooth. It is with the development of the underlying circular muscle layer that longitudinal ridges and buckling of the epithelium occurs9, 11. These ridges go on to form zigzags, in chick, which ultimately resolve into villi with further growth of the longitudinal muscle layer9, 11. The growth of these muscle layers provides a compressive force limiting any outward development driving buckling of the luminal surface11. These changes in geometry have been successfully modeled mathematically and found to hold true across several vertebrate species with slight variations in progression11.
Not only does the gut experience an array of mechanical cues and forces during embryonic and fetal development, but is also undergoing mechanical manipulations post-natally. On a daily basis during the digestive process, the gut is experiencing internal and external forces. Both segmentation and peristalsis result in physical forces. Segmentation, attributed to focused coordinated contractions of the surrounding circular muscle, results in compressive force32. Peristaltic contractions proximal to the food bolus coupled with distal relaxation are then responsible for its forward movement, of which generates an internal sheer force and radial pressure on the epithelium32, 33. Furthermore, it has been shown that an absence of mechano-luminal forces impact the gut homeostasis in humans34. It is logical to believe the mechanical forces of digestion generated by the smooth muscle in coordination with the enteric nervous system play a role in the homeostasis of gut architecture, though this interplay has not been well described34–36.
With the advent of microfluidic devices, like the various “on-a-chip” bioreactors, we are able to both model and modulate specific mechanical forces present in native tissues37, 38. This category of devices may serve as a basis for a more developmental approach in constructing what we might term as organogenesis chips, as they grow to include the biological, mechanical, and electrical facets required. Perhaps the well-described patterning protocols for deriving various organoids can be translated within a context of dynamic forces3, 39–41. We would speculate that as our mimicry of natural developmental processes improves, the tissues yielded will better approximate native ones.
These initial experiments support the importance of creating interdisciplinary marriages between the biological and mechanical developmental processes in tissue engineering. In order for significant advances to be made in the scaling of organoid structures into organs, we must work to better mimic the mechanics of development within their biological context. We feel that the present study provides a technology that can be translated to bigger animal models to engineer more mature, larger-scale human-intestinal-tissue segments, with the hopeful application of transplantation.
METHODS
Human Tissue
Human tissue collection was performed with the prior approval of Cincinnati Children’s Hospital Medical Center’s (CCHMC) Institutional Review Board (Tissue Characterization, Study No. 2014-0427). Surgical samples of pathologically normal adult human jejunum were obtained from patients undergoing bariatric procedures between the ages of 16 and 25 years old. Informed consent or assent was obtained from all patients and/or parent/legal guardians as appropriate. Pathologically normal infant jejunum surgical samples and/or paraffin sections of ages three months and younger were obtained as de-identified samples from the Cincinnati Biobank, part of Cincinnati Children’s Research Foundation as well as the Better Outcomes for Children Biorepository.
Animals
Adult immune-deficient NSG mice with ages between day of life 56 and 84 were used in all experiments (Comprehensive Mouse and Cancer Core Facility, Cincinnati, Ohio). Mice were housed in CCHMC’s pathogen-free animal vivarium and handled humanely in accordance with the NIH Guide for the Care and Use of Laboratory Animals. NSG mice were fed antibiotic chow (275 p.p.m. Sulfamethoxazole and 1,365 p.p.m. Trimethoprim; Test Diet). Both food and water were provided ad libitum before and after surgeries. All experiments were performed with the prior approval of CCHMC’s Institutional Animal Care and Use Committee (Signaling Pathways associated with Intestinal Stem Cell Expansion, Protocol No. 2016-0014).
Generation of Human Intestinal Organoids
HIOs were generated and maintained as previously described2–4. Briefly, line H1 embryonic stem cells (WiCell Research Institute, Inc.) were grown in feeder-free conditions in Matrigel (BD Biosciences) coated six-well Nunclon surface plates (Nunc) and maintained in mTESR1 media (Stem Cell Technologies). For induction of definitive endoderm (DE), cells were passaged with Accutase (Stem Cell Technologies) and plated at a density of 65,000 cells per well in 24-well Nunc plates. Cells were allowed to grow in mTESR1 media for two days before treatment with 100 ng/ml of Activin A for three days as previously described. DE was then treated with hindgut induction medium (RPMI 1640, 100x NEAA, 2% dFCS,) for four days with 100 ng/ml FGF4 (R&D) and 3 µM Chiron 99021 (Tocris) to induce formation of mid-hindgut spheroids. Spheroids were then plated in Growth Factor Reduced (GFR) Matrigel and maintained in intestinal growth medium (Advanced DMEM/F-12, N2 supplement, B27 supplement, 15 mM HEPES, 2 mM L-glutamine, penicillin-streptomycin) supplemented with 100 ng/ml EGF (R&D) to generate human intestinal organoids (HIOs). Media was changed twice weekly thereafter. HIOs were replated in fresh Matrigel every 14 days. HIOs were utilized for surgical transplantation between days 28 and 36.
Generation of Nitinol Springs
NiTi springs were formed as previously described20. Briefly, NiTi wires of diameter 0.152 mm were wrapped around a mandrel and heat set in order to impart the spring’s desired geometry (Nitinol Devices & Components). The resulting springs were then cut down to have a relaxed length of approximately 12 mm. Springs were compressed to half their relaxed length prior to implantation and placed within a gelatin capsule (Torpac, Inc.) which was subsequently double coated with C-A-P (Eastman Chemical Company) as previously described14.
Transplantation of Human Intestinal Organoids and Implantation of Springs
HIOs were prepared for transplantation as previously described4, 5, 42. Briefly, single matrigel embedded HIOs were transplanted into the mesentery of the mice at the most distal arcade before the ileocecal junction. Mice were anesthetized with 2% inhaled isoflurane (Butler Schein), and the abdomen shaved and prepped in sterile fashion using isopropyl alcohol and povidine-iodine. A 2 cm midline incision was made and approximately 4 cm of the intestines pulled out. A small pocket was created in the mesentery and the HIO placed within. The abdominal cavity was irrigated with normal saline with Zosyn (2 mg/ml; Pfizer Inc.) and the intestine placed back within the abdominal cavity. The abdominal wall muscles and skin were then closed in a double layer fashion and the mice were given a subcutaneous injection of Buprenex (0.05 mg/kg; Midwest Veterinary Supply) for pain management. Ten to twelve weeks following engraftment, the mice then underwent a secondary surgery with similar preparations. During this procedure, the tHIO was incised along a length of approximately 3 mm using Vannas scissors to gain access to the lumen. Directionality of the incision varied between tHIOs based on their shape, vascularization and proximity to the mouse’s bowel. Any mucous plugs were manually removed. Then, using an 18 G blunt tip fill needle positioned at the incision of the lumen, the tHIO was irrigated with normal saline. Care was taken during this process to avoid damaging the epithelium. With the luminal space cleared, the gelatin capsule was placed within the lumen of the tHIO to allow for spring deployment parallel to the adjacent mouse intestine. This was done to reduce the risk of obstruction. The tHIO was then closed using a 9–0 silk suture in a simple interrupted fashion. Sham operated mice underwent the same aforementioned procedure except that the C-A-P coated, gelatin capsule was empty, without spring. Mice were sacrificed and tissue harvested 14d postoperatively.
MicroCT Imaging System
An IVIS Spectrum (PerkinElmer) coupled to a computer with Living Image software (Caliper Life Sciences) was used in the acquisition of microCT images. Briefly, live mice were anesthetized with 2% inhaled isoflurane prior to and during imaging which was used to monitor post-operative spring deployment.
Tissue Processing, Immunohistochemistry, and Light Microscopy
Segments of human jejunum, tHIO and tHIO+S grafts were fixed overnight in 4% paraformaldehyde (PFA), processed and embedded in paraffin. Sections of 5µm thickness were deparaffinized and either stained immediately with a histological kit (Pentachrome, Newcomer Supply) or subjected to antigen retrieval, and stained. For both primary and secondary antibodies, incubations took place at 4°C overnight in 1% bovine serum albumin in phosphate buffered saline (PBS). Antibodies, their respective dilutions are listed in Table S1. The Vectastain ABC system (Vector Laboratories) was used for amplification. The following kits were used for signal detection: diaminobenzidine substrate kit, Immpact SG substrate kit, and Vector Red substrate kit (Vector Laboratories). Lillie-Mayer’s Hematoxylin (Dako North America, Inc.) or Nuclear Fast Red (PolySciences, Inc.) was used as a counterstain. Images were acquired using a Nikon Eclipse Ti microscope and analyzed using Nikon Elements Imaging Software (Nikon).
Electron Microscopy
Scanning Electron Microscopy
Segments of human jejunum, tHIO and tHIO+S grafts were fixed overnight in 3% gluteraldehyde in 0.175 M sodium cacodylate buffer with pH 7.4. Samples were then buffer rinsed and post fixed in 1% osmium tetroxide in 0.175 M cacodylate buffer for 1 hour at 4°C. After another buffer rinse, samples were put through a graded ethanol series (25, 50, 75, 95, 3x 100%) for dehydration. Specimens were then critical point dried in a Leica EM CPD300, stub mounted and sputter coated 10 nm thick with 60/40 gold palladium using a Leica EM ACE600. A Hitachi SU8010 transmission electron microscope was used to image samples.
Transmission Electron Microscopy
Segments of human jejunum, tHIO and tHIO+S grafts were fixed and dehydrated in the same fashion as those for scanning electron microscopy. Infiltration was performed with 2X propylene oxide followed by a graded infiltration with LX-112. Samples were polymerized into blocks overnight at 37°C and subsequently kept at 60°C for 3 days. Thicks were sectioned and stained with Toluidine Blue to verify tissue orientation. Blocks were then sectioned at 10 nm, skipping 10 µm between grids. Sections were stained with urinyl acetate and lead citrate before imaging with a Hitachi H7600 transmission electron microscope.
Morphometric Analysis, Cell Type Quantification
Morphometric analysis was performed on hematoxylin and eosin stained tissue sections. Crypt depth, crypt width, villus height, villus width, and mucosal thickness were measured for a minimum of 20 well-oriented crypt-villus units per tissue sample and then averaged using Nikon NIS imaging software (Nikon). Microvilli were measured using ImageJ; at least 150 microvilli per sample across three different grids of at least 10 µm apart were quantified. Profiles of proliferation by position within the crypt were determined using dual MKI67 and CDH1 immunostaining. A minimum of 10 intact crypts were analyzed per sample and averaged. For quantification of cell types along the crypt-villus axis the following were utilized: CHGA immunostaining for quantification of endoendocrine cells, Alcian Blue pH 2.5 staining for goblet cells, LYZ for Paneth cells. For semi-quantitative analysis of the stem cell compartment OLFM4 immunostaining was used. In each case a minimum of 20 well-oriented crypt-villus units per tissue sample were quantified and averaged.
Statistics and Reproducibility
For analysis of proliferative profiles, Gaussian curves were fit to each data set and a comparison of fits was performed using Prism software (GraphPad). For comparisons between three or more groups, two-way ANOVAs and post-hoc Tukey’s tests were completed using Prism software as appropriate. For statistics comparing two groups, like the case of normalized FPKMs, either a student t test or Mann-Whitney test was used according to data distribution. The statistical significance cutoff was p < 0.05 and confidence 95%.
Ex vivo Epithelial Permeability
The epitheliums of freshly harvested tHIO, tHIO+S grafts and human surgical samples were carefully dissected through a technique similar to seromuscular stripping23, 43. tHIOs and fresh surgical samples were opened longitudinally. All further dissection was done in ice cold Kreb’s buffer (NaCl, 117 mM; KCl, 4.7 mM; MgCl2, 1.2mM; NaH2PO4, 1.2 mM; NaHCO3, 25 mM; CaCl2, 2.5 mM and glucose, 11 mM). Tissue segments were then pinned in a dish containing 0.5 cm thick cured Sylgard (Electron Microscopy Sciences). Unlike mouse tissue, the seromusculature layers (serosa, longitudinal, and circular smooth muscle) are relatively thick in both tHIOs and human jejunum. These layers are then micro-dissected as one unit from the epithelium using Dumont #5 and #7 forceps along with Vannas scissors taking care to only handle the edges of the tissue (Fine Science Tools, Inc.). The layers are gently separated and cut in small increments. After separation, gross tissue integrity was assessed using the stereoscope’s bottom lighting for evenness and uniformity in appearance. The edges of the tissue, experiencing the majority of manipulation, were discarded along with any portions of the epithelium that appeared damaged. The dissection set up can be found in Supplemental Method 1. Two examples of tissues dissected in this fashion can be found in Supplemental Method 1 after completion of the Ussing assay. Some remnant subepithelial mucosa remained after dissection. Epithelial segments were mounted between the hemi-chambers of an Ussing apparatus (Physiologic Instruments), and 0.008 cm2 of tissue was exposed to 3 mL of oxygenated Krebs buffer at 37 °C. The transepithelial potential difference was detected with two paired electrodes containing 4% agar in 3 M KCl. The electrodes were connected to a VVC MC8 voltage clamp amplifier (Physiologic Instruments, San Diego). Electrode potential difference and fluid resistance values were offset before tissue segments were mounted in the chamber. A 30 min monitoring period was allowed for the establishment of equilibrium in the chamber. Then, tissues were voltage-clamped at 0 mV while continuously measuring the short circuit current (Isc). For FITC-dextran permeability, 2.2 mg/ml FITC-dextran was added into apical side, and a sample was taken from the basolateral side every 30 minutes for 3 hours, replacing the same amount of fresh modified Kreb’s buffer in the basolateral side. Finally, samples were read with a plate-reader (Synergy 2, BioTek).
Ex vivo Muscle Strength
tHIO and tHIO+S grafts were harvested and placed in ice-cold Hank’s Balanced Salt Solution (HBSS). Human surgical samples were maintained overnight at 4°C in HBSS prior to assaying in order to minimize the effects of anesthetics. Muscle strips (4–6 mm in length and 1–2 mm in width) were dissected from the samples. Strips were then suspended vertically in an organ bath chamber (Radnoti) filled with freshly prepared Krebs-Ringer solution (Sigma; supplemented with 2.5 mM CaCl2 and 15 mM NaHCO3; pH 7.4), warmed to 37°C and gassed with 95% O2 + 5% CO2. Segments were allowed to equilibrate for an hour at an initial tension of 0.5 g. The contractile response of the muscle was continuously recorded, using 4-chamber tissue-organ bath chambers with isometric force transducers (0–25g; AD Instruments) coupled to a multi-channel bridge amplifiers and data recorder (AD Instruments; PowerLab 4/35) linked to a computer equipped with LabChart Pro software (AD Instruments). A logarithmic dose response to Carbamyl-β-methylcholine chloride (Bethanechol; Sigma-Aldrich) was obtained through the administration of exponential doses with concentrations of 1 nM to 10 mM at 2 minute intervals before the administration of 10 µM scopolamine (Tocris Bioscience).
RNA Isolation and Sequencing
RNA was extracted using an RNeasy Plus Micro Kit (Qiagen) according to manufacturer guidelines. Samples were then quantified and submitted to CCHMC’s DNA Sequencing and Genotyping core for Next Generation Sequencing. All samples were assayed to have RNA integrity numbers greater than eight. After quality control, a cDNA library was created and sequenced using an IlluminaHiSeq2000 (Illumina) with 20 million paired-end reads per sample.
RNA sequencing bioinformatics
Pre-processing
Data pre-processing was performed in Galaxy public server (https://usegalaxy.org). fastq datasets were encoded in Phred+33 to fastqsanger under the attributes for each dataset (FASTQ Groomer, Galaxy v1.0.4). Read quality was assessed using FastQC (Galaxy, v0.67). Adapter and low quality (< 20) sequences were removed using Trimmomatic (SLIDINGWINDOW) (Galaxy v0.32.3). Reads were mapped against the human genome GRCh37\Hg19 using Tophat2/Bowtie2 with default settings (Galaxy v2.1.0). The number of reads uniquely mapping to each gene was counted using feature Counts (Galaxy v1.4.6.p5) using the Illumina iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn) annotation file. Alternatively, each sample was independently processed with Cufflinks44 in order to generate an initial transcriptome. We used the Cuffmerge tool to merge the private transcriptomes into a single reference, and at the same time annotated known genes and extended partial transcripts45. This common transcriptome was used in a second pass with Cufflinks to quantify each transcript and gene (known or new) in each sample46. The reference annotation used was based on the UCSC knownGenes table47.
Analysis of RNA-seq data
RNA-seq analysis was performed using Strand NGS 2.9 software (Strand Life Sciences). Count tables generated in Galaxy were processed and normalized using the DESeq2 package48 within the Strand NGS 2.9 R console. We performed a PCA using Rstudio software v1.0.14. From the PCA, the gene with the highest loadings for each principal component were extracted and plotted using the standalone hi_loadings function in pcaExplorer package. A functional enrichment analysis using the limmaquickpca2go routine provided by the limma package was performed on the 10000 genes with the highest loadings for each principal component. ANOVA and moderated t-Test were used to performed statistical analysis amongst samples and between groups. Functional enrichment analysis was performed in the ToppGene suite (https://toppgene.cchmc.org). Plots were generated using ggplot2 v2.2.1 and GOplot v1.02 in Rstudio v1.0.14.
Analysis of RNA-seq data with publicly available datasets
Publicly available datasets of fetal and adult full thickness intestinal tissues were downloaded from EBI-AE and NCBI-GEO databases using the Galaxy public server (Table 2). Datasets were selected upon RNA extraction and library preparation methods and sequencing technology (Illumina HiSeq). Only forward reads were used from paired-end files. Datasets were encoded to fastqsanger and read quality was assessed using FastQC (Galaxy, v0.67). Adapter and low quality (< 20) sequences were removed using Trimmomatic (SLIDINGWINDOW). Reads were mapped against the human genome GRChb37\Hg19 using HISAT2 with default settings (Galaxy v2.0.5.1)49. Count tables were generated in Galaxy using feature Counts using the Illumina iGenomes annotation file. The count matrix was processed in R 3.4.1 using the functions exported by the pcaExplorer package50 for a standalone usage. Briefly, the count matrix was transformed and normalized using a variance stabilizing transformation (VST) to the count data. We visualized the sample PCA using the pcaplot function. The genes with the highest variance were selected to compute the PCA i.e. from 500 to 10000 genes. From the PCA, the gene with the highest loadings for each principal component were extracted and plotted using the hi_loadings function. A functional enrichment analysis using the limmaquickpca2go routine provided by the limma package was performed on the 10000 genes with the highest loadings for each principal component.
The detailed analysis and R scripts are provided in the following Github repository (www.github.com\maximemahe\Poling2017). The accession number for data generated for this paper is ArrayExpress: E-MTAB-6017 and includes the transplanted HIO, with or without lengthening device, the human adult and infant data shown in Figure 3 and supplementary Figure 2. The additional RNA-seq datasets including HIOs derived from H1 and H9 ES cell lines, transplanted HIOs, Fetal, newborn and adult small intestine were downloaded from public databases (Table 2).
Protein Phosphorylation Arrays
C-Series phosphorylation arrays were performed according to manufacturer recommendations (RayBiotech, Inc). Briefly, protein was extracted from flash frozen full thickness tHIO and tHIO+S tissues, quantified, and normalized between samples. Protein was incubated on antibody array nitrocellulose membranes, followed by horseradish peroxidase based amplification and detection. Chemiluminescent readings were taken using a ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc.) and densitometry data extracted using ImageJ software. Readings were normalized to the positive loading controls and membrane background signal subtracted.
Data Availability
The detailed transcriptome analysis and R scripts are provided in the following Github repository (www.github.com\maximemahe\Poling2017). The accession number for data generated for this paper is ArrayExpress: E-MTAB-6017. The authors declare that all remaining data supporting the findings of this study are available within the paper and its supplementary information.
Supplementary Material
Acknowledgments
The authors thank Gary L. Keller, DVM and Veterinary Services staff for their support in completing animal work, J. Matthew Kofron, PhD for his kind assistance with IVIS Spectrum microCT imaging, and Bryan Donnelly for assistance with protein work. This work was funded in part by NIH grants P30 DK078392 (DHC Pilot and Feasability Award to M.M.M.; DNA sequencing and iPSCs core facilities) and 1K99DK110414-02 (M.M.M.); an AGA – Athena Blackburn Research Scholar Award in neuroenteric diseases from the American Gastroenterology Association (M.M.M.) and a “New Team” grant (BOGUS to M.M.M.) from the Bioregate Regenerative Medicine Cluster, University of Nantes and Pays de la Loire Region. The Dunn, Helmrath and Wells laboratories are members of the Intestinal Stem Cell Consortium, supported by NIDDK and NIAID. This research was also supported in part by the Cincinnati Children’s Research Foundation and the Cincinnati Biobank, as well as the Better Outcomes for Children Biorepository.
Footnotes
Author Contributions
H.M.P. and M.M.M. conceived and designed the study; H.M.P., D.W., T.A.H., M.A.H., and M.M.M. performed experiments and collected data; N.B. generated HIOs; N. H. and J.C.Y.D. manufactured springs; H.M.P., D.W., S.C., and M.M.M. analyzed data; H.M.P., M.B., S.P.H., J.M.W., M.A.H., and M.M.M. interpreted experimental findings; H.M.P. and M.M.M. prepared figures and drafted manuscript; H.M.P., S.P.H., J.M.W., M.A.H., and M.M.M. made critical revisions to the manuscript; all authors approved the manuscript’s final version.
Competing Interests
The authors do not have any competing or financial interests.
References
- 1.Wells JM, Spence JR. How to make an intestine. Development. 2014;141:752–760. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCracken KW, Howell JC, Wells JM, Spence JR. Generating human intestinal tissue from pluripotent stem cells in vitro. Nature protocols. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nature medicine. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Workman MJ, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nature medicine. 2017;23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkbeiner SR, et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem cell reports. 2015 doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology. 2016;150:1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, et al. The Contributions of Human Mini-Intestines to the Study of Intestinal Physiology and Pathophysiology. Annu Rev Physiol. 2017;79:291–312. doi: 10.1146/annurev-physiol-021115-105211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson CM. On Buckling Morphogenesis. J Biomech Eng. 2016;138:021005. doi: 10.1115/1.4032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurpios NA, et al. The direction of gut looping is established by changes in the extracellular matrix and in cell:cell adhesion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8499–8506. doi: 10.1073/pnas.0803578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shyer AE, et al. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161:569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savin T, et al. On the growth and form of the gut. Nature. 2011;476:57–62. doi: 10.1038/nature10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark R, Panduranga M, Carman G, Dunn JC. Development of an endoluminal intestinal lengthening capsule. Journal of pediatric surgery. 2012;47:136–141. doi: 10.1016/j.jpedsurg.2011.10.031. [DOI] [PubMed] [Google Scholar]
- 15.Rouch JD, et al. Scalability of an endoluminal spring for distraction enterogenesis. Journal of pediatric surgery. 2016;51:1988–1992. doi: 10.1016/j.jpedsurg.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Demehri FR, et al. Development of an endoluminal intestinal attachment for a clinically applicable distraction enterogenesis device. Journal of pediatric surgery. 2016;51:101–106. doi: 10.1016/j.jpedsurg.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demehri FR, Freeman JJ, Fukatsu Y, Luntz J, Teitelbaum DH. Development of an endoluminal intestinal lengthening device using a geometric intestinal attachment approach. Surgery. 2015;158:802–811. doi: 10.1016/j.surg.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luntz J, Brei D, Teitelbaum D, Spencer A. Mechanical Extension Implants for Short-Bowel Syndrome. Proc SPIE Int Soc Opt Eng. 2006;6173:617309. doi: 10.1117/12.659112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allard J, et al. Immunohistochemical toolkit for tracking and quantifying xenotransplanted human stem cells. Regenerative medicine. 2014;9:437–452. doi: 10.2217/rme.14.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shekherdimian S, Panduranga MK, Carman GP, Dunn JC. The feasibility of using an endoluminal device for intestinal lengthening. Journal of pediatric surgery. 2010;45:1575–1580. doi: 10.1016/j.jpedsurg.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Marsh MN, Swift JA. A study of the small intestinal mucosa using the scanning electron microscope. Gut. 1969;10:940–949. doi: 10.1136/gut.10.11.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooton D, Lentle R, Monro J, Wickham M, Simpson R. The Secretion and Action of Brush Border Enzymes in the Mammalian Small Intestine. Rev Physiol Biochem Pharmacol. 2015;168:59–118. doi: 10.1007/112_2015_24. [DOI] [PubMed] [Google Scholar]
- 23.Clarke LL. A guide to Ussing chamber studies of mouse intestine. American journal of physiology. Gastrointestinal and liver physiology. 2009;296:G1151–1166. doi: 10.1152/ajpgi.90649.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Pinilla PJ, et al. Ano1 is a selective marker of interstitial cells of Cajal in the human and mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1370–1381. doi: 10.1152/ajpgi.00074.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mammoto A, Mammoto T, Ingber DE. Mechanosensitive mechanisms in transcriptional regulation. Journal of cell science. 2012;125:3061–3073. doi: 10.1242/jcs.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burn SF, Hill RE. Left-right asymmetry in gut development: what happens next? Bioessays. 2009;31:1026–1037. doi: 10.1002/bies.200900056. [DOI] [PubMed] [Google Scholar]
- 27.Sherwood RI, Chen TY, Melton DA. Transcriptional dynamics of endodermal organ formation. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spence JR, Lauf R, Shroyer NF. Vertebrate intestinal endoderm development. Developmental dynamics : an official publication of the American Association of Anatomists. 2011;240:501–520. doi: 10.1002/dvdy.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shahbazi MN, et al. Self-organization of the human embryo in the absence of maternal tissues. Nature cell biology. 2016;18:700–708. doi: 10.1038/ncb3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soffers JHM, Hikspoors JPJM, Mekonen HK, Koehler SE, Lamers WH. The growth pattern of the human intestine and its mesentery. Bmc Dev Biol. 2015;15 doi: 10.1186/s12861-015-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran K, Brun R, Kuo B. Evaluation of regional and whole gut motility using the wireless motility capsule: relevance in clinical practice. Therap Adv Gastroenterol. 2012;5:249–260. doi: 10.1177/1756283X12437874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engmann J, Burbidge AS. Fluid mechanics of eating, swallowing and digestion - overview and perspectives. Food & function. 2013;4:443–447. doi: 10.1039/c2fo30184a. [DOI] [PubMed] [Google Scholar]
- 34.Wieck MM, et al. Prolonged Absence of Mechanoluminal Stimulation in Human Intestine Alters the Transcriptome and Intestinal Stem Cell Niche. Cell Mol Gastroenterol Hepatol. 2017;3:367–388. e361. doi: 10.1016/j.jcmgh.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terry BS, Lyle AB, Schoen JA, Rentschler ME. Preliminary mechanical characterization of the small bowel for in vivo robotic mobility. J Biomech Eng. 2011;133:091010. doi: 10.1115/1.4005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregersen H, Kassab GS, Fung YC. The zero-stress state of the gastrointestinal tract: biomechanical and functional implications. Digestive diseases and sciences. 2000;45:2271–2281. doi: 10.1023/a:1005649520386. [DOI] [PubMed] [Google Scholar]
- 37.Low LA, Tagle DA. Organs-on-chips: Progress, challenges, and future directions. Exp Biol Med (Maywood) 2017 doi: 10.1177/1535370217700523. 1535370217700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christoffersson J, van Noort DV, Mandenius CF. Developing organ-on-a-chip concepts using bio-mechatronic design methodology. Biofabrication. 2017 doi: 10.1088/1758-5090/aa71ca. [DOI] [PubMed] [Google Scholar]
- 39.Sato T, Clevers H. SnapShot: Growing Organoids from Stem Cells. Cell. 2015;161 doi: 10.1016/j.cell.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 40.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516 doi: 10.1038/nature13863. 400-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grapin-Botton A. Three-dimensional pancreas organogenesis models. Diabetes Obes Metab. 2016;18:33–40. doi: 10.1111/dom.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahe MM, Brown NE, Poling HM, Helmrath MA. In Vivo Model of Small Intestine. Methods Mol Biol. 2017;1597:229–245. doi: 10.1007/978-1-4939-6949-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giles DA, et al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nature medicine. 2017;23:829–838. doi: 10.1038/nm.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 46.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marini F. pcaExplorer: Interactive Visualization of RNA-seq Data Using a Principal Components Approach. R package version 2.3.0. 2017 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The detailed transcriptome analysis and R scripts are provided in the following Github repository (www.github.com\maximemahe\Poling2017). The accession number for data generated for this paper is ArrayExpress: E-MTAB-6017. The authors declare that all remaining data supporting the findings of this study are available within the paper and its supplementary information.