Abstract
Background:
Aging is related to an increased risk of morbidity and mortality and is affected by environmental factors. Exposure to polycyclic aromatic hydrocarbons (PAHs) is associated with adverse health outcomes; but the association of such exposure with DNA methylation aging, a novel aging marker, is unclear.
Objectives:
Our aim was to investigate the association of PAH exposure with methylation aging.
Methods:
We trained and validated a methylation age predictor suitable for Chinese populations using whole blood methylation data in 989 Chinese and 160 Caucasians. We defined two aging indicators: , as methylation age minus chronological age; and aging rate, the ratio of methylation to chronological age. The association of PAH exposure with aging indicators was evaluated using linear regressions in three panels of healthy Chinese participants (, among the aforementioned 989 Chinese participants) whose exposure levels were assessed by 10 urinary monohydroxy-PAH metabolites.
Results:
We developed a methylation age predictor providing accurate predictions in both Chinese individuals and Caucasian persons (, ). Among the 10 urinary metabolites that we measured, 1-hydroxypyrene and 9-hydroxyphenanthrene were associated with methylation aging independently of other OH-PAHs and risk factors; 1-unit increase in 1-hydroxypyrene was associated with a 0.53-y increase in [95% confidence interval (CI): 0.18, 0.88; false discovery rate (FDR) ] and 1.17% increase in aging rate (95% CI: 0.36, 1.98; ), whereas for 9-hydroxyphenanthrene, the increase was 0.54-y for (95% CI: 0.17, 0.91; ), and 1.15% for aging rate (95% CI: 0.31, 1.99; ). The association direction was consistent across the three Chinese panels with the association magnitude correlating with the panels’ exposure levels; the association was validated by methylation data of purified leukocytes. Several cytosine-phosphoguanines, including those located on FHL2 and ELOVL2, were found associated with both aging indicators and monohydroxy-PAH levels.
Conclusions:
We developed a methylation age predictor specific for Chinese populations but also accurate for Caucasian populations. Our findings suggest that exposure to PAHs may be associated with an adverse impact on human aging and epigenetic alterations in Chinese populations. https://doi.org/10.1289/EHP2773
Introduction
Aging is related to a progressive decline of physical, psychological, and social functions and a great risk of many disabilities and diseases. Human bodies age at different rates. Both genetic and environmental factors are among the determinants of aging (de Magalhães et al. 2012). In recent years, a growing body of evidence suggested that the state of DNA methylation, an epigenetic modification that plays crucial roles in gene regulation and genomic stability (Jones 2012), was closely related to age over long time periods (Christensen et al. 2009; Fraga and Esteller 2007; Hannum et al. 2013; Horvath 2013; Oberdoerffer and Sinclair 2007; Richardson 2003), and was associated with a range of age-related diseases, including neurodegenerative diseases and cancers (De Jager et al. 2014; Esteller 2008). Using genome-wide technologies, pioneering studies based on Caucasian/Hispanic populations found that DNA methylation at certain age-related genes could capture aspects of biological age (i.e., the methylation age) (Hannum et al. 2013; Horvath et al. 2012; Horvath 2013) and serve to detect human aging status under different health conditions (Hannum et al. 2013; Horvath 2013). Recent studies have reported significant methylation age accelerations in patients with cancers or Down syndrome (Hannum et al. 2013; Horvath 2013; Horvath et al. 2015). Methylation age of liver was associated with body-mass index (BMI) (Horvath et al. 2014), and methylation age of whole blood was associated with the performance of physical and cognitive fitness and all-cause mortality in elderly populations (Marioni et al. 2015a, 2015b). These novel findings highlighted the importance of DNA methylation in longevity and the potential usage of epigenetic age in evaluation of the biological “clock.”
Because DNA methylation could be altered by genetic and environmental factors and can integrate the impact of both factors on a phenotype or disease (Schadt 2009), DNA methylation age may likewise reflect the environmentally induced effects on aging.
Polycyclic aromatic hydrocarbons (PAHs) are a group of ubiquitous pollutants that are generated during the incomplete combustion or pyrolysis of organic matter (Samanta et al. 2002). PAH exposure regularly occurs for humans – from ambient air (e.g., municipal and industrial waste incineration, coal/wood/waste burning, or vehicle exhausts) and indoor air (e.g., smoking or cooking) to foods and water (e.g., grills/barbecues or some crops, contaminated water) (WHO 2010). PAHs have various toxic effects on multiple organs and have been related to the risk of cancers and cardiovascular diseases (Burstyn et al. 2005; Kim et al. 2013). Prior studies have observed associations between such exposure with aging indicators such as telomere length (Pavanello et al. 2010) and skin aging scores (Vierkötter et al. 2010), and with global or gene-specific DNA methylation alterations (Herbstman et al. 2012; Tang et al. 2012). However, the relation between PAH exposure and DNA methylation aging is still unclear.
In the present study, we hypothesized that exposure to higher levels of PAHs may be associated with increased age accelerations. We first built a methylation age predictor that performed accurately for Chinese populations using genome-wide methylome data and defined two indicators of methylation aging: (suggesting the extra years a person aged) and aging rate (suggesting the speed of the aging process). Then, using 10 urinary monohydroxy-PAH (OH-PAH) metabolites as internal exposure biomarkers accessing total PAH exposure from all sources, we quantitatively assessed the association between exposure to PAHs and DNA methylation aging (Figure S1).
Methods
Study Population
Our study was primarily conducted in three panels of Chinese populations with a total of 539 healthy participants. Detailed information of these participants has been described in a previous study (Zhu et al. 2016) and is briefly mentioned below.
The coke-oven workers (COW) panel.
A Coke Oven Cohort was established in 2010 in a coke-oven plant in Wuhan, China (Li et al. 2012). A total of 1,628 coke-oven workers who have worked in this plant at the top, side, bottom, or adjunct of coke ovens, or in the management offices for more than 1 y, and who signed the participation agreement, were enrolled at study baseline. We randomly selected 144 workers for the genotyping and methylation assays among those participants who met the following criteria: (1) donated both blood and urine samples at baseline; (2) had a BMI between 18.0 and 30.5; (3) worked in the same plant for ; (4) did not report any disease at baseline; (5) did not report having fever or infectious disease in the 2 weeks before the time of blood collection, and did not take any medication in the 4 weeks before the time of blood collection; and (6) had baseline total urinary OH-PAH () levels in the highest tertile. After the quality controls (QC) of methylation and genotyping data, 137 workers who passed QC were included in the final analyses.
The Wuhan–Zhuhai (WHZH) panel.
Established in 2011, the WHZH cohort recruited 3,053 residents from two urban communities in Wuhan, China, and 1,759 residents from two urban communities in Zhuhai, China (Song et al. 2014). The four communities were sampled using a stratified, cluster sampling approach; all residents age 18–80 y and without severe illnesses had lived in the same communities for more than 5 y and agreed to participate were enrolled (Song et al. 2014). Among the cohort participants who (1) donated both blood and urine samples at baseline; (2) neither reported any disease at baseline interview, nor showed any abnormalities (not including limb/visual/hearing disability due to previous work injuries) at baseline examinations; (3) did not report having fever or infectious disease in 2 weeks before the time of blood collection, and did not take any medication in 4 weeks before the time of blood collection, 180 Wuhan residents were selected as matched controls for a case–control study of cardiovascular disease (, matching with the cases by age, sex, and BMI) (Li et al. 2017) and/or matched controls for the aforementioned COWs (selected from individuals whose urinary in the lowest tertile, and matching with the COWs by age, sex, and BMI; ; the cardiovascular study and COW study shared 64 controls) (Zhu et al. 2016). Another 103 Zhuhai residents were also selected as matched controls for the aforementioned cardiovascular case–control study, matching with the cases by age, sex, and BMI (Li et al. 2017). Among the selected 180 Wuhan residents and 103 Zhuhai residents, 162 Wuhan and 99 Zhuhai samples passed QC for both genotyping and methylation assays and were initially included in our methylation age prediction. Three Wuhan residents were later found to be extreme outliers of and aging rate and were excluded from the study (Figure S2), leaving 258 subjects remaining in final analyses.
The Shiyan (SY) panel.
To investigate the effects of genetic, epigenetic, and environmental factors on gene expression, we recruited 144 individuals who participated in the annual physical examinations at the Health Examination Center of Dongfeng Central Hospital (Dongfeng Motor Corporation and Hubei University of Medicine) in Shiyan, Hubei, China, from April to May 2015. Subjects were enrolled if they (1) aged 20–70 y; (2) agreed to participate; (3) donated both blood and urine samples; (4) reported no chronic diseases, and (5) reported no fever/infectious diseases in 2 weeks before the time of blood collection and no medication prescribed in 4 weeks before the time of blood collection. We applied the genome-wide genotyping, methylation, and expression assays on the 144 participants; all subjects passed QC of these data and were included in the final analyses.
In addition, participants in the above three panels were asked to eat a bland diet and then fast for at least 12 h before sample collection. All biological samples were collected, processed, and stored following the same protocols under similar conditions in the same lab.
The methylation dataset with sorted leukocyte subsets.
We enrolled eight healthy men who attended the annual medical examinations at Tongji Medical College (Wuhan, China) in February 2015, aiming to assess the cellular heterogeneity of DNA methylation in purified leukocyte subsets. The recruitment criteria were the same as the SY panel, with an addition of willingness to donate blood. Protocols have been described previously (Li et al. 2017).
External validation sets for the age prediction model.
To seek a separated validation of our age prediction model, we acquired two additional methylation datasets, the Twins Methylation Study (TwinS) with 450 Chinese adults (i.e., 225 pairs of twins) from The Chinese National Twin Registry (Wang et al. 2015), and an asthmatic family panel from the Saguenay-Lac-Saint-Jean region in Québec (SLSJ) with 160 Canadian children and adults (Liang et al. 2015). Detailed information has been described in prior publications (Laprise 2014; Liang et al. 2015; Wang et al. 2015).
Our study was approved by the Institutional Review Boards (respectively the Ethics Committee of Tongji Medical College, the Biomedical Ethics Committee of Peking University, and the Ethics Committee of the Centre de santé et de services sociaux de Saguenay). Informed written consent was obtained from each participant prior to the study.
Laboratory Assays
Genome-wide DNA methylation and gene expression assays.
Genome-wide methylation assays for the WHZH, SY, COW, and TwinS panels were conducted in the same lab and has been described previously (Li et al. 2017; Wang et al. 2015; Zhu et al. 2016). Briefly, DNA was extracted from whole blood using standard procedures. Bisulfite conversion was performed using Zymo EZ DNA Methylation kit (Zymo Research). In each original study, we randomized all samples/pairs/blocks (case–control pairs, different samples from the same subjects) across plates and beadchips. DNA methylation levels at cytosine-phosphoguanine (CpG) sites were quantified using Infinium HumanMethylation450 BeadChip (Illumina) as per instructions from Illumina. IDAT files generated from iScan (Illumina) system were processed in to R using the minfi package (Aryee et al. 2014). CpG probes were excluded if they: (1) assay single nucleotide polymorphisms (SNPs) rather than CpGs; (2) had a missing rate (for a probe of a certain sample, missing was defined when detection or bead counts ); or (3) potentially contained or extended on SNPs with minor allele frequency (MAF) in the 1000 Genomes Project 20110521 release for ASN population, or possibly cross-hybridized to other genomic locations (41,296 probes). Samples were removed if they: (1) were outliers from test of Multidimensional Scaling; (2) were mixed-up samples; (3) had a missing rate across probes; or (4) failed quality controls of genome-wide genotyping data, including sex discrepancies (based on sex chromosomes), heterozygosity outliers (felt out of range of ), unexpected duplicates or relatives ( in identity by descent analysis), and individual genetic call rate . After QC, raw methylation data from all three panels were normalized together using the dasen method in the wateRmelon package (Pidsley et al. 2013).
In the SLSJ, genome-wide methylation of leukocyte nuclear pellets DNA was assayed using Infinium HumanMethylation450 BeadChip (Illumina), with the details described previously (Liang et al. 2015). Genome-wide methylation data were then processed using the Touleimat and Tost analysis pipeline (Touleimat and Tost 2012). Samples were removed if they had a missing rate across probes; probes were removed if they: (1) assay SNPs rather than CpGs; and (2) overlapped with any SNPs with MAF (in 1,000 Genomes Project phase 1 EUR population) in the probe sequence or in position or of the query site. The lumi package was used for background and color bias correction (Du et al. 2008). BeadChip ID numbers and position on chip were included as categorical covariates to account for potential batch effects. Quantile normalization across samples was applied to probes within each functional category (CpG island, shelf, shore, etc.) separately to correct the shift of methylation values between Infinium I and Infinium II probes.
Genome-wide expression assay for the SY panel has been described previously (Li et al. 2017). Briefly, total RNA of SY was extracted from blood leukocytes within 2 h after blood draw using the TRIzol® LS Reagent (Life Technologies) according to standard protocols. Expressions profiles were measured using HumanHT-12 version 4 Expression BeadChip (Illumina) per instructions from Illumina. IDAT files generated from iScan (Illumina) system were processed using GenomeStudio GE module version 1.9.0 and quantile–quantile normalized using beadarray (Dunning et al. 2007) package.
Leukocyte subsets separation and purification.
Leukocyte subsets separation and purification were performed on blood samples of the eight healthy men recruited from Wuhan, China. The detailed laboratory procedures have been reported in our previous study (Li et al. 2017). Briefly, for each subject, a total of blood was collected; was used for cell counting (Roche Cobas 8000; BD FACSCanto II Flow Cytometer) and the remaining was processed to cell separation immediately. We first separated peripheral blood mononuclear cells (PBMCs) and granulocytes by density centrifuge using Ficoll-Paque Plus™ (GE Healthcare). Monocytes, and , , B, and Natural Killer (NK) cells were then separated and purified from washed PBMCs using paramagnetic microbeads coupled with anti-CD14, anti-CD4, anti-CD8, anti-CD19, or anti-CD56 antibodies (Miltenyi Biotec), and a MidiMACS™ separator (Miltenyi Biotec) following standard protocols of magnetic-activated cell sorting (MACS). Granulocytes were collected by density gradient from cell pellet, washed, and resuspended. Neutrophils were separated and purified from granulocytes by paramagnetic microbeads with anti-CD16 antibodies and a MidiMACS™ separator following MACS protocols. A duplicate of cells of each purified cell subtype was used for purity validation on a BD™ LSR II flow cytometer (BD Biosciences). DNA of purified leukocyte subsets were isolated and stored at until Beadchip assay.
Urinary creatinine and OH-PAH metabolites.
Baseline urinary levels of creatinine and 12 OH-PAH metabolites in the coke oven cohort and the WHZH-cohort have been reported in detail in our previous studies (Deng et al. 2014; Kuang et al. 2013; Li et al. 2012; Zhou et al. 2016). The same methods were applied to the SY panel and the eight men from cell-sorting to measure the same urinary biomarkers. Though measured separately, OH-PAH metabolites in each study cohort/panel were quantified using the same protocol and platform, and thus the limits of quantification (LOQ) were the same across panels, ranging from (Table S1). Of the 12 metabolites, 10 noncarcinogenic markers, namely 1-OH-naphthalene, 2-OH-naphthalene, 2-OH-fluorene, 9-OH-fluorene, 1-OH-phenanthrene, 2-OH-phenanthrene, 3-OH-phenanthrene, 4-OH-phenanthrene, 9-OH-phenanthrene, and 1-OH-pyrene, were mostly detectable thus included in the analyses (Table S1). Measurements below the LOQ is imputed using 50% of the LOQ. The other 2 carcinogenic markers, namely 6-OH-chrysene and 3-OH-benzo[a]pyrene, were all below the LOQ and futile for the study (Kuang et al. 2013; Li et al. 2012; Zhou et al. 2016). The molar concentrations of OH-PAHs were then calibrated by urinary creatinine and presented as micromoles per millimole creatinine. were the sum of the 10 detectable OH-PAH metabolites, after half-LOQ-imputation and creatinine-recalibration.
Data Collection and Definition of Covariates
Baseline data of the COW, WHZH, and SY panels, including demographic data, lifestyle and occupation information, and disease histories were collected from standard questionnaires. Clinical tests results, including proportions of neutrophil, lymphocyte, and intermediate cells (sum of monocyte, eosinophils and basophils) in whole blood for the same tubes of blood samples used for methylation/expression assays were acquired from clinical recodes. BMI was calculated as the body mass divided by the square of body height and was expressed in unit of . Current smokers were defined as individuals who smoked cigarettes/day over the past 6 mo; ex-smokers were those who quitted smoking for more than 6 mo; and never-smokers were subjects who never smoked. Cigarette pack-years was calculated by multiplying the number of packs of cigarettes smoked per day by the number of years the person had smoked. Current alcohol drinkers were defined as individuals who drank over the past 6 mo; ex-drinkers were those who quit drinking for more than 6 mo; and never-drinkers were those who never drank any liquor. Marriage status referred to whether the subjects were single, married, separated, divorced, or widowed. Education level indicated if a participant had a college education or not. In our cohorts, employment status defined whether the participants were employed, unemployed, retired, or off duty due to work injuries. Frequency of vegetables and fruits consumption ranged from , 4–12, 12–20, and 20–30 d (that the person had eaten vegetables or fruits) per month. Subjects regularly exercised for were defined as physically active, and otherwise as inactive. Covariates were missing for a small number of samples; in order not to lose statistical power for the missing covariates in association analyses, mean (for continuous variables normally distributed), median (for continuous variables not normally distributed), or mode (for categorical variables) were used to impute the missing covariates.
Statistical Analyses
Methylation age prediction.
We used the WHZH panel as the training set and used the COW and SY panel as the primary testing datasets. The WHZH panel was chosen as the training set because (1) the WHZH participants had a larger sample size and a wider range and standard deviation (SD) of age in comparison with the other primary panels (, age , and in the WHZH; , age , and in the SY; and , age , and in the COW panel); these parameters were major determinants of the prediction accuracy (Horvath 2013); and (2) participants in the WHZH panel were sampled from general urban communities, whereas the other two panels were sampled either from a coke-oven plant (i.e., COW) or from a community within an automobile company (i.e., SY); therefore, individuals in the WHZH panel were more representative of the general populations.
To build the age predictor in WHZH, we applied elastic net, a penalized regression model, (Friedman et al. 2010) to regress the chronological age on 420,771 quality-controlled autosome CpGs together with gender, smoking, drinking, BMI, and cellular proportions. The lambda value of the elastic net model was chosen using a 10-fold cross-validation approach. We then applied this age predictor to the testing datasets SY and COW to acquire the predicted age (the methylation age) in these two panels. The methylation age was computed as the weighted sum of selected CpGs with weights equal to the regression coefficients from the prediction model, plus a model intercept. The selected CpGs and coefficients captured the variations of methylation age, whereas the model intercept may capture specific batch effects and population features. Therefore, after applying the age predictor to the testing datasets, we fitted a linear regression of in the WHZH, COW, and SY panels, separately; we then recalibrated the model intercept of SY and COW to make the population intercept of SY and COW the same as the population intercept of WHZH (referred to as intercept equalization in the following text). The coefficients of CpGs were unchanged when applying the age predictor. Through this method, we equalized the systematic shifts of methylation values and thus increased the comparability across panels. A similar recalibration method was used in prior studies (Hannum et al. 2013).
As for the training set (the WHZH panel), we used the leave-one-out cross-validation (LOOCV) approach to obtain an unbiased methylation age. In each run, the elastic net regression was applied to all but one sample (the training set), and the model obtained was applied to the left-out sample (the testing set). A similar method was used in a previous study to acquire unbiased methylation age suitable for subsequent analyses (Horvath 2013).
Pearson’s correlation coefficient (R) and root-mean-square errors (RMSE) between the chronological age and methylation age were calculated to evaluate the prediction accuracy.
When applying the age predictor to the methylation dataset of eight healthy men from leukocytes subsets cell-sorting, intercept calibration was not performed due to limited sample size and cellular heterogeneity.
Validation of the age prediction model using external datasets.
To seek for an additional validation of the prediction accuracy, we applied our age predictor to 2 external datasets – TwinS and SLSJ. Because methylation of the TwinS was assayed in the same lab at the same time with our training set, we recalibrated the intercept using the aforementioned intercept equalization method. For the SLSJ, the methylation data was assayed in a different lab, and 5 CpGs in our age predictor were unavailable due to failure in QC; therefore, to minimize the shift, we recalibrated the model intercept to achieve the least RMSE between methylation and chronological age. The model coefficients of CpGs were unchanged during the intercept recalibration, and thus the correlation between methylation and chronological age was unaffected. We compared prediction parameters (R and RMSE) in the validation panels with those in the primary panels to evaluate the model accuracy.
In addition, we calculated the methylation age using the existing Horvath predictor (Horvath 2013) in our resident panels (i.e., , unexposed to coke-oven emissions), TwinS, and SLSJ. The model intercept of the WHZH, SY, and TwinS were recalibrated using the intercept equalization method and in SLSJ, using the least RMSE method. We compared R and RMSE of our predictor with the Horvath predictor within the same panels.
Association of PAH exposure and methylation aging.
The association analyses were conducted in our three primary panels, in which urinary OH-PAH measurements were available (Figure S1). was calculated as the chronological age minus methylation age; aging rate was the ratio of chronological age to methylation age; both indicators were normally distributed. OH-PAH metabolites were natural-log-transformed; subjects without sufficient urine samples or with measurements outliers () in each panel were excluded before analyses (Table S1).
To evaluate the associations of each OH-PAH metabolite with aging, we fitted multivariate linear regressions with (or aging rate) as the dependent variable and each OH-PAH metabolite (continuous variable) as the indicator. Covariates that were adjusted for in the regression included age, gender, BMI, smoking status, cigarette pack-years, alcohol drinking, geographical regions, and microarray operation date (Model 1). We further adjusted for cellular proportions (including neutrophil, lymphocyte and intermediate cells; Model 2) and marriage status, education, employment status, frequency of vegetables and fruits intake, and physical activity to account for other confounding (Model 3). The association analyses were conducted in the WHZH, SY, and COW separately and combined using a fixed-effect meta-analysis to obtain robust association estimations. Associations of metabolites with aging indicators were considered significant if false discovery rate (FDR) .
Correlations between OH-PAHs were tested using Pearson’s correlation coefficients on a nature-log transformed scale. To identify the OH-PAH metabolites independently associated with age acceleration, we fitted a stepwise regression, in which all covariates were forced in, whereas the significant OH-PAH metabolites (those with in individual metabolites analysis) were first included and then backward selected (significance level to ).
In a secondary analysis, we pooled WHZH, SY, and COW together to obtain the quartile cut-off points of the 9-OH-phenanthrene and 1-OH-pyrene and categorized the participants from all three panels into quartiles using the same cut-offs. We then compared the age acceleration of higher quartiles with the lowest quartile using the maximally adjusted model (plus adjusting for the study panels) by linear regressions, using the pooled dataset of the three panels (to increase sample size in each quartile). To quantify a linear trend, we assigned the median value of metabolites within each quartile and modeled this variable continuously.
In another secondary analysis, to test whether our significant findings of whole blood methylation can be replicated by cell-specific methylation, we examined the relation between 9-OH-phenanthrene or 1-OH-pyrene exposure and age acceleration in each leukocyte subtype in eight healthy men. Due to the limited sample size and the non-normal distribution of both aging indicators and OH-PAHs, we categorized the eight participants into high- vs. low-exposure groups according to the inflection point on the curve of the metabolite’s level and compared the methylation age acceleration in high- vs. low-exposure groups using the Mann-Whitney U-test. Box plots were used to visualize the data; no covariates were adjusted for.
Association of CpGs with aging indicators and OH-PAH metabolites.
To eliminate outliers and archive a normal distribution for linear regression, methylation values of the 239 CpGs were inverse-normal transformed (to a normal distribution with a mean value of 0 and a SD of 1) before this analysis. The association of each CpG with methylation aging ( or aging rate) was evaluated using linear regressions, adjusted for age, gender, BMI, smoking status, cigarette pack-years, alcohol drinking, geographical regions, microarray operation date, and cellular proportions; a (Bonferroni corrected for 239 CpGs) was considered as statistically significant. We then assessed the association of 1-OH-pyrene or 9-OH-phenanthrene with aging-related CpGs using linear regression, adjusting for the aforementioned covariates, with a considered as statistically significant. All analysis was conducted separately in the WHZH, SY, and COW, and combined using a meta-analysis. For the CpGs of interest, the association of methylation levels with the expression of corresponding genes were evaluated in the SY panel using linear regressions with adjustment for age and gender. All analyses were performed using R (version 3.1.2; R Core Team).
Results
Characteristics of the Study Participants
In terms of our primary study panels, the WHZH included 258 healthy community residents age 26–89 y (mean age 53.8 y; 79.5% men); the SY included 144 healthy residents age 22–71 y (mean age 41.3 y; 74.3% men); and the COW included 137 healthy coke-oven workers age 26–60 y (mean age 46.5 y; 78.1% men) (Table 1). Individuals in the COW panel had the highest OH-PAHs levels (median creatinine), followed by those in the WHZH panel (median creatinine), whereas participants in the SY panel had the lowest OH-PAHs levels (median creatinine) (Table 2).
Table 1.
Characteristics of the study population.
| Variables | WHZH () | SY () | COW () |
|---|---|---|---|
| Age variables | |||
| Age, years | 53.8 (12.9) | 41.3 (10.3) | 46.5 (8.9) |
| Methylation age, years | 53.9 (11.8) | 43.8 (9.7) | 50.2 (9.0) |
| , years | 0.09 (4.05) | 2.45 (3.50) | 3.55 (4.00) |
| Aging rate | 1.01 (0.08) | 1.07 (0.09) | 1.08 (0.09) |
| Characteristics | |||
| Men | 205 (79.5%) | 107 (74.3%) | 107 (78.1%) |
| Smoking status | |||
| Current-smokers | 108 (41.9%) | 45 (31.2%) | 84 (61.3%) |
| Ex-smokers | 25 (9.7%) | 2 (1.4%) | 3 (2.2%) |
| Never-smokers | 125 (48.4%) | 97 (67.4%) | 50 (36.5%) |
| Cigarette pack-yearsa | 18 (8, 29) | 20 (5, 31) | 21 (14, 35) |
| Alcohol drinking status | |||
| Current-drinkers | 81 (31.4%) | 55 (38.2%) | 49 (35.8%) |
| Ex-drinkers | 5 (1.9%) | 1 (0.7%) | 4 (2.9%) |
| Never drinkers | 172 (66.7%) | 88 (61.1%) | 84 (61.3%) |
| Employment status | |||
| Employed | 137 (53.1%) | 140 (97.2%) | 137 (100%) |
| Unemployed at working age | 41 (15.9%) | 0 | 0 |
| Retired | 72 (27.9%) | 4 (2.8%) | 0 |
| Physically disabled by work injuries | 8 (3.1%) | 0 | 0 |
| Marriage status | |||
| Single | 3 (1.2%) | 19 (13.2%) | 4 (2.9%) |
| Married | 241 (93.4%) | 125 (86.8%) | 132 (96.4%) |
| Divorced or widowed | 14 (5.4%) | 0 | 1 (0.7%) |
| Education level | |||
| High school or lower | 219 (84.9%) | 94 (65.3%) | 89 (65.0%) |
| Bachelor or above | 39 (15.1%) | 50 (34.7%) | 48 (35.0%) |
| Physical activity | |||
| Physically active | 128 (49.6%) | 79 (54.9%) | 74 (54.0%) |
| Physically inactive | 130 (50.4%) | 65 (45.1%) | 63 (46.0%) |
| BMI, | 23.4 (2.8) | 24.2 (2.7) | 23.5 (2.7) |
| White blood cell counts, | 6.0 (1.6) | 6.0 (1.4) | 6.7 (1.5) |
| Neutrophils proportion (%) | 55.8 (8.3) | 58.0 (8.2) | 59.6 (7.3) |
| Lymphocytes proportion (%) | 37.4 (8.2) | 33.7 (7.5) | 36.8 (7.1) |
| Intermediate cells proportion (%) | 6.9 (3.8) | 8.4 (2.7) | 3.6 (1.2) |
Note: Continuous variables were presented as mean (SD) or median (25th percentile, 75th percentile). Categorical variables were presented as n (%). Data were complete for each variable after processing as described in the Methods. Intermediate cell was the sum of monocytes, eosinophils and basophils. WHZH, study subjects selected from the WHZH-cohort; SY, participants recruited from Shiyan, China; COW, study subjects selected from the cohort of coke-oven workers.
Among ever smokers.
Table 2.
Creatinine-calibrated concentrations of urinary OH-PAH metabolites ( creatinine) in each study panel.
| OH-PAH metabolites | WHZH | SY | COW | |||
|---|---|---|---|---|---|---|
| a | Median (25th, 75th) | a | Median (25th, 75th) | a | Median (25th, 75th) | |
| 1-OH-Naphthalene () | 232 | 0.31 (0.22, 0.5) | 139 | 0.04 (0.02, 0.11) | 137 | 2.45 (1.73, 4.23) |
| 2-OH-Naphthalene () | 232 | 0.56 (0.40, 0.93) | 136 | 0.12 (0.04, 0.42) | 136 | 2.28 (1.48, 3.54) |
| 2-OH-Fluorene () | 231 | 1.20 (0.85, 1.63) | 139 | 0.12 (0.04, 0.63) | 136 | 9.25 (6.48, 15.44) |
| 9-OH-Fluorene () | 234 | 2.98 (1.95, 5.10) | 140 | 0.88 (0.17, 3.34) | 134 | 6.28 (3.78, 13.01) |
| 1-OH-Phenanthrene () | 234 | 0.87 (0.66, 1.38) | 140 | 0.11 (0.06, 0.25) | 137 | 7.99 (5.55, 13.57) |
| 2-OH-Phenanthrene () | 230 | 0.62 (0.44, 0.91) | 139 | 0.36 (0.14, 0.83) | 136 | 3.37 (2.17, 5.50) |
| 3-OH-Phenanthrene () | 230 | 1.11 (0.75, 1.44) | 138 | 0.07 (0.04, 0.18) | 137 | 3.83 (1.60, 6.23) |
| 4-OH-Phenanthrene () | 230 | 1.07 (0.80, 1.53) | 138 | 0.86 (0.36, 1.67) | 136 | 2.89 (1.47, 6.14) |
| 9-OH-Phenanthrene () | 232 | 2.26 (1.60, 3.22) | 139 | 0.73 (0.31, 1.33) | 133 | 8.18 (4.79, 14.13) |
| 1-OH-Pyrene () | 231 | 0.26 (0.17, 0.37) | 140 | 0.16 (0.04, 0.34) | 137 | 3.30 (2.53, 4.96) |
| Total OH-PAHs () | 233 | 2.44 (1.88, 3.12) | 136 | 1.13 (0.65, 1.79) | 137 | 13.31 (10.5, 18.86) |
Note: WHZH, study subjects selected from the WHZH-cohort; SY, participants recruited from Shiyan, China; COW, study subjects selected from the cohort of coke oven workers.
Summary data were calculated among subjects with sufficient and qualified urine samples, after half-LOQ imputation and outlier ( at the ln-transformed scale) removal.
With respect to the two validation panels for the age predictor, the TwinS contained 450 Chinese individuals age 18–81 y (mean age 44.8 y; 65.6% men), and the SLSJ contained 160 Caucasians age 5–79 y (mean age 29.0 y; 20% men).
A Methylation Age Predictor with Improved Accuracy in Chinese Populations
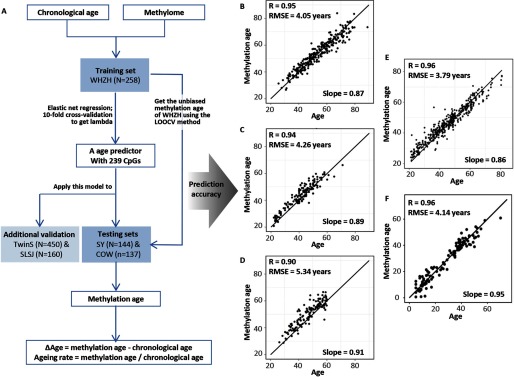
Based on an elastic net regression in the WHZH panel (the training dataset), a total of 239 quality-controlled autosome CpGs were selected into the age predictor (Figure 1 and Table S2), which achieved a high accuracy of age prediction in the SY and COW panel (the testing datasets). The correlation between chronological age and methylation age in SY and COW was 0.94 and 0.90, respectively, and the RMSE was 4.26 and 5.34, respectively (Figure 1). The unbiased methylation age acquired using LOOCV in the WHZH panel showed a similar prediction accuracy with a correlation coefficient of 0.95 and a RMSE of 4.05 (Figure 1).
Figure 1.
Methylation age prediction. (A): the procedures of methylation age prediction; (B), (C), and (D) present orderly the correlation and RMSE between methylation age and chronological age in our primary study panels – WHZH, SY, and COW; (E) and (F) present orderly the correlation and RMSE in the external validation populations – TwinS and SLSJ. Note: WHZH, study subjects selected from the WHZH-cohort; SY, participants recruited from Shiyan, China; COW, study subjects selected from the cohort of coke oven workers; TwinS, the Twins Methylation Studies; and SLSJ, the asthmatic family panel from the Saguenay-Lac-Saint-Jean region in Québec. RMSE, the root-mean-square error.
The high accuracy of our age predictor was further validated in an external Chinese population (in TwinS: , ) and a Caucasian population (in SLSJ: , ); both R and RMSE were similar with those in our primary panels (Figure 1).
When applying the existing Horvath predictor to the , TwinS, and SLSJ panels, the correlation between chronological age and methylation age was 0.83, 0.92, and 0.96, respectively, with an RMSE of 7.48, 9.85, and 6.46. When fitting a linear regression of the chronological age against methylation age, a shift in the slope was also observed with the Horvath model (Figure S3). In comparison with the Horvath predictor, our age predictor showed a higher accuracy and less error in Chinese populations and a similar accuracy in Caucasian populations.
Exposure to PAHs and Accelerated Methylation Aging
The mean values of in our primary study panels WHZH, SY, and COW were 0.09, 2.45, and 3.55 y, respectively, and the mean aging rate was 1.01, 1.07, and 1.08, respectively (Table 1). As expected, the COW showed an increased and aging rate (). We then assessed whether exposure to PAHs was associated with accelerated methylation aging.
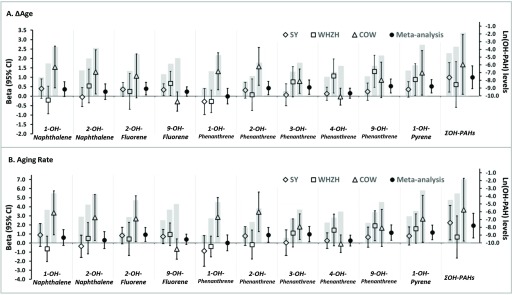
After adjusting for multiple risk factors, total urinary level was positively associated with both and aging rate; the association was slightly strengthened when further adjusting for leukocytes types and socioeconomic factors, with 1 unit (nature-log transformed scale) increment in associated with a 1.0-y increase in (95% CI: 0.4, 1.6; ) and a 1.9% increase in aging rate (95% CI: 0.6, 3.3; ) (Table 3). When analyzing each OH-PAH metabolite individually, urinary 2-OH-phenanthrene, 3-OH-phenanthrene, 9-OH-phenanthrene and 1-OH-pyrene were positively associated with , whereas 9-OH-phenanthrene and 1-OH-pyrene were positively associated with aging rate () in the maximally adjusted model (Table 3 Model 3). For these significant OH-PAH metabolites, the association direction was generally consistent across the three Chinese panels, with COW having the highest metabolites levels and showing the largest age acceleration (per metabolite increment), and SY having the lowest metabolites levels and showing the smallest age acceleration (Figure 2); this result suggested a potential dose–response relationship between PAH exposure and methylation aging at the population level.
Table 3.
Estimated mean difference in methylation aging indicators ( in years, and aging rate %) in association with a 1-unit increment in ln-transformed urinary OH-PAH metabolite concentrations.
| OH-PAH metabolites | Model 1 | Model 2 | Model 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta (95% CI) | FDR | Beta (95% CI) | FDR | Beta (95% CI) | FDR | ||||
| Association with | |||||||||
| 1-OH-Naphthalene | 0.37 | (, 0.75) | 0.07 | 0.41 | (0.01, 0.80) | 0.06 | 0.37 | (, 0.77) | 0.08 |
| 2-OH-Naphthalene | 0.29 | (, 0.69) | 0.06 | 0.28 | (, 0.70) | 0.06 | 0.24 | (, 0.65) | 0.08 |
| 2-OH-Fluorene | 0.38 | (0.05, 0.71) | 0.06 | 0.39 | (0.06, 0.73) | 0.06 | 0.40 | (0.07, 0.73) | 0.05 |
| 9-OH-Fluorene | 0.21 | (, 0.46) | 0.08 | 0.21 | (, 0.46) | 0.09 | 0.24 | (, 0.49) | 0.08 |
| 1-OH-Phenanthrene | (, 0.32) | 0.94 | (, 0.34) | 0.98 | 0.00 | (, 0.41) | 0.76 | ||
| 2-OH-Phenanthrene | 0.35 | (0.01, 0.70) | 0.05 | 0.38 | (0.03, 0.74) | 0.04 | 0.43 | (0.08, 0.79) | 0.04 |
| 3-OH-Phenanthrene | 0.44 | (0.07, 0.81) | 0.03 | 0.47 | (0.09, 0.85) | 0.03 | 0.48 | (0.10, 0.86) | 0.03 |
| 4-OH-Phenanthrene | 0.16 | (, 0.42) | 0.06 | 0.16 | (, 0.42) | 0.06 | 0.16 | (, 0.42) | 0.08 |
| 9-OH-Phenanthrene | 0.52 | (0.16, 0.87) | 0.001 | 0.55 | (0.18, 0.92) | 0.001 | 0.54 | (0.17, 0.91) | 0.004 |
| 1-OH-Pyrene | 0.60 | (0.27, 0.94) | 0.001 | 0.62 | (0.27, 0.97) | 0.001 | 0.53 | (0.18, 0.88) | 0.004 |
| 0.94 | (0.34, 1.53) | 0.002a | 0.98 | (0.38, 1.58) | 0.002a | 1.00 | (0.40, 1.61) | 0.002a | |
| Association with aging rate | |||||||||
| 1-OH-Naphthalene | 0.60 | (, 1.44) | 0.19 | 0.70 | (, 1.56) | 0.15 | 0.61 | (, 1.48) | 0.23 |
| 2-OH-Naphthalene | 0.37 | (, 1.28) | 0.22 | 0.38 | (, 1.29) | 0.22 | 0.32 | (, 1.25) | 0.25 |
| 2-OH-Fluorene | 0.87 | (0.12, 1.62) | 0.08 | 0.90 | (0.14, 1.67) | 0.06 | 0.93 | (0.16, 1.70) | 0.06 |
| 9-OH-Fluorene | 0.37 | (, 0.93) | 0.22 | 0.37 | (, 0.93) | 0.22 | 0.40 | (, 0.97) | 0.23 |
| 1-OH-Phenanthrene | (, 0.71) | 0.97 | (, 0.76) | 0.91 | 0.03 | (, 0.89) | 0.71 | ||
| 2-OH-Phenanthrene | 0.75 | (, 1.53) | 0.08 | 0.83 | (0.04, 1.62) | 0.06 | 0.90 | (0.09, 1.70) | 0.06 |
| 3-OH-Phenanthrene | 0.84 | (0.04, 1.64) | 0.08 | 0.92 | (0.10, 1.75) | 0.06 | 0.98 | (0.14, 1.82) | 0.06 |
| 4-OH-Phenanthrene | 0.29 | (, 0.87) | 0.19 | 0.31 | (, 0.90) | 0.17 | 0.27 | (, 0.87) | 0.23 |
| 9-OH-Phenanthrene | 1.07 | (0.27, 1.87) | 0.01 | 1.17 | (0.35, 1.99) | 0.009 | 1.15 | (0.31, 1.99) | 0.02 |
| 1-OH-Pyrene | 1.22 | (0.45, 1.98) | 0.01 | 1.23 | (0.45, 2.01) | 0.009 | 1.17 | (0.36, 1.98) | 0.02 |
| 1.78 | (0.48, 3.08) | 0.01a | 1.89 | (0.57, 3.21) | 0.008a | 1.92 | (0.57, 3.28) | 0.008a | |
Note: Urinary OH-PAH metabolites were natural-log transformed before analysis. Association analyses were conducted separately in WHZH, SY, and COW, and combined using a meta-analysis. CI, confidence interval. Model 1: adjusted for age, gender, smoking status, cigarette pack-years, drinking status, BMI, geographic regions (for individuals from the WHZH-cohort), and array operating date. Model 2: further adjusted for cellular proportions (neutrophils, lymphocytes and intermediated cells). Model 3: further adjusted for marriage status, education, employment status, frequency of vegetables and fruits intake, and physical activity.
Different from each OH-PAH metabolite for which we presented the FDR values, the association significance for were presented as un-multiple corrected P values.
Figure 2.
Associations of urinary OH-PAH metabolites with methylation aging indicators. Results of all 10 urinary OH-PAH metabolites and were presented. The left y-axis and the effect lines show the associations of urinary OH-PAH metabolites with (A) and aging rate (%) (B). Note: , beta in the SY panel; , the WHZH panel; , the COW panel; , the meta-analysis of the three panels. The right y-axis and gray bars depict mean urinary OH-PAH metabolites levels in the nature-log scale. Note: WHZH, study subjects selected from the WHZH-cohort; SY, participants recruited from Shiyan, China; COW, study subjects selected from the cohort of coke-oven workers.
The 10 OH-PAH metabolites were correlated with each other (Figure S4). Using a stepwise regression, we found that 9-OH-phenanthrene and 1-OH-pyrene were significantly associated with both and aging rate independently of other OH-PAHs, risk factors, leukocyte types, as well as socioeconomic factors (Table S3). Each unit increment in the ln-transformed 9-OH-phenanthrene (corresponded to 1.32, 1.74, and 0.76-SD in the COW, WHZH, and SY panels) were associated with an 0.54-y increase in (95% CI: 0.17, 0.91; ) and 1.15% increase in aging rate (95% CI: 0.31, 1.99; ); whereas the numbers for 1-unit increment of 1-OH-pyrene (corresponded to 1.66, 1.65, and 0.69-SD in the COW, WHZH, and SY panels) were 0.53-y for (95% CI: 0.18, 0.88; ) and 1.17% for aging rate (95% CI: 0.36, 1.98; ) (Table 3 and Figure S5). In the pooled analysis of the three panels, the age acceleration gradually increased with the increasing quartiles of the 2 metabolites (Table S4), suggesting a linear relationship between PAH exposure and methylation aging.
Replication of the PAH-aging Association Using Cell Subtype Specific Methylation
When replicating the association of 9-OH-phenanthrene and 1-OH-pyrene with methylation aging using cell-specific methylation (i.e., neutrophils, monocytes, , , B and NK cells) in the eight healthy men (Table S5), we observed a notable trend: A higher level of 9-OH-phenanthrene was related to a higher methylation age. especially in , and NK cells, whereas a higher level of 1-OH-pyrene was related to a higher methylation age in and cells (Figure S6).
CpGs Associated with Age Acceleration and PAH Exposure
We examined the association of the 239 CpGs in our age predictor with aging indicators and OH-PAH metabolites. Among the CpGs significantly associated with and aging rate (Bonferroni corrected ), we identified 3 CpGs associated with urinary 1-OH-pyrene levels (located on FHL2 and BOK; ), and 6 CpGs associated with 9-OH-phenanthrene levels (located on ELOVL2, TRIM59, ACSS3, RNF170, and 3q25.31; ) (Table S6).
Of these aging-, PAH-related CpGs, methylation level at cg22454769 was positively correlated with the expression of FHL2 (). In comparison with individuals who did not express ELOVL2 in blood leukocytes, individuals who expressed the gene had a lower cg24724428 methylation level (, ); and among these subjects, methylation of cg24724428 was inversely correlated with the ELOVL2 expression () (Figure S7).
Association of Other Risk Factors with DNA Methylation Aging
In our study populations, BMI showed a mild U-shape relation with methylation aging, and individuals with limb/visual/hearing disability due to previous work injuries (but without other diseases) had a higher (eight cases in WHZH, ) (Table S3 and Figure S8) in the maximally adjusted model. We did not find other risk factors or traits associated with methylation aging (Table S3 and Figure S8).
Discussion
Not everyone’s biological “clock” ticks in the same way. Environmental factors were believed to be contributors of human aging, but their influences and underlying mechanisms are largely unclear. Here, adopting a methylation aging marker established especially for Chinese populations, we found that exposure to PAHs was associated with accelerated methylation aging independently of known risk factors. To our knowledge, our study is the first to build a methylation age predictor specifically for Chinese populations and to assess the link between PAH exposure and methylation aging.
Findings from the current study were consistent with previous reports that coke-oven workers occupationally exposed to PAHs had a shorter telomere length in comparison with the less-exposed employees (Pavanello et al. 2010), and that long-term skin exposure to PM-bound PAHs could lead to higher skin aging scores (Vierkötter et al. 2010). Moreover, our study extended beyond the previous evidence. First, we used the objectively measured OH-PAH metabolites to quantify the total exposure to PAHs from a variety of sources (including diet, cooking, smoking, heating, traffic, refuse combustion, and occupational exposures). The use of OH-PAH metabolites not only improved the measurement accuracy of the exposure, but also enabled us to separate the effect of different PAH species. For instance, similar to previous studies (Gao et al. 2016; Horvath et al. 2014), we did not observe significant association of tobacco smoking with methylation aging; this observation might be partially because tobacco smoking greatly contributes to the exposure to naphthalene but not to phenanthrene and pyrene (Zhu et al. 2016). Second, in comparion with the previous cellular aging indicators, methylation age provided additional information – it has a similar scale with chronological age and can be linked to the epigenetic behavior of certain age-relevant genes. The value of DNA methylation age has been underscored by recent studies examining its associations with BMI (Horvath et al. 2014), cancers (Hannum et al. 2013; Horvath 2013), Down syndrome (Horvath et al. 2015), and physical and cognitive fitness in elderly populations (Marioni et al. 2015b). Notably, one study found that a 5-y increase in was associated with a 21% higher mortality risk in later life (Marioni et al. 2015a), highlighting the promising value of methylation age in studies of longevity. Our results, coupled with previous evidence, suggested that exposure to PAHs may be a contributor to accelerated aging.
Prior studies suggested that methylation age measured the cumulative work of the epigenetic maintenance system, in which a greater power was needed to maintain the epigenetic stability during stressful times, leading to a high tick rate of the biological clock (Horvath 2013). Exposure to PAHs could stimulate a chain of internal responses including xenobiotics metabolism (Shimada and Fujii-Kuriyama 2004), oxidative stress and DNA damages (Kuang et al. 2013), leading to adverse health effects (Kim et al. 2013), cancers (Boffetta et al. 1997; Bosetti et al. 2007), cardiovascular diseases (Burstyn et al. 2005), or neurological diseases (Ionescu et al. 2011). Both PAH exposure and its related damages (e.g., oxidative stress) have been associated with DNA methylation alterations (Franco et al. 2008; Herbstman et al. 2012). Therefore, PAH exposure may disturb the internal environment, which requires a greater power to reinstate the epigenetic stability, and thus appeared as association with accelerated methylation aging.
Notably, CpGs located on FHL2 and ELOVL2 were associated with both aging indicators and exposure to PAH and were correlated with gene expressions in the present study. These CpGs have been previously used as age predictors (Hannum et al. 2013). Intriguingly, FHL2 is a coregulator for AHR (Kollara and Brown 2010), which encodes a nuclear receptor that regulates the xenobiotic-metabolism enzymes for PAHs (Billiard et al. 2002) and the multi-functional pathway of ERK2 signaling (Purcell et al. 2004); it could affect the self-renewal, quiescence, and survival of hematopoietic cells (Hou et al. 2015). Methylation of FHL2 might play a role in the association of PAH with aging through its function in PAH metabolism, cell signaling, and cell survival. ELOVL2 encodes an polyunsaturated fatty acids elongase essential in lipid homeostasis regulation (Pauter et al. 2014). Interestingly, PAH exposure has been related to lipid oxidation damages and disturbed lipid homeostasis (Kuang et al. 2013), and lipid oxidation is a culprit of aging (Kregel and Zhang 2007). Therefore, methylation of ELOVL2 might participate in the PAH-aging association via the regulation of lipid homeostasis. Our data suggest that targeted epigenetic alteration may play a part in the association between PAH exposure and aging. The mechanisms underlying the association of PAH exposure with accelerated aging are still speculative and warrant further investigations.
It is worthwhile to note that the use of an accurate age predictor is critical for association studies examining methylation age accelerations. Previous studies have established several age predictors using data from non-Asian populations (Hannum et al. 2013; Horvath 2013), but no age predictor has been developed for Asian populations. Despite the nice performance in Caucasians, the widely used Horvath predictor exhibited larger errors (, ) and systematic shifts when applied to Chinese populations. In contrast, our age predictor provided a more accurate prediction in Chinese populations and a similar accuracy in a Caucasian population (, ). Of note, , instead of methylation age, was the most frequently used indicators in association studies and is more clinically relevant. Although the application of the Horvath predictor to our study populations might initially seem acceptable (e.g., in the testing set SY, the R of the Horvath age with chronological age was 0.83, and with methylation age from our predictor, 0.85), the correlation between from the Horvath predictor and our predictor was only 0.452 due to the variation in chronological age (Figure S9 A and B), indicating a substantial reduction in accuracy. Our simulations also confirmed that the inaccuracy in methylation age can cause a rapid reduction in the accuracy of (Figure S9 C). When applying the Horvath predictor to assess the association of PAH exposure with age acceleration, we observed a generally consistent association direction, but weaker effect coefficients and statistical power when comparing to those from our predictor (Figure S9 D), probably due to bias and loss of power from less accuracy. Although these data do not support a statistically significant validation, they do not rule out the possibility of replication. Indeed, for , 9-OH-phenanthrene, and 1-OH-pyrene, which we found significantly associated with , when correcting the association estimates from the Horvath by regression calibration (Hardin et al. 2003) (i.e., dividing by between the two ), the effect estimates became similar to what we observed using our age predictor (Figure S9 D). However, the regression recalibration does not provide higher precision of the estimate, therefore P values remain unchanged. The similar effect estimate after regression calibration suggests a potential replication if the predictor achieved sufficient prediction accuracy. We acknowledge that our association findings warrant further validation and generalization. An accurate methylation age prediction is essential to ensure an unbiased and more powerful association analysis of age accelerations; therefore, future methylation aging studies should consider prediction accuracy in the data interpretation.
Ethnic difference might be a factor limiting the accuracy of methylation age predictors and the generalization of the association results across populations/studies. Previous studies suggest that individuals of different ethnicities may exhibit different methylation profiles, rates of aging, and association effects (Diez Roux et al. 2009; Galanter et al. 2017; Horvath et al. 2016). Such differences could be due to intrinsic/genetic mechanisms, differences in habitual diets and environmental exposures, as well as differences in epigenetic/biological responses to lifetime exposures, hence the need for a methylation age predictor that is suitable and accurate for the study population. However, this aspect does not implicate the need for a different aging predictor in each study population, but to highlight the need for more systematic study of DNA methylation aging across populations and a more refined methylation age predictor that is accurate and suitable in multiethnic populations (including Chinese). Currently, our methylation age predictor could be particularly useful for future methylation aging studies in Chinese populations and may be suitable for studies comparing Chinese with Caucasian populations. Of note, our age predictor was established based on methylation of blood leukocytes. Additional data are needed to develop a multitissue age predictor that fits Chinese populations. Another factor that limited the accuracy of the predictor is the range of age in the training set. However, this factor is less likely to be the reason why the Horvath predictor is less accurate in our population as the Horvath predictor was trained in populations ranging in age from 0 to100 y old.
DNA methylation of blood leukocytes is appropriate for the current study for two reasons. First, whole blood is the most widely used and easy-to-access tissue in epidemiological studies. Although the Horvath age predictor can be applied to multiple tissues, the robustness and clinical relevance of methylation age were mostly examined using whole blood (Chen et al. 2016; Gao et al. 2016; Marioni et al. 2015a, 2015b). Second, because blood is an important carrier of internal PAHs, circulating leukocytes have constant and direct contact with the internal exposures, making them a direct target tissue of PAH exposure. Although our primary analyses were conducted in whole blood, our major findings remained consistent after adjusting for cellular compositions and were replicated using cell subtype-specific methylation data, suggesting that these findings were not significantly confounded by the mixed cellular nature of whole blood. Future studies using sorted blood cells or other purified target tissues might offer further insights.
The strengths of the current study include the establishment of an accurate methylation age predictor specific for Chinese populations but also suitable for Caucasian populations, the use of the objectively measured OH-PAH metabolites, replication in several populations, adjustment of many potential confounders, and the replication of whole blood results in purified leukocytes subsets. Several limitations of our study warrant discussion. First, because our association analyses were conducted in several Chinese populations and the methylation aging was measured only in whole blood, our association results may not be generalized to other populations or methylation aging of other tissues. Although our methylation age predictor performed well in a Caucasian population, we were unable to test the association of PAH exposure with methylation aging due to lack of PAH data in the Caucasian population. Second, the present study used a cross-sectional design, with both PAH exposure and methylation aging measured using samples collected at the same time; therefore, the temporal order between PAH exposure, methylation alteration, and aging acceleration cannot be inferred. However, it should be noted that, as a toxic xenobiotic that human bodies do not synthesize and utilize, PAHs are more likely to be driven by unintentional, external exposures rather than DNA methylation levels. Nevertheless, our study provided novel insights into the association of exposure to pollutants with human aging. Future studies of other populations, on other tissues, and using longitudinal design and repeated measurements, are needed to expand upon our findings, providing more evidence for pollutant control and prevention of age acceleration.
Conclusion
In summary, our study reported that exposure to PAHs was associated with methylation age acceleration, underscoring the negative impact of PAH exposure on aging and extending the study of the methylation clock into the area of environmental health. Our study also established a methylation age predictor specific for Chinese populations but also suitable for Caucasian populations. Future studies are warranted to investigate the health and clinical significance of such environment-related aging effects.
Supplemental Material
Acknowledgments
We thank all study participants, research staff, and students who participated in this work. This work was funded by the National Natural Science Foundation of China (91643202 and 81390542), the National Key Program of Research and Development of China (2016YFC0900800), the 111 Project and the Program for Changjiang Scholars, the Innovative Research Team in University to T.W., and the Fundamental Research Funds for the Central Universities (HUST: 2016YXZD045).
Authors' contributions: T.W., J.L., and L. Liang designed the study. T.W., X. Zhang, H.G., and J.L. managed the project. J. L., X. Zhu, K.Y., X.L., H.J., Y.Z., S.D., Y.B., X.H., B.L., and H.S. prepared DNA samples and/or performed beadchip experiments.Q.D., J.H., H.S., Y.Z. and H.J. performed labrotary measurememts of urinary samples. T.W., X. Zhang, H.G., W.C., J.Y, C.L., Y.G., B.Z., Z.Z., H.S., J.L., H.J., K.Y., and Y.Z. managed or participated in participants enrolling, clinical and epidemiological data collection and cleaning, and sample collection. J.L., X. Zhu, L. Liang, and H.J. performed data preprocessing and quality controls. For the TwinS study, W.G., and L.Li designed the study, participated epidemiological investigations, and managed the funding; B.W. prepared the DNA samples and preprocesed the methylation data. For the SLSJ study, C.L., and M.L. designed the study, perfromed clinical studies, prepared the DNA samples, and managed the funding. L.Liang, T.W. and F.B.H. directed the analysis and made major comments/edits to the manuscript. J.L. analyzed the data, wrote the first draft and made major revisions to the manuscript. All authors made contributions in commenting and revising the paper.
References
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. 2014. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10):1363–1369, PMID: 24478339, 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard SM, Hahn ME, Franks DG, Peterson RE, Bols NC, Hodson PV. 2002. Binding of polycyclic aromatic hydrocarbons (PAHS) to teleost aryl hydrocarbon receptors (AHRS). Comp Biochem Physiol B, Biochem Mol Biol 133(1):55–68, PMID: 12223212. [DOI] [PubMed] [Google Scholar]
- Boffetta P, Jourenkova N, Gustavsson P. 1997. Cancer risk from occupational and environmental exposure to polycyclic aromatic hydrocarbons. Cancer Causes Control 8(3):444–472, PMID: 9498904. [DOI] [PubMed] [Google Scholar]
- Bosetti C, Boffetta P, La Vecchia C. 2007. Occupational exposures to polycyclic aromatic hydrocarbons, and respiratory and urinary tract cancers: a quantitative review to 2005. Ann Oncol 18(3):431–446, PMID: 16936186, 10.1093/annonc/mdl172. [DOI] [PubMed] [Google Scholar]
- Burstyn I, Kromhout H, Partanen T, Svane O, Langard S, Ahrens W, et al. 2005. Polycyclic aromatic hydrocarbons and fatal ischemic heart disease. Epidemiology 16(6):744–750, PMID: 16222163. [DOI] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. 2016. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 8(9):1844–1865, PMID: 27690265, 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. 2009. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5(8):e1000602, PMID: 19680444, 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. 2014. Alzheimer's disease: Early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 17(9):1156–1163, PMID: 25129075, 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Wuttke D, Wood SH, Plank M, Vora C. 2012. Genome-environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol Rev 64(1):88–101, PMID: 22090473, 10.1124/pr.110.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Huang S, Zhang X, Zhang W, Feng J, Wang T, et al. 2014. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect 122(7):719–725, PMID: 24633190, 10.1289/ehp.1307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez Roux AV, Ranjit N, Jenny NS, Shea S, Cushman M, Fitzpatrick A, et al. 2009. Race/ethnicity and telomere length in the multi-ethnic study of atherosclerosis. Aging Cell 8(3):251–257, PMID: 19302371, 10.1111/j.1474-9726.2009.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. 2008. Lumi: a pipeline for processing Illumina microarray. Bioinformatics 24(13):1547–1548, PMID: 18467348, 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, Tavaré S. 2007. Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics 23(16):2183–2184, PMID: 17586828, 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- Esteller M. 2008. Epigenetics in cancer. N Engl J Med 358(11):1148–1159, PMID: 18337604, 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. 2007. Epigenetics and aging: the targets and the marks. Trends Genet 23(8):413–418, PMID: 17559965, 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. 2008. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 266(1):6–11, PMID: 18372104, 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22, PMID: 20808728. [PMC free article] [PubMed] [Google Scholar]
- Galanter JM, Gignoux CR, Oh SS, Torgerson D, Pino-Yanes M, Thakur N, et al. 2017. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. Elife 6, 10.7554/eLife.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Breitling LP, Brenner H. 2016. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget 7(30):46878–46889, PMID: 27276709, 10.18632/oncotarget.9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49(2):359–367, PMID: 23177740, 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J, Schmeidiche H, Carroll R. 2003. The regression-calibration method for fitting generalized linear models with additive measurement error. The Stata Journal 3:373–385. [Google Scholar]
- Herbstman JB, Tang D, Zhu D, Qu L, Sjödin A, Li Z, et al. 2012. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect 120(5):733–738, PMID: 22256332, 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, et al. 2012. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol 13(10):R97, PMID: 23034122, 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol 14(10):R115, PMID: 24138928, 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, et al. 2014. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA 111(43):15538–15543, PMID: 25313081, 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Garagnani P, Bacalini MG, Pirazzini C, Salvioli S, Gentilini D, et al. 2015. Accelerated epigenetic aging in Down syndrome. Aging Cell 14(3):491–495, PMID: 25678027, 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. 2016. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 17(1):171, PMID: 27511193, 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Wang X, Li L, Fan R, Chen J, Zhu T, et al. 2015. Fhl2 regulates hematopoietic stem cell functions under stress conditions. Leukemia 29(3):615–624, PMID: 25179730, 10.1038/leu.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu R, Broza Y, Shaltieli H, Sadeh D, Zilberman Y, Feng X, et al. 2011. Detection of multiple sclerosis from exhaled breath using bilayers of polycyclic aromatic hydrocarbons and single-wall carbon nanotubes. ACS Chem Neurosci 2(12):687–693, PMID: 22860162, 10.1021/cn2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492, PMID: 22641018, 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kim KH, Jahan SA, Kabir E, Brown RJ. 2013. A review of airborne polycyclic aromatic hydrocarbons (PAHS) and their human health effects. Environ Int 60:71–80, PMID: 24013021, 10.1016/j.envint.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Kollara A, Brown TJ. 2010. Four and a half LIM domain 2 alters the impact of aryl hydrocarbon receptor on androgen receptor transcriptional activity. J Steroid Biochem Mol Biol 118(1-2):51–58, PMID: 19815066, 10.1016/j.jsbmb.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Kregel KC, Zhang HJ. 2007. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol 292(1):R18–R36, PMID: 16917020, 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- Kuang D, Zhang W, Deng Q, Zhang X, Huang K, Guan L, et al. 2013. Dose-response relationships of polycyclic aromatic hydrocarbons exposure and oxidative damage to DNA and lipid in coke oven workers. Environ Sci Technol 47(13):7446–7456, PMID: 23745771, 10.1021/es401639x. [DOI] [PubMed] [Google Scholar]
- Laprise C. 2014. The Saguenay-Lac-Saint-Jean asthma familial collection: The genetics of asthma in a young founder population. Genes Immun 15(4):247–255, PMID: 24646526, 10.1038/gene.2014.12. [DOI] [PubMed] [Google Scholar]
- Li J, Zhu X, Yu K, Jiang H, Zhang Y, Deng S, et al. 2017. Genome-wide analysis of DNA methylation and acute coronary syndrome. Circ Res 120(11):1754–1767, PMID: 28348007, 10.1161/CIRCRESAHA.116.310324. [DOI] [PubMed] [Google Scholar]
- Li X, Feng Y, Deng H, Zhang W, Kuang D, Deng Q, et al. 2012. The dose-response decrease in heart rate variability: any association with the metabolites of polycyclic aromatic hydrocarbons in coke oven workers? PLoS One 7(9):e44562, PMID: 23024753, 10.1371/journal.pone.0044562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Willis-Owen SAG, Laprise C, Wong KCC, Davies GA, Hudson TJ, et al. 2015. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature 520(7549):670–674, PMID: 25707804, 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. 2015a. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 16:25, PMID: 25633388, 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, et al. 2015b. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 44:1388–1396, PMID: 25617346, 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Sinclair DA. 2007. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol 8(9):692–702, PMID: 17700626, 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- Pauter AM, Olsson P, Asadi A, Herslöf B, Csikasz RI, Zadravec D, et al. 2014. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J Lipid Res 55(4):718–728, PMID: 24489111, 10.1194/jlr.M046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavanello S, Pesatori AC, Dioni L, Hoxha M, Bollati V, Siwinska E, et al. 2010. Shorter telomere length in peripheral blood lymphocytes of workers exposed to polycyclic aromatic hydrocarbons. Carcinogenesis 31(2):216–221, PMID: 19892797, 10.1093/carcin/bgp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. 2013. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14:293, PMID: 23631413, 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell NH, Darwis D, Bueno OF, Müller JM, Schule R, Molkentin JD. 2004. Extracellular signal-regulated kinase 2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. Mol Cell Biol 24(3):1081–1095, PMID: 14729955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. 2003. Impact of aging on DNA methylation. Ageing Res Rev 2(3):245–261, PMID: 12726774. [DOI] [PubMed] [Google Scholar]
- Samanta SK, Singh OV, Jain RK. 2002. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol 20(6):243–248, PMID: 12007492. [DOI] [PubMed] [Google Scholar]
- Schadt EE. 2009. Molecular networks as sensors and drivers of common human diseases. Nature 461(7261):218–223, PMID: 19741703, 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujii-Kuriyama Y. 2004. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci 95(1):1–6, PMID: 14720319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Hou J, Huang X, Zhang X, Tan A, Rong Y, et al. 2014. The Wuhan-Zhuhai (WHZH) cohort study of environmental air particulate matter and the pathogenesis of cardiopulmonary diseases: study design, methods and baseline characteristics of the cohort. BMC Public Health 14:994, PMID: 25252923, 10.1186/1471-2458-14-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, et al. 2012. Maternal exposure to polycyclic aromatic hydrocarbons and 5'-CpG methylation of interferon-γ in cord white blood cells. Environ Health Perspect 120(8):1195–1200, PMID: 22562770, 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touleimat N, Tost J. 2012. Complete pipeline for Infinium(®) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 4(3):325–341, PMID: 22690668, 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- Vierkötter A, Schikowski T, Ranft U, Sugiri D, Matsui M, Krämer U, et al. 2010. Airborne particle exposure and extrinsic skin aging. J Invest Dermatol 130(12):2719–2726, PMID: 20664556, 10.1038/jid.2010.204. [DOI] [PubMed] [Google Scholar]
- Wang B, Gao W, Yu C, Cao W, Lv J, Wang S, et al. 2015. Determination of zygosity in adult Chinese twins using the 450K methylation array versus questionnaire data. PLoS One 10(4):e0123992, PMID: 25927701, 10.1371/journal.pone.0123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization). 2010. WHO guidelines for indoor air quality: selected pollutants. In: WHO Guidelines for Indoor Air Quality: Selected Pollutants. Geneva, Switzerland:World Health Organization. [Google Scholar]
- Zhou Y, Sun H, Xie J, Song Y, Liu Y, Huang X, et al. 2016. Urinary polycyclic aromatic hydrocarbon metabolites and altered lung function in Wuhan, china. Am J Respir Crit Care Med 193(8):835–846, PMID: 26636906, 10.1164/rccm.201412-2279OC. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li J, Deng S, Yu K, Liu X, Deng Q, et al. 2016. Genome-wide analysis of DNA methylation and cigarette smoking in a Chinese population. Environ Health Perspect 124(7):966–973, PMID: 26756918, 10.1289/ehp.1509834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.