Abstract
Site-1 protease (S1P) cleaves membrane-bound sterol regulatory element-binding proteins (SREBPs), allowing their transcription-stimulating domains to translocate to the nucleus where they activate genes governing lipid synthesis. S1P is a potential target for lipid-lowering drugs, but the effect of S1P blockade in animals is unknown. Here, we disrupt the S1P gene in mice. Homozygous germ-line disruptions of S1P were embryonically lethal. To disrupt the gene inducibly in liver, we generated mice homozygous for a floxed S1P allele and heterozygous for a transgene encoding Cre recombinase under control of the IFN-inducible MX1 promoter. When IFN was produced, 70–90% of S1P alleles in liver were inactivated, and S1P mRNA and protein were reduced. Nuclear SREBPs declined, as did mRNAs for SREBP target genes. Cholesterol and fatty acid biosynthesis in hepatocytes declined by 75%. Low density lipoprotein (LDL) receptor mRNA declined by 50%, as did the clearance of 125I-labeled LDL from plasma, but plasma cholesterol fell, suggesting that LDL production was reduced. These data raise the possibility that S1P inhibitors may be effective lipid-lowering agents, but they suggest that nearly complete inhibition will be required.
Keywords: sterol regulatory element-binding proteins‖cholesterol‖fatty acids‖knockout mice
The proteolytic release of sterol regulatory element-binding proteins (SREBPs) from cell membranes stimulates lipid synthesis in hepatocytes and other cells. Inhibition of this proteolytic release in liver might lead to reduced lipid synthesis and reduced lipid accumulation in liver, blood, and other organs. The consequences of blocking hepatic SREBP proteolysis can now be investigated as a result of the recent molecular identification of the proteins that mediate this process (1).
SREBPs are synthesized as membrane-bound precursors of ≈1,150 aa in length (1). The NH2 terminal domain of ≈480 aa is a transcription factor of the basic helix–loop–helix leucine zipper family. This domain is followed by a hairpin membrane-attachment domain of ≈80 aa, which consists of two transmembrane helices separated by a short 30-aa hydrophilic loop that projects into the lumen of the endoplasmic reticulum. The COOH-terminal domain of ≈590 aa faces the cytosol where it performs a regulatory function.
Immediately after their synthesis, SREBPs form complexes with SREBP cleavage-activating protein (SCAP), an endoplasmic reticulum protein that contains eight membrane-spanning helices followed by a cytoplasmic domain of 545 aa (2). The binding of SREBP to SCAP is mediated through interactions between the cytoplasmic COOH-terminal domains of both proteins. In sterol-depleted cells the SCAP/SREBP complex leaves the endoplasmic reticulum and reaches the Golgi complex where two membrane-bound proteases act sequentially to release the NH2-terminal domain of the SREBPs so that they can enter the nucleus to activate transcription of genes involved in lipid synthesis and uptake (3, 4). When cells are overloaded with sterols, the SCAP/SREBP complex fails to exit the endoplasmic reticulum, the SREBPs are not cleaved, and lipid synthesis declines. By this means, cholesterol controls its own synthesis as well as that of fatty acids (5).
The two proteases that process SREBPs were identified through somatic cell genetic studies in cultured Chinese hamster ovary cells (1). The first cleavage is mediated by Site-1 protease (S1P), a membrane-bound serine protease of the subtilisin family (6). S1P cleaves SREBPs following the leucine of the sequence Arg-Xaa-Xaa-Leu-Ser (RXXLS) that is found near the midpoint of the hydrophilic luminal loop of the SREBPs (7). This cleavage is required to permit the action of the second protease (Site-2 protease, S2P), which cleaves within the first transmembrane helix of the SREBPs to liberate the NH2-terminal fragment so that it can enter the nucleus (8, 9).
Like other members of the subtilisin family, S1P is synthesized as an inactive precursor (1,052 aa) with a cleaved signal sequence, called the A form (1,030 aa). The A form cleaves itself sequentially at two sites to create B and C forms, releasing NH2-terminal fragments of 115 and 49 aa, respectively. Both B and C forms are enzymatically active in cleaving peptides that fit the RXXL consensus (10, 11).
In cultured Chinese hamster ovary cells, SCAP, S1P, and S2P are all essential for the synthesis of cholesterol and unsaturated fatty acids. Mutant cell lines that lack any one of these proteins require exogenous sources of cholesterol and unsaturated fatty acids to grow (8, 12, 13). The roles of these proteins are less well established in liver, which synthesizes most of the cholesterol and fatty acids in vivo.
Hepatocytes, like most other cells, produce three forms of SREBP, designated SREBP-1a, -1c, and -2 (5). SREBP-1a and -1c are produced from a single gene through the use of alternate promoters that give rise to alternate first exons that splice into a common second exon. In liver, the SREBP-1c transcript predominates, which is opposite to the situation in cultured cells (14). SREBP-1c mRNA in liver is markedly induced by insulin, and it declines when insulin declines, as in fasting (15, 16). SREBP-1c increases the synthesis of fatty acids by enhancing transcription of genes encoding acetyl CoA carboxylase, fatty acid synthase, and stearoyl CoA desaturase-1 (15, 17). SREBP-2 is produced from an independent gene, and its major role is to stimulate the synthesis of cholesterol by increasing the mRNAs for all known enzymes of the cholesterol biosynthetic pathway (18).
In liver, as in cultured cells, SREBPs are synthesized as membrane-bound precursors that are proteolytically cleaved to nuclear forms (19). To determine the requirement for SCAP in this cleavage, a strain of SCAP-deficient mice was created in which a crucial region of the SCAP gene was flanked by loxP sites (20), which are targets for the bacteriophage Cre recombinase (21). These mice were bred to transgenic mice that express the Cre recombinase in liver under the control of the IFN-inducible MX1 promoter (21, 22). When these mice were treated with polyinosinic/polycytidylic acid (pIpC), an inducer of IFN, 90–95% of the hepatic SCAP genes were inactivated (20). The content of SREBPs in hepatic nuclei fell markedly, and the rates of synthesis of cholesterol and fatty acids declined by 80%, owing to a decline in all SREBP target mRNAs. The mRNAs encoding the SREBPs also declined, apparently because these genes are activated by the SREBPs themselves through direct or indirect feed-forward mechanisms (23–25).
In the current experiments, we used the IFN-inducible system of Cre-mediated recombination to explore the requirement for S1P in the processing of SREBPs in the livers of mice. The results indicate a major, but not absolute, requirement for S1P. They also suggest that S1P inhibitors must block nearly 100% of enzyme activity to disrupt SREBP function in liver.
Materials and Methods
We obtained pIpC (catalogue no. P1530) from Sigma. Measurement of cholesterol and triglycerides, immunoblot analyses, RNase protection assays, and lipid synthesis and secretion in mouse hepatocytes all were carried out as described (20, 26).
Cloning of Mouse S1P Gene.
A genomic mouse S1P clone was isolated from a 129S6/SvEv mouse strain bacteriophage λ FIX II genomic library (provided by Alan Bradley, Baylor College of Medicine, Houston). The library was screened with a S1P cDNA probe prepared by PCR from mouse liver first-strand cDNA by using the following primers: 5′ primer, 5′-AGATGGAGAAGAAGCGGAGAAAGAAATGAAAGCCTCT-3′, and 3′ primer, 5′-GAGAATTCCACCTTCAAAGTCAGGTGGGAA-3′. The corresponding genomic DNA insert was excised from the bacteriophage vector and subcloned into the NotI site of pBluescript II KS (+) (Stratagene), resulting in plasmid pKS-mS1P. This clone covered a portion of intron 1, exons 2–6, and a portion of intron 6 (Fig. 1a).
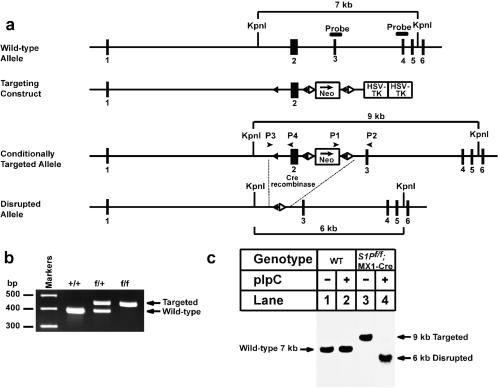
Figure 1.
Generation and characterization of a floxed S1P allele. (a) Schematic of gene-targeting strategy. Excision of the sequences between the loxP sites by the Cre recombinase deletes exon 2, which includes the first initiator methionine, the signal sequence (amino acids 1–22), and the preprosegment (amino acids 23–54) of S1P (6). The location of the probe used for Southern analysis (exons 3 + 4) is denoted by a filled box. The positions of primers (P1 and P2, P3 and P4) used for PCR detection of homologous recombination are denoted by arrowheads. (b) Genotype analysis of the conditionally targeted S1P mice by PCR of tail-derived DNA with primers P3 and P4 as described in Materials and Methods. (c) Southern blot analysis of KpnI-digested genomic DNA from livers of wild-type (WT) and S1Pf/f;MX1-Cre transgenic mice treated with pIpC as described in Materials and Methods.
Construction of Targeting Vector for Conditional Disruption of S1P.
A conditional targeting vector of a replacement type was produced by inserting a loxP site into intron 1 and a loxP;frt-flanked pgk-neo-pA cassette into intron 2. Cre-mediated recombinase removes exon 2, which encodes the signal sequence (amino acids 1–22) and a portion of the prosegment (amino acids 23–54) of S1P (6, 10). Details of the targeting vector construction are available on request.
Embryonic Stem (ES) Cell Culture for Disruption of S1P.
Passage 8 SM-1 ES cells were electroporated with the S1P targeting vector as described (27). Recombined clones were identified by PCR using primers P1 (5′-GATTGGGAAGACAATAGCAGGCATGC-3′ from the 3′ untranslated region of the neo gene) and P2 (5′-TTCCACTTCACTGCTTTTCAGAGCAC-3′ from exon 3 of the S1P gene outside of the targeting vector). The loxP site in intron 1 was confirmed by PCR with primers P3 (5′-GAGAGCTGCAGATGACAGGGGACACAG-3′ located ≈300 bp upstream of exon 2) and P4 (5′-GCCCAATCCACCGCTCTGTAGCGGAC-3′ located in exon 2). All targeted clones were confirmed by Southern blot analysis using a cDNA probe containing exons 3 and 4 of S1P as described (27).
Generation of S1P+/−, S1Pf/+, and S1Pf/f;MX1-Cre Mice.
Three targeted ES clones were injected separately into C57BL/6J blastocysts, yielding chimeric males whose coat color (agouti) indicated a contribution of ES cells from 25% to 100%. All 24 chimeric males were fertile, four of which produced offspring that carried the S1Pflox allele through the germ line. One line was established and used for further breeding.
To generate knockout mice that lacked S1P in all tissues, mice heterozygous for the S1Pflox allele (referred to as S1Pf/+ mice) were bred with protamine-1 promoter (Prm)-Cre transgenic mice (28) to produce S1Pf/+;Prm-Cre male mice. These mice were bred with wild-type mice (C57BL/6J;129S6/SvEv strain) to segregate the recombined S1P− allele, which had a deletion of exon 2 and flanking intronic sequences (referred to as S1P− allele). Heterozygous S1P+/− mice were then bred together in an attempt to produce homozygous S1P−/− mice.
To generate tissue-specific S1P knockout mice, S1Pf/+ mice were bred with MX1-Cre transgenic mice of the C57BL/6J;SJL strain (22) to produce S1Pf/+;MX1-Cre mice. The S1Pf/+;MX1-Cre mice were bred with S1Pf/+ mice to generate S1Pf/f;MX1-Cre mice. PCR was used to genotype the mice with primers P3 and P4 (30 cycles, 94°C, 30 s; 60°C, 30 s; 72°C, 2 min). The wild-type allele produced a PCR product of 380 bp, and the floxed allele produced a product of 434 bp. Cre expression was induced by i.p. injection of the IFN inducer, pIpC (21, 22). Each mouse received four or five 300-μl injections of a 1 mg/ml solution of pIpC in water every 48 h (20). Mice were analyzed 7–14 days after the final injection of pIpC. Mice were housed in colony cages and maintained on a 12-h light/12-h dark cycle and fed Teklad Mouse/Rat Diet 7002 from Harlan Teklad Premier Laboratory Diets (Madison, WI).
Real-Time Reverse Transcriptase–PCR (RT-PCR).
Total RNA was prepared from livers with an RNA STAT-60 kit (Tel-Test, Friendswood, TX). cDNA was synthesized from 5 μg of DNase I-treated total RNA (RNase free, FPLC pure, Amersham Pharmacia) using the SuperScript First-Strand Synthesis System (catalogue no. 11904–018, GIBCO/BRL) and random hexamer primers. Specific primers for each gene were designed by using primer express software (Perkin–Elmer) (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). The real-time RT-PCR contained, in a final volume of 30 μl, 50 ng of reverse-transcribed total RNA, 167 nM of the forward and reverse primers, and 15 μl of 2× SYBR Green PCR Master Mix. PCRs were carried out by using the Applied Biosystems Prism 7700 Sequence Detection System. All reactions were done in triplicate. The relative amounts of all mRNAs were calculated by using the comparative CT method (User Bulletin no. 2, Perkin–Elmer). Glyceraldehyde-3-phosphate dehydrogenase mRNA was used as the invariant control.
Plasma Clearance of 125I-Low Density Lipoprotein (LDL).
LDL (density, 1.019–1.063 g/ml) was prepared from plasma of LDL receptor knockout mice (LDLR−/−) (26) by sequential ultracentrifugation (29) and radiolabeled with sodium 125I as described (29). Recipient mice were fasted 2 h and anesthetized with sodium pentobarbital (60 mg/kg). Each mouse received an i.v. bolus of 0.2 ml of 0.15 M NaCl containing 30 μg of 125I-labeled LDL via the right external jugular vein. Blood was collected from the left jugular vein 1 min after the 125I-labeled LDL injection and at the indicated times thereafter. Total 125I-labeled apolipoprotein B (apoB) (apoB-48 plus apoB-100) in plasma was measured by isopropanol precipitation (30).
Results
Fig. 1a shows the vector and the targeting strategy that was used to disrupt S1P. The bacterial neomycin resistance gene (neo) flanked by loxP and flp recombinase sites was inserted into the second intron of the S1P gene. An additional loxP site was inserted into the first intron. Cre-mediated recombination between the loxP sites removes exon 2 of S1P, which contains the initiator methionine, the entire signal peptide sequence, and 32 aa of the preprosegment (10). Mice homozygous for the floxed S1P allele, designated S1Pf/f, were bred to two lines of transgenic mice that express Cre recombinase driven by: (i) the mouse Prm (Prm-Cre) (28), and (ii) the MX1 promoter (22). Prm-Cre mice express Cre in the testis, resulting in the germ-line deletion of S1P. Heterozygous mice derived from this cross are designated S1P+/−. MX1-Cre mice express Cre in hepatocytes when induced by treatment with pIpC. Mice derived from this cross (designated S1Pf/f;MX1-Cre) have a conditional deletion.
Fig. 1b shows the PCR analysis that was used to genotype the S1Pf/f mice. DNA derived from mouse tails was amplified with primers P3 and P4 (Fig. 1a). Genomic DNA from mice homozygous for the floxed allele (f/f) produced a single 434-bp PCR product. Fig. 1c shows a Southern blot analysis that was used to detect Cre-mediated recombination in DNA from livers of S1Pf/f;MX1-Cre mice. The DNA was digested with KpnI and blotted with a cDNA probe that corresponds to exons 3 and 4 of the S1P gene (as shown in Fig. 1a). In wild-type mice we visualized a 7-kb fragment that was unaffected by pIpC injections (Fig. 1c, lanes 1 and 2). In S1Pf/f;MX1-Cre mice, we obtained a 9-kb band, which corresponds to the targeted allele (Fig. 1c, lane 3). Five injections of pIpC resulted in a marked reduction in the 9-kb band and the appearance of a 6-kb band, which results from recombination between the 5′ and 3′ loxP sites (Fig. 1c, lane 4).
We first attempted to obtain mice with homozygous germ-line deletions of S1P by breeding S1P+/− mice with each other. These crosses did not produce any offspring homozygous for the disrupted S1P allele (Table 4, which is published as supporting information on the PNAS web site). We did not investigate the cause or developmental stage of this embryonic lethality. However, using a gene-trap approach to randomly inactivate multiple genes in mouse ES cells, Mitchell et al. (31) found that homozygous disruption of S1P prevented normal epiblast formation and subsequent implantation (before day 4).
To study the metabolic effects of deleting S1P from liver, we examined five S1Pf/f;MX1-Cre mice that were injected with pIpC to induce expression of Cre recombinase. Two weeks after the fifth pIpC injection, the mice were killed, and the results are presented in Table 1, Fig. 2, and Table 2, experiment A. Liver DNA was prepared from each of the five animals and the amount of Cre-mediated recombination was found to vary from 77% to 90%, as determined by the Southern blotting method illustrated in Fig. 1c. Body weight, liver weight, and liver cholesterol content were not affected by pIpC injection either in wild-type or S1Pf/f;MX1-Cre mice (Table 1). The S1Pf/f;MX1-Cre mice injected with pIpC showed a 36% decrease in the plasma concentration of cholesterol and a 50% decrease in plasma triglycerides. The content of triglyceride in liver also decreased.
Table 1.
Phenotypic comparison of wild-type and S1Pf/f;MX1-Cre mice treated with or without pIpC
| Parameter | Wild type
|
S1Pf/f;MX1-Cre
|
||
|---|---|---|---|---|
| −pIpC | +pIpC | −pIpC | +pIpC | |
| Number and sex | 5 males | 5 males | 5 males | 5 males |
| Body weight, g | 25.4 ± 2.9 | 27.8 ± 1.6 | 26.0 ± 1.2 | 26.5 ± 2.2 |
| Liver weight, g | 1.3 ± 0.2 | 1.4 ± 0.1 | 1.4 ± 0.1 | 1.4 ± 0.1 |
| Liver cholesterol content, mg/g | 1.9 ± 0.1 | 2.2 ± 0.14 | 2.5 ± 0.1 | 2.3 ± 0.2 |
| Liver triglyceride content, mg/g | 4.4 ± 1.1 | 6.6 ± 2.5 | 5.0 ± 1.1 | 3.4 ± 0.8* |
| Total plasma cholesterol, mg/dl | 94 ± 7.7 | 114 ± 14 | 100 ± 15 | 64 ± 4.9** |
| Total plasma triglyceride, mg/dl | 123 ± 46 | 86 ± 20 | 98 ± 17 | 49 ± 12** |
Male mice (7–8 weeks of age) were injected five times i.p. with either PBS (−pIpC) or pIpC (300 μg/injection) as described in Materials and Methods. Fourteen days after the last injection, blood and tissues were obtained. Each value represents mean ± SEM of five mice. Wild-type mice were littermates of S1Pf/f mice. ∗ denote level of statistical significance (Student's t test) between wild-type (+pIpC) and S1Pf/f;MX1-Cre mice treated with pIpC.
, P < 0.05;
, P < 0.01; and
, P < 0.001.
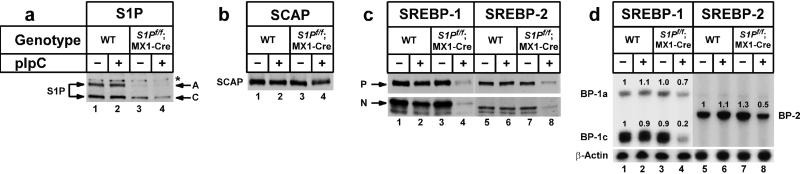
Figure 2.
Levels of proteins and mRNAs in livers of wild-type (WT) and S1Pf/f;MX1-Cre mice with or without injection of pIpC. The mice used in these experiments are described in Table 1. (a) Immunoblot analysis of precursor (A) and cleaved active form (C) of S1P (10). * denotes a nonspecific band. (b) Immunoblot analysis of SCAP. (c) Immunoblot analysis of SREBP-1 and -2 precursor (P) and nuclear (N) forms. (a–c) After treatment with or without pIpC, livers from the four groups of mice described in Table 1 were pooled, and aliquots of the membrane (50 μg protein) (a and b and the upper gel in c) and nuclear extract fractions (30 μg protein) (lower gel in c) were subjected to SDS/PAGE. Immunoblot analysis was performed as described (10, 20). Filters were exposed to Kodak X-Omat Blue XB-1 film 10–30 s at room temperature. (d) RNase protection assay of SREBP-1a, -1c, and -2 mRNAs. Total RNA isolated from livers of mice (Table 1) was pooled, and 15-μg aliquots were subjected to RNase protection assay. After RNase digestion, protected fragments were separated by gel electrophoresis and exposed to film for 4 h at −80°C and quantified as described in Materials and Methods. Intensity of each band relative to lane 1 is denoted above the band.
Table 2.
Relative amounts of mRNAs in livers from wild-type and S1Pf/f;MX1-Cre mice treated with or without pIpC
| Gene | Experiment A
|
Experiment B
|
||||||
|---|---|---|---|---|---|---|---|---|
| Wild type
|
S1Pf/f;MX1-Cre
|
Wild type
|
S1Pf/f;MX1-Cre
|
|||||
| −pIpC | +pIpC | −pIpC | +pIpC | −pIpC | +pIpC | −pIpC | +pIpC | |
| S1P | 1 | 1.28 | 0.14 | 0.07 | 1 | 1.07 | 0.17 | 0.05 |
| SREBP-1a | 1 | 1.07 | 0.99 | 0.61 (62%)* | 1 | 1.15 | 1.13 | 0.82 (73%)* |
| SREBP-1c | 1 | 0.89 | 0.85 | 0.20 (24%) | 1 | 0.96 | 0.79 | 0.07 (9%) |
| SREBP-2 | 1 | 1.37 | 1.06 | 0.55 (52%) | 1 | 0.85 | 1.30 | 0.55 (42%) |
| SCAP | 1 | 1.05 | 0.87 | 0.85 (98%) | 1 | 1.08 | 1.11 | 1.21 (109%) |
| LDL receptor | 1 | 1.25 | 1.17 | 0.55 (47%) | 1 | 0.96 | 1.11 | 0.59 (53%) |
| HMG CoA synthase | 1 | 1.05 | 1.13 | 0.55 (49%) | 1 | 1.29 | 1.97 | 0.68 (35%) |
| HMG CoA reductase | 1 | 1.25 | 1.16 | 0.77 (66%) | 1 | 0.82 | 1.26 | 0.59 (47%) |
| Farnesyl diphosphate synthase | 1 | 0.96 | 0.91 | 0.32 (35%) | 1 | 1.24 | 1.29 | 0.30 (23%) |
| Squalene synthase | 1 | 1.24 | 1.24 | 0.56 (45%) | 1 | 1.16 | 1.33 | 0.64 (48%) |
| Acetyl CoA carboxylase | 1 | 0.84 | 0.74 | 0.49 (66%) | 1 | 1.19 | 1.12 | 0.53 (47%) |
| Fatty acid synthase | 1 | 0.98 | 1.04 | 0.41 (39%) | 1 | 0.98 | 1.12 | 0.37 (33%) |
| Stearoyl CoA desaturase-1 | 1 | 0.63 | 0.76 | 0.43 (57%) | 1 | 0.86 | 0.73 | 0.20 (27%) |
| ApoB | 1 | 1.13 | 0.88 | 0.93 (106%) | 1 | 0.95 | 0.99 | 0.91 (92%) |
| ApoE | 1 | 1.02 | 1.01 | 1.12 (111%) | 1 | 0.87 | 0.95 | 0.93 (98%) |
Mice used for experiment A are the same mice used in Table 1 and Fig. 2. Mice used for experiment B are from a different experiment in which five male mice of each genotype were treated exactly as described in Table 1. For experiments A and B, total RNA from livers of five mice was pooled and subjected to real-time RT–PCR quantification as described in Materials and Methods. Values represent the amount of mRNA relative to those in the untreated wild-type mice (−pIpC), which are arbitrarily assigned a value of 1.
Values in parenthesis for S1Pf/f;MX1-Cre mice denote percentage of value in the absence of pIpC relative to that in the presence of pIpC.
Fig. 2 shows immunoblot analyses of S1P, SCAP, SREBP-1, and SREBP-2 proteins in the pooled livers from the groups of mice described in Table 1. In wild-type mice, the immunoblot for S1P showed two bands. The upper band corresponds to the full-length inactive precursor form of S1P, designated the A form. The lower band corresponds in size to the smallest cleaved active form of S1P, designated the C form (10). In wild-type mice, pIpC injection did not alter the amount of the A or C form of S1P (Fig. 2a, lanes 1 and 2). The livers of S1Pf/f;MX1-Cre mice exhibited a marked decrease in the A form and a less pronounced reduction in the C form of S1P, even without pIpC injections (Fig. 2a, lane 3). When the S1Pf/f;MX1-Cre mice were injected with pIpC, the amounts of S1P A and C forms were reduced to a level that is estimated to be 90–95% less than that observed in livers of wild-type mice (Fig. 2a, lane 4). The amount of SCAP protein was not significantly affected under any condition (Fig. 2b).
The precursor and nuclear forms of SREBP-1 and -2 were unaltered in livers of wild-type mice injected with pIpC (Fig. 2c, lanes 1 and 2, lanes 5 and 6). In the absence of pIpC injection, the amounts of precursor and nuclear SREBP-1 and -2 in liver were the same as that observed in wild-type mice (Fig. 2c, lanes 1 and 3, lanes 5 and 7). After pIpC injection in the S1Pf/f;MX1-Cre mice, the SREBP-1 and -2 precursors and nuclear forms both declined markedly (Fig. 2c, lanes 4 and 8).
Fig. 2d shows the results of an RNase protection assay that measures the mRNA levels of SREBP-1a, -1c, and -2 in the livers of the same animals used in Fig. 2c. After pIpC injection in the S1Pf/f;MX1-Cre mice, there was a significant decline in the mRNA for SREBP-1c and -2 and a slight decline in the mRNA for SREBP-1a (Fig. 2d, lanes 4 and 8). These findings are consistent with studies showing that SREBPs themselves are target genes for SREBP action (23–25, 32, 33).
Table 2, experiment A shows the results of quantitative PCR assays designed to measure mRNA levels in livers of the mice studied in Fig. 2 and Table 1. Experiment B shows results from a second identical experiment. Total hepatic RNA was subjected to real-time quantitative RT-PCR using the gene-specific primers. In the absence of pIpC injection, the S1P mRNA was reduced by ≈85% in S1Pf/f;MX1-Cre mice, suggesting that the neo cassette decreased S1P mRNA expression level. When the S1Pf/f;MX1-Cre mice were injected with pIpC, the mRNA for S1P declined by a further 50% (Table 2, experiment A) or 66% (Table 2, experiment B) for a total reduction of 95% as compared with wild-type levels. Under these conditions, the mRNAs for SREBP-1a, -1c, and -2 also declined with the largest decrease occurring in SREBP-1c mRNA. The mRNA levels of SREBP target genes involved in cholesterol biosynthesis and uptake [3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) synthase, farnesyl diphosphate synthase, squalene synthase, and LDL receptor] were reduced by 45–77%. A 34–53% decrease was measured in the mRNA for HMG CoA reductase. The mRNAs for SREBP target genes responsible for fatty acid biosynthesis (acetyl CoA carboxylase, fatty acid synthase, and stearoyl CoA desaturase-1) were decreased by 44–73%. The level of SCAP mRNA was not altered significantly in any of the mice. ApoB and apoE mRNA levels were also unchanged.
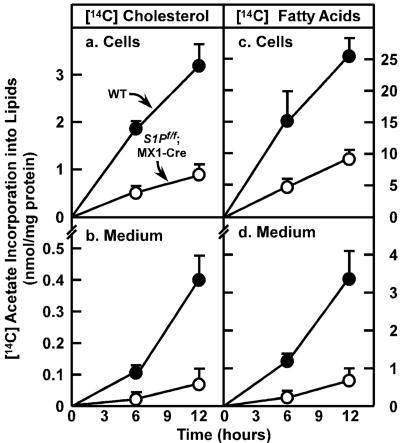
To determine the effects of S1P deletion on the overall rates of lipid synthesis, we measured the incorporation of [14C]acetate into cellular and secreted cholesterol and fatty acids in freshly prepared primary hepatocytes from wild-type and S1Pf/f;MX1-Cre mice injected with pIpC (Fig. 3). In hepatocytes from pIpC-injected S1Pf/f;MX1-Cre mice, the rates of 14C-labeled acetate incorporation into cellular [14C]cholesterol and [14C]fatty acids were decreased by 74% and 64%, respectively (Fig. 3 a and c). There was also a marked decrease in the secretion of labeled cholesterol and total fatty acids into the culture medium (decreases of 83% and 78%, respectively) (Fig. 3 b and d).
Figure 3.
Rates of lipid synthesis and secretion by primary hepatocytes from wild-type mice and S1Pf/f;MX1-Cre mice. Male mice (12 weeks of age) were injected i.p. with pIpC. Fourteen days after the last injection, primary hepatocytes were prepared as described in Materials and Methods. After a 2-h attachment period, hepatocytes were incubated with 0.5 mM sodium [14C]acetate (52 dpm/pmol), and medium and cells were harvested at the indicated time. Content of 14C-labeled cholesterol and fatty acids in the cells and medium was quantified as described in Materials and Methods. Each value is mean ± SEM of duplicate incubations from three wild-type and three S1Pf/f;MXI-Cre mice. Similar results were obtained in two independent experiments.
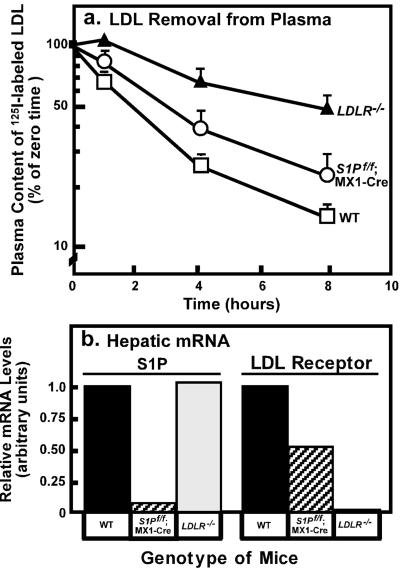
S1Pf/f;MX1-Cre mice injected with pIpC had a decrease of ≈50% in the expression of LDL receptor mRNA (Table 2). To determine whether this reduction would alter the clearance of apoB-containing lipoproteins, we measured the ability of wild-type, S1Pf/f;MX1-Cre, and LDL receptor knockout mice, all injected with pIpC, to clear 125I-labeled LDL from plasma. For this purpose, we isolated LDL from LDL receptor knockout mice and labeled the apoB with 125I. We injected the 125I-LDL into pIpC-treated wild-type mice, S1Pf/f;MX1-Cre mice, and LDL receptor knockout mice. The latter mice served as positive controls to show the rate of clearance when the LDL receptor is absent. As shown in Fig. 4a, there was a partial reduction in the clearance of 125I-labeled apoB in S1Pf/f;MX1-Cre mice injected with pIpC. The rate of clearance was intermediate between that of the wild-type and LDL receptor knockout mice. Fig. 4b shows that the level of LDL receptor mRNA was decreased by ≈50% in S1Pf/f;MX1-Cre mice.
Figure 4.
(a) Plasma clearance of 125I-labeled LDL in wild-type (WT, n = 6), S1Pf/f;MX1-Cre (n = 5), and LDLR−/− (n = 4) mice. Male mice (7–8 weeks of age) were injected i.p. with pIpC five times as described in Materials and Methods. Seven days after the last injection, 125I-labeled LDL (30 μg protein, 300 cpm/ng apoB protein) was injected i.v. into mice of the indicated genotype. Blood was obtained at the indicated time for quantification of plasma content of 125I-labeled total apoB. Data are plotted as percent of zero time value. Each value represents mean ± SEM of data from 4–6 mice. (b) Quantification of LDL receptor mRNA in mouse liver. Liver RNA was prepared from mice used in a, and real-time RT-PCR quantification was performed as described in Materials and Methods. Values represent the relative mRNA levels for mice of the indicated genotype (normalized to glyceraldehyde-3-phosphate dehydrogenase expression). Similar results were obtained in a second independent experiment.
Discussion
The current data provide evidence that S1P plays a crucial role in the processing of SREBPs in liver and is thus necessary for normal rates of lipid synthesis. Disruption of S1P in liver, mediated by an inducible Cre recombinase, led to a decline in nuclear SREBPs, a decline in the mRNAs of the SREBP target genes, and a decrease in the rates of synthesis of cholesterol and fatty acids as measured in hepatocytes isolated from the gene-disrupted animals. However, these declines were not as complete as the ones observed previously when the hepatic SCAP gene was disrupted by the same inducible Cre recombinase (20). The partial nature of the declines may indicate that the disruption of S1P was not complete enough to abolish S1P function, or they may indicate that another protease can partially substitute for S1P in liver.
In a previous study from our laboratory, SCAP was conditionally disrupted after injection of pIpC into transgenic animals bearing the same MX1-Cre gene used in the current studies (20). The degree of recombination of SCAP was more than 95%, the decline in SCAP mRNA was at least 80%, and the mRNAs for the target genes declined by at least 70% (LDL receptor and HMG CoA synthase) and as much as 95% (HMG CoA reductase, acetyl CoA carboxylase, fatty acid synthase, stearoyl CoA desaturase-1). In hepatocytes isolated from these mice, the rates of incorporation of [14C]acetate into cellular [14C]cholesterol and fatty acids declined by 84–87%, and the secretion of [14C]cholesterol and fatty acids into the culture medium declined by 90–93% (20).
The first indication of a different result in S1P-deficient animals came from an examination of S1Pf/f;MX1-Cre mice that were not injected with pIpC. In these animals there was no evidence of recombination, yet the amount of S1P mRNA was reduced by 80–85% when compared with wild-type mice (Table 2). We postulate that the reduction in S1P mRNA was caused by an interference in gene transcription caused by the presence of the neo cassette. The amount of the active C form of S1P protein was also reduced, but it did not decline as much as the precursor A form (Fig. 2a, lane 3). Despite this reduction in S1P, the amounts of nuclear SREBPs were normal (Fig. 2c, lanes 3 and 7), and the amounts of the target mRNAs did not decline (Table 2). These data could be explained if active S1P is normally present in excess, and a 50–80% reduction still leaves sufficient enzyme to cleave SREBPs.
When S1Pf/f;MX1-Cre mice were treated with pIpC, the degree of recombination ranged between 77% and 90%, even after multiple injections. This finding contrasts with the recombination frequency of >95% that was obtained for SCAP. Recombination in S1P reduced S1P mRNA levels by an additional 50–66%, resulting in an overall decline of 95% (Table 2). The active C form of S1P also declined (Fig. 2a, lane 4), and we observed a definite decline in the precursor and nuclear forms of SREBP-1 and -2 (Fig. 2c, lanes 4 and 8). There was also a decline in the mRNAs for SREBP-1c and -2, and a slight decline in the mRNA for SREBP-1a (Fig. 2d and Table 2). The decline in SREBP mRNAs is likely a consequence of the decline in nuclear SREBPs because the promoters for both of these genes contain functional SRE elements (24, 25, 32). In addition, transcription of the SREBP-1c gene requires LXR, a nuclear hormone receptor that is activated by an oxysterol, the level of which decreases when cholesterol synthesis declines (23, 33).
As a result of the decline in nuclear SREBPs, there was a decline in all of the mRNAs for SREBP target genes. However, in contrast to the decline observed with SCAP disruption (70–95%), the declines with S1P disruption varied from about 40% (HMG CoA reductase) to about 70% (farnesyl diphosphate synthase) (Table 2). Moreover, the rates of cholesterol and fatty acid synthesis, as measured in isolated hepatocytes declined by only 64–83% (Fig. 3) as contrasted with the 84–93% reduction observed previously in the SCAP knockout mice (20). The partial nature of these declines indicates that some functional SREBPs were present in the nucleus of the S1P knockout mice.
The difference in phenotypic severity between the SCAP and S1P knockouts may relate to the different roles of the two proteins in SREBP processing. SCAP forms complexes with SREBPs, and therefore it acts stoichiometrically (6). A 90% reduction in SCAP would lead directly to a 90% reduction in SCAP/SREBP complexes, which are the substrates for S1P. On the other hand, S1P acts catalytically. Like most other enzymes, it is likely that S1P is present in great excess and that SREBP processing is not reduced severely until the amount of S1P declines below 5% of the wild-type value. Previous studies of enzyme deficiency states reveal that as little as 5% of residual activity can prevent abnormal phenotypes in some diseases (34). The impact of SCAP deficiency is heightened by the instability of the SREBPs in the absence of SCAP. Thus, in SCAP knockout mice the levels of the SREBP precursors were reduced even more than would be predicted from the decline in SREBP mRNAs (20).
The partial effects of Cre-mediated recombination of S1P in liver contrast with the complete effects observed in mutant Chinese hamster ovary cells that fail to express S1P (12). These mutant cells synthesize virtually no cholesterol, and they must be supplied with this substance exogenously. This difference may reflect incomplete disruption of the hepatic S1P gene, or it may indicate that hepatocytes, but not Chinese hamster ovary cells, express another protease that can substitute for SREBP cleavage.
Disruption of S1P reduced the amount of LDL receptor mRNA by ≈50% (Table 2 and Fig. 4b). This was reflected by a decline of ≈50% in the receptor-mediated clearance of i.v. injected 125I-LDL from plasma (Fig. 4a). Despite this reduction in LDL clearance, the total concentration of cholesterol fell when S1P was disrupted (Table 1). FPLC analysis revealed that this decline was mostly attributable to a fall in HDL levels (data not shown). There was also a decline in the small amount of LDL that is normally present in mouse plasma. A decline in LDL levels in the face of reduced clearance suggests that the production of LDL was reduced in the mice with the disrupted S1P. LDL metabolism is quantitatively very different in humans and mice (26, 30). Nevertheless, if humans respond to S1P deficiency in the same way that mice respond, the current data predict a beneficial LDL-lowering effect of inhibitors of S1P, provided that a near-complete inhibition of enzyme activity can be obtained.
Supplementary Material
Acknowledgments
We thank Richard Gibson, Scott Clark, Liz Lummus, Norma Anderson, Amy Cox, Judy Sanchez, and Jeff Cormier for excellent technical assistance. This work was supported by grants from the National Institutes of Health (HL-20948), the Perot Family Foundation, the Moss Heart Foundation, and the W. M. Keck Foundation. J.D.H. is a Pew Scholar in the Biomedical Sciences and is the recipient of an Established Investigator Grant from the American Heart Association and a Research Scholar Award from the American Digestive Health Foundation Industry.
Abbreviations
- apoB
apolipoprotein B
- ES
embryonic stem
- HMG CoA
3-hydroxy-3-methylglutaryl coenzyme A
- LDL
low density lipoprotein
- pIpC
polyinosinic/polycytidylic acid
- Prm
protamine-1 promoter
- RT-PCR
reverse transcriptase–PCR
- S1P
Site-1 protease
- SCAP
SREBP cleavage-activating protein
- SREBP
sterol regulatory element-binding protein
References
- 1.Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakai J, Nohturfft A, Goldstein J L, Brown M S. J Biol Chem. 1998;273:5785–5793. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 3.DeBose-Boyd R A, Brown M S, Li W-P, Nohturfft A, Goldstein J L, Espenshade P J. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 4.Nohturfft A, Yabe D, Goldstein J L, Brown M S, Espenshade P J. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 5.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 6.Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 7.Duncan E A, Brown M S, Goldstein J L, Sakai J. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- 8.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M T, Chang T-Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 9.Duncan E A, Davé U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 10.Espenshade P J, Cheng D, Goldstein J L, Brown M S. J Biol Chem. 1999;274:22795–22804. doi: 10.1074/jbc.274.32.22795. [DOI] [PubMed] [Google Scholar]
- 11.Cheng D, Espenshade P J, Slaughter C A, Jaen J C, Brown M S, Goldstein J L. J Biol Chem. 1999;274:22805–22812. doi: 10.1074/jbc.274.32.22805. [DOI] [PubMed] [Google Scholar]
- 12.Rawson R B, Cheng D, Brown M S, Goldstein J L. J Biol Chem. 1998;273:28261–28269. doi: 10.1074/jbc.273.43.28261. [DOI] [PubMed] [Google Scholar]
- 13.Rawson R B, DeBose-Boyd R, Goldstein J L, Brown M S. J Biol Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 14.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M S. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimomura I, Bashmakov Y, Ikemoto S, Horton J D, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton J D, Shimomura I, Brown M S, Hammer R E, Goldstein J L, Shimano H. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng Z, Otani H, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda M, Korn B S, Hammer R E, Moon Y-A, Komuro R, Horton J D, Goldstein J L, Brown M S, Shimomura I. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 22.Rohlmann A, Gotthardt M, Hammer R E, Herz J. J Clin Invest. 1998;101:689–695. doi: 10.1172/JCI1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBose-Boyd R A, Ou J, Goldstein J L, Brown M S. Proc Natl Acad Sci USA. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 25.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty A H, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 26.Ishibashi S, Brown M S, Goldstein J L, Gerard R D, Hammer R E, Herz J. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimano H, Shimomura I, Hammer R E, Herz J, Goldstein J L, Brown M S, Horton J D. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Gorman S, Dagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 30.Horton J D, Shimano H, Hamilton R L, Brown M S, Goldstein J L. J Clin Invest. 1999;103:1067–1076. doi: 10.1172/JCI6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell K J, Pinson K I, Kelly O G, Brennan J, Zupicich J, Scherz P, Leighton P A, Goodrich L V, Lu X, Avery B J, et al. Nat Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- 32.Miserez A R, Cao G, Probst L, Hobbs H H. Genomics. 1997;40:31–40. doi: 10.1006/geno.1996.4525. [DOI] [PubMed] [Google Scholar]
- 33.Repa J J, Liang G, Ou J, Bashmakov Y, Lobaccaro J-M A, Shimomura I, Shan B, Brown M S, Goldstein J L, Mangelsdorf D J. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaudet A L, Scriver C R, Sly W S, Valle D. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 3–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.