Abstract
A large number of functional hepatocytes is required for bioartificial liver therapy. Simian virus 40 T-antigen (SV40T) has been previously reported to improve the immortalized proliferation of primary hepatocytes to generate a sufficient number of cells; however, these long-term immortalized hepatocytes may induce further malignant transformation in vivo. In the present study, the SV40T immortalization gene and two suicide genes, herpes simplex virus thymidine kinase (HSV-tk) and cytosine deaminase (CD), were transducted into primary hepatocytes to construct a novel type of Cre/LoxP-mediated reversible immortalized hepatocyte line. Polymerase chain reaction analysis and western blotting confirmed that the SV40T, HSV-tk and CD genes were successfully inserted into hepatic progenitor cells and their expression was controlled by Cre/LoxP recombination. Total removal of SV40T could be achieved via the ganciclovir (GCV)/HSV-tk suicide system. Cells maintained their biosafety in vivo with CD gene expression and 5-fluoro-cytosine (5-FC) induced cell death. Following transplantation into the carbon tetrachloride (CCl4) model group, the majority of cells had survived after 14 days post-implantation and a number of the cells had transported into the liver parenchyma. When compared with the CCl4 model group, the transplanted cells repaired the liver biochemical index and pathological structure markedly. Thus, the present study reports a novel reversible immortalized hepatocyte with double suicide genes, which exhibited the cellular phenotype and recovery function of normal liver cells. This method maximally guaranteed the biological safety of immortalized hepatocytes for in vivo application, providing a reliable, safe and ideal cell material for the artificial liver technique.
Keywords: immortalization, simian virus 40 T-antigen, suicide gene, herpes simplex virus thymidine kinase, cytosine deaminase
Introduction
Severe liver failure is the most common type of liver disease syndrome; it is associated with a high mortality rate and is therefore a serious threat to human health. The most effective treatment for liver failure is liver transplantation (1,2). However, <10% of patients have the opportunity to receive liver transplantation due to the shortage of donor livers available. As the liver function of the patients markedly deteriorates over time, a number of patients perish while waiting for a donor liver (3). Bioartificial liver (BAL) is an extracorporeal bioarti-ficial organ device based on functional liver cell culture, which is able to temporarily replace liver detoxification, synthesis, secretion and conversion among other functions. Patients who receive BAL treatment may have an increased survival rate while waiting for liver transplantation and may also have improved liver regeneration (4–6).
In BAL, a large number of hepatocytes is required and the cells used should have a mature function of protein synthesis, metabolism and detoxification. However, primary hepatocytes do not generate enough cells and hepatic cells derived from stem cells have limited liver abilities. Therefore, current research has focused on how to promote the in vitro proliferation and differentiation of hepatocytes. Simian virus 40 T-antigen (SV40T) is known to improve the immortalized proliferation of primary hepatocytes in order to produce a sufficient number of cells; however, long-term immortalized hepatocytes may induce further malignant transformation in vivo. In recent years, many studies have focused on how to decrease the risks associated with the application of SV40T-immortalized hepa-tocytes in cell therapy (7,8).
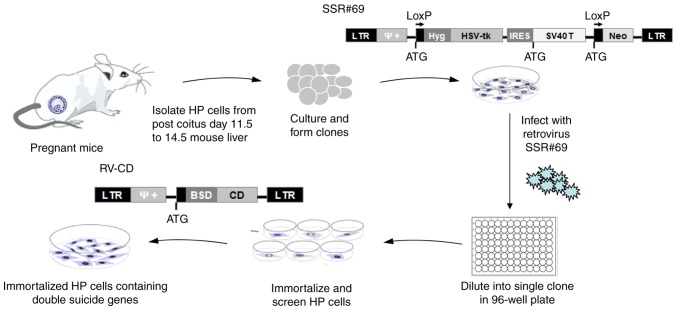
In view of the issues associated with current reversible immortalization methods, the aim of the present study was to transduct the SV40T immortalization gene and two suicide genes, herpes simplex virus thymidine kinase (HSV-tk) and cytosine deaminase (CD), into primary hepatocytes using a retrovirus to construct a new type of Cre/LoxP-mediated reversible immortalized hepatocyte cell line (Fig. 1). In this system, Cre-mediated site-specific recombination technology was used to control the expression of the SV40T and HSV-tk genes. This cell line is able to proliferate constantly in vitro, exhibiting the normal phenotype and function of liver cells; in addition, the SV40T immortalized gene, which is suspected to be carcinogenic and an inhibitor of liver cell function, may be knocked out before in vivo application. Removal of SV40T may be achieved via the HSV-tk/ganci-clovir (GCV) system (9). In addition, exogenous cells in vivo may be selectively targeted by the CD/5-fluorocytosine (5-FC) system to induce cell death in order to avoid malignant transformation (10). Thus, the technique of the present study may provide a stable, secure and reliable source of liver cells for BAL technology.
Figure 1.
Flow diagram of the experimental procedure to produce the reversibly immortalized HP cells containing double suicide genes. HP, hepatic progenitor; LTR, long terminal repeat; Hyg, hygromycin; SV40T, simian virus 40 T-antigen; HSV-tk, herpes simplex virus thymidine kinase; BSD, blasticidin S; CD, cytosine deaminase; IRES, internal ribosome entry site; Neo, neomycin; RV-CD, retrovirus containing CD gene; SSR#69, retroviral vector expressing SV40T and Hyg-resistance genes flanked by paired LoxP recombination targets (12).
Materials and methods
Cell culture and chemicals
The hepatic progenitor HP14-19 cell line expressing the HSV-tk suicide gene and SV40T immortalized gene was constructed previously (11,12). Cells were cultured in complete Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C and 5% CO2.
Construction of the pSEB-CD plasmid
A ~1,300 bp fragment of the full-length CD gene from Escherichia coli JM107 genomic DNA was amplified using the polymerase chain reaction (PCR). A BamHI restriction site was added to the 5′-end of the forward primer, and a SalI restriction site was added to the 5′-end of the reverse primer. The primer sequences were as follows: 5′-CGAGGATCCATGTCGAATAACGCTTTAC-3′ (forward) and 5′-GCTGTCGACTCAACGTTTGTAATCGATG-3′ (reverse). The target CD gene fragment was amplified by touch-down PCR with a reaction system consisting of 4 µg DNA template, 2 µl 330 µmol/l upstream and downstream primers, 25 µl 2X high-fidelity master mix (Novoprotein Technology Corporation, Shanghai, China), and double-distilled water to 50 µl, and the following thermocycling conditions: 95°C for 3 min, followed by 10 cycles of 92°C for 20 sec, 65°C for 20 sec and 72°C for 120 sec (with 1°C decrease per cycle), and then 25 cycles of 92°C for 20 sec, 55°C for 20 sec and 72°C for 120 sec, and finally 72°C for 3 min. Following detection by 1% agarose gel electrophoresis, the PCR products were digested with BamHI/SalI and then directionally cloned into the BglII/SalI-digested pSEB-C3F retroviral vector (Molecular Oncology Laboratory, The University of Chicago Medical Center, Chicago, IL, USA). The constructed pSEB-CD plasmid was selected by colony PCR, and confirmed by full-length PCR, double enzyme digestion (EcoRI and SalI) and sequencing (Huada Gene, Inc., Shenzhen, China). All enzymes were purchased from Takara Biotechnology Co., Ltd., Dalian, China.
Establishment of the HP14-19-CD stable cell line
The pSEB-CD plasmid expressing a blasticidin S (BSD) selection marker and pCLAmpho mammalian expression vector (Molecular Oncology Laboratory, The University of Chicago Medical Center) were co-transfected into 293 cells to package the retrovirus at 37°C and 5% CO2 for 7 days. HP14-19 cells were then seeded into a T-25 flask and infected with retrovirus at 37°C and 5% CO2 for 7 days; they were then selected in the presence of 3 µg/ml BSD (Invitrogen; Thermo Fisher Scientific, Inc.) for 14 days.
Semi-quantitative (sq)-PCR analysis
Total RNA from HP14-19 cells and HP-14-19-CD cells was extracted using an RNA Extraction kit (BioTeke Corporation, Beijing, China) at the indicated times and reverse-transcribed into cDNA using Superscript II reverse transcriptase (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The cDNA samples were diluted 5–10-fold and subjected to PCR amplifi-cation with a reaction system consisting of 2 µg DNA template, 1 µl 330 µmol/l upstream and downstream primers, 7.5 µl 2X Taq master mix (Novoprotein Technology Corporation), and double-distilled water to 15 µl. PCR primers were designed using the Primer3 program (Table I). A touch-down PCR protocol was performed with the following thermocycling conditions: 95°C for 3 min, then 10 cycles of 92°C for 20 sec, 65°C for 20 sec and 72°C for 20 sec (with 1°C decrease per cycle), followed by 30 cycles of 92°C for 20 sec, 55°C for 20 sec and 72°C for 20 sec, and finally 72°C for 3 min. The PCR products were electrophoresed on a 1.5% agarose gel. All samples were normalized to the expression level of GAPDH.
Table I.
Primers for the reverse transcription-polymerase chain reaction.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| GAPDH | TTCCAGGAGCGAGACCC | CCACAGCTTTCCAGAGGG |
| SV40T | ATTTGCCTTCAGGTCAGG | ACTCCAATTCCCATAGCC |
| HSV-tk | CATCTACACCACACAACACC | ATGCTGCCCATAAGGTATC |
| CD | CTGGATGCCGAACAAGG C | GAAACATCGACATGGGTAC |
SV40T, simian virus 40 T-antigen; HSV-tk, herpes simplex virus thymidine kinase; CD, cytosine deaminase.
Western blot analysis
Cells were lysed in radioimmunopre-cipitation assay buffer with phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology, Haimen, China). The protein concentrations were determined with a Bicinchoninic Acid Protein assay kit (Beyotime Institute of Biotechnology). Total protein (20 µg/group) was separated by SDS-PAGE (SV40T and HSV-tk protein were separated on a 10% gel; CD and β-actin proteins were separated on a 15% gel). Beyotime Institute of Biotechnology) and then transferred onto a poly-vinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). The membrane was blocked with 5% non-fat milk (Beyotime Institute of Biotechnology) in Tris-buffered saline containing 0.1% Tween-20 (TBST) at room temperature for 1 h and then incubated at 4°C overnight with primary antibodies against SV40T (1:200; cat. no. sc-148; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), HSV-tk (1:200; cat. no. sc-28037; Santa Cruz Biotechnology, Inc.), CD (1:1,000; cat. no. ab35251; Abcam, Cambridge, MA, USA) and β-actin (1:200; cat. no. sc-47718; Santa Cruz Biotechnology, Inc.). Following washing with TBST, the membrane was incubated with the appropriate secondary antibody conjugated to horseradish peroxidase (goat anti-mouse, 1:1,000, cat. no. sc-2302 and goat anti-rabbit, 1:1,000, cat no. sc-2004; both from Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. Finally, the protein expression levels were determined using an enhanced chemiluminescent substrate (Nanjing Kaiji Biotechnology Co., Nanjing, China) and exposed under the Syngene G-Box Imaging system (Syngene Europe, Cambridge, UK).
Cell viability assay
HP14-19-CD cells were infected with adenovirus Ad-Cre (12) to remove the HSV-tk gene, then cultured in complete Dulbecco's modified Eagle's medium with 10% FBS containing various concentrations (0, 0.1, 0.2, 0.5, 1, 2, 5 and 10 µg/ml) of GCV (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). HP14-19 and HP14-19-CD cells were treated with various concentrations (0, 10, 20, 40 and 80 µg/ml) of 5-FC (Sigma-Aldrich; Merck KGaA). HP14-19-CD cells which were infected with Ad-green fluorescent protein served as control group. Crystal violet staining was performed in order to assess cell viability. Cells were incubated in 24-well plates at 2.0×104 cells/well and analyzed after 1 week. Briefly, the culture medium was removed and the cells were fixed with 4% paraformaldehyde at room temperature for 10 min, then stained with 0.05% crystal violet for 30 min. Following washing with tap water, cells were dried on filter paper. The blue dye was dissolved in 500 µl methanol and determined at an excitation wavelength of 540 nm using a Multimode Microplate Reader (Thermo Fisher Scientific, Inc.). Three independent experiments were performed in triplicate. The mean and standard deviation were calculated.
Cell implantation and in vivo imaging
The use and care of animals was approved by the Institutional Animal Care and Use Committee of The Children's Hospital of Chongqing Medical University (Chongqing, China). Nude mice (3 males and 3 females, 5–6 weeks of age, 22–23 g) were purchased from Tengxin Institute of Biotechnology (Chongqing, China). The animals were kept at room temperature between 22 and 26°C with 40–60% relative humidity and a 12-h light/12-h dark cycle with free access to food and water. HP14-19 and HP14-19-CD cells were labelled with adenovirus (Ad)-firefly luciferase and then subcutaneously inoculated in the front and rear notum on the left and/or right side of the 6 nude mice (1×106 cells per injection). 5-FC (300 mg/kg per day) was intragastrically administered to the nude mice in vivo every 2 days following cell transplantation. At 0, 5 and 10 days after implantation, mice were intraperitoneally injected with 0.1 ml D-luciferin (Gold Biotechnology, Inc., St Louis, MO, USA) at 2 mg/ml, and visualized using the IVIS-200 optical in vivo imaging system (Xenogen Corporation, Alameda, CA, USA) to dynamically observe the luciferase signals and determine the survival rate of cells.
Liver index and blood biochemical detection
A total of 21 nude mice (all male, 5–6 weeks of age, 22–23 g) were purchased from Tengxin Institute of Biotechnology. The animals were kept at room temperature between 22 and 26°C with 40–60% relative humidity and a 12-h light/12-h dark cycle, and were randomly divided into a normal group (n=3), a 2% carbon tetrachloride (CCl4) group (n=9) and a CCl4+cells group (n=9). A total of 18 nude mice were used to construct an acute liver injury model established via 2% CCl4 gavage. Considering the large amount of haemorrhagia during the procedure and a typically low survival rate following portal vein injection, it was elected to transplant cells via the splenic vein (13). Cells were pre-labeled with Hoechst 33342 (Beyotime Institute of Biotechnology) 24 h after liver injury (14,15). The liver index (liver wet weight/body weight ×100%), serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in each group were detected using assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) at the indicated time points.
Histochemical staining
Following sacrifice of the mice, liver tissue specimens were obtained and fixed in 4% para-formaldehyde, embedded in paraffin following dehydration, and serially cut into 5-µm sections. Serial sections were then stained with 0.2% hematoxylin and 0.5% eosin (H&E) at room temperature, and images were captured at ×200 magnification under a light microscope (Nikon Corporation, Tokyo, Japan).
Statistical analysis
All data are presented as the mean ± standard deviation and were analyzed using SPSS software (version 19.0; IBM Corp., Armonk, NY, USA). Statistical analysis was performed using the two-tailed Student's t-test to determine significant differences between two groups, and one-way analysis of variance followed by Tukey's honest significant difference post hoc test was performed to determined significant differences among at least three groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Construction of pSEB-CD plasmid and HP14-19-CD stable cell line
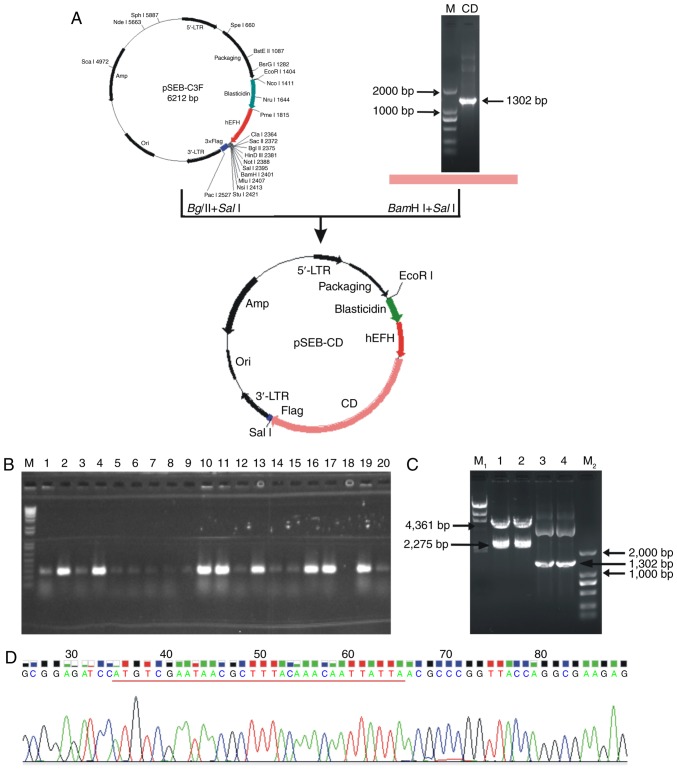
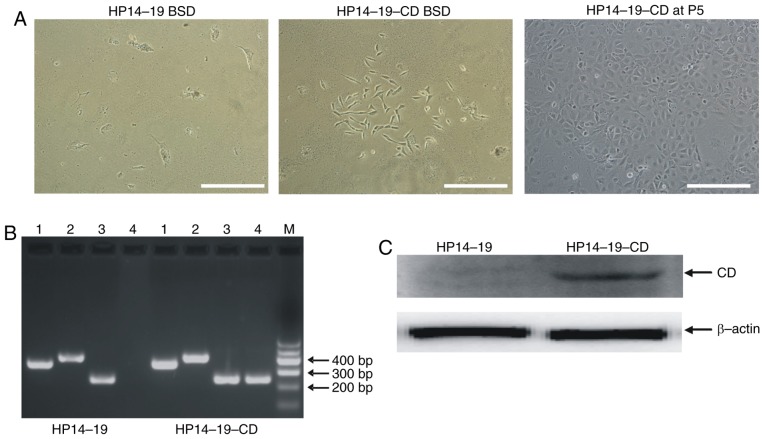
A ~1,300 bp fragment of the full-length CD gene from E. coli JM107 genomic DNA was amplified using PCR. This ~1,300 bp DNA fragment could be removed from the constructed pSEB-CD plasmid by digestion with SalI and BamHI restriction enzymes. In addition, the target genes were confirmed to be correctly cloned in the pSEB-C3F vector by gene sequencing and matched the CD sequence in GenBank® (www.ncbi.nlm.nih.gov/genbank) (Fig. 2). The HP14-19 cell line contains the hygromycin-resistant/HSV-tk fusion gene and immortalized SV40T, which can be removed by Cre/LoxP recombination. As the pSEB-C3F vector contained the BSD resistance gene, following infection with the pSEB-CD-produced retrovirus, the cells also acquired the BSD resistance gene and thus were able to survive in the presence of BSD (Fig. 3A). HP14-19 and HP14-19-CD cells expressed the SV40T and HSV-tk genes; however, only HP14-19-CD cells stably exhibited CD mRNA and protein expression (Fig. 3B). Therefore, the procedure of the present study successfully established long-term cell culture of hepatic progenitor cells containing immortalizing SV40T and double suicide genes; the HSV-tk and stable CD genes could also be subsequently knocked out.
Figure 2.
Successful construction of the pSEB-CD plasmid. (A) Construction process and structure of the pSEB-CD plasmid. A ~1,300 bp PCR product of full-length CD gene was digested with BamHI/SalI and then directionally cloned into the BglII/SalI-digested pSEB-C3F retroviral vector to construct pSEB-CD plasmid. (B) Positive recombinants were selected by colony PCR, colonies with positive specific PCR products were amplified for plasmid extraction. Lane M, DNA marker; lane 2, 4, 10, 11, 13, 16, 17, 19, positive PCR products specific for CD genes. (C) The constructed pSEB-CD plasmids were confirmed by full-length PCR and enzyme digestion. Lane M1, λ-HindIII marker; lanes 1 and 2, pSEB-CD plasmid was digested with SalI and EcoRI; lanes 3 and 4, full-length CD gene was amplified from pSEB-CD plasmid; lane M2, D2000 marker. (D) The target gene was confirmed to be successfully cloned in the pSEB-C3F vector using gene sequencing. CD, cytosine deaminase; PCR, polymerase chain reaction.
Figure 3.
Establishment of the HP14-19-CD stable cell line. (A) The pSEB-CD plasmid expressing a blasticidin selection marker and pCLAmpho mammalian expression vector were co-transfected into 293 cells to package the retrovirus. HP14-19 cells were seeded into a T-25 flask and infected with retrovirus for 7 days, prior to selection in the presence of 3 µg/ml BSD for 14 days. Scale bar, 200 µm. (B) Polymerase chain reaction analysis of the suicide-associated genes. At least three independent PCR experiments were performed to detect the expression of genes of interest. Lane 1, GAPDH; lane 2, SV40T; lane 3, HSV-tk; lane 4, CD; lane M, DNA marker. Representative PCR results are presented. (C) Western blot analysis of CD expression, normalized to β-actin. Representative results are presented. BSD, blasticidin S; CD, cytosine deaminase; P5, fifth generation.
Separate functions are exerted by the two suicide genes
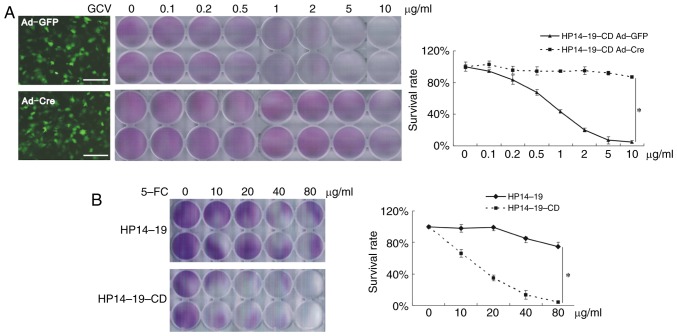
As the HP14-19-CD cells contained the HSV-tk suicide gene, cell death was dose-dependently induced by the addition of GCV. HSV-tk expression was controlled by Cre-LoxP site-specific recombination. Ad-mediated Cre expression was able to inhibit the expression of HSV-tk, thus Ad-Cre-infected cells were able to survive, even in the presence of high-dose GCV medium (P<0.05; Fig. 4A). The CD suicide gene codes for cytosine deaminase, which converts the non-toxic compound 5-FC into 5-fluorouracil to induce cell death. As presented in Fig. 3B, when exposed to various doses of 5-FC, the rate of HP14-19-CD cell death was significantly increased compared with that of HP14-19 cells (P<0.05; Fig. 4B).
Figure 4.
Crystal violet staining was conducted to assess cell viability. (A) Ad-Cre- and Ad-GFP-infected cells were stained with crystal violet solution for 10 min, and the absorbance was measured at 540 nm following dissolution of the dye. (B) HP14-19 and HP14-19-CD cells were stained with crystal violet solution for 10 min, and the absorbance was measured at 540 nm following dissolution of the dye. Scale bar, 200 µm. Three independent experiments were performed in duplicate, and representative results are presented. *P<0.05. Ad, adenovirus; GFP, green fluorescent protein; CD, cytosine deaminase; 5-FC, 5-fluorocytosine; GCV, ganciclovir.
The efficiency of Cre-LoxP-mediated recombination was not 100%, thus Ad-Cre-infected HP14-19-CD cells still expressed SV40T and HSV-tk mRNA and protein at a low level. However, using the negative selection with GCV, all cells expressing the SV40T immortalization and HSV-tk genes were removed, and the selected cells still expressed the CD gene and protein (Fig. 5A and B).
Figure 5.
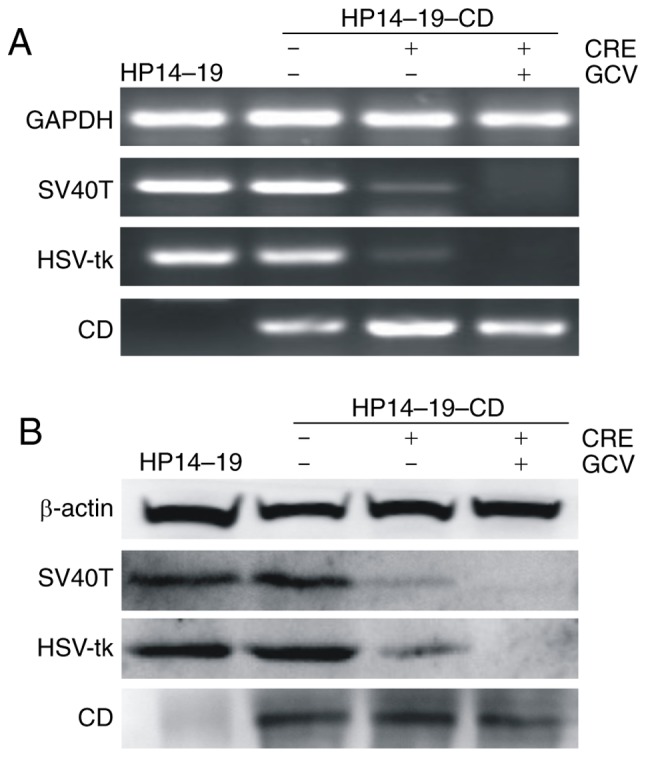
Cell selection. (A) mRNA expression of SV40T, HSV-tk and CD genes were exhibited by polymerase chain reaction analysis, and representative results are presented. (B) Protein levels of SV40T, HSV-tk and CD were detected by western blot analysis. Representative results are presented. SV40T, simian virus 40 T-antigen; HSV-tk, herpes simplex virus thymidine kinase; CD, cytosine deaminase; GCV, ganciclovir.
Next, the effect of the CD gene on cell suicide was investigated in vivo. HP14-19 and HP14-19-CD cells were labelled with Ad-firefly luciferase and then subcutaneously inoculated into nude mice. Optical in vivo imaging was employed to observe luciferase signaling in surviving cells. As presented in Fig. 6, the original luciferase signal of HP14-19 and HP14-19-CD cells was markedly detectable on day 0; the luciferase signal of HP14-19 cells decreased slowly and remained easily detectable on day 10. By contrast, the luciferase signal exhibited by HP14-19-CD cells was weaker compared with that of HP14-19 cells, and was almost impossible to detect on day 10. These results demonstrated that the immortalization of HP14-19-CD cells could be successfully altered, while maintaining its biosafety in vivo with CD gene expression.
Figure 6.
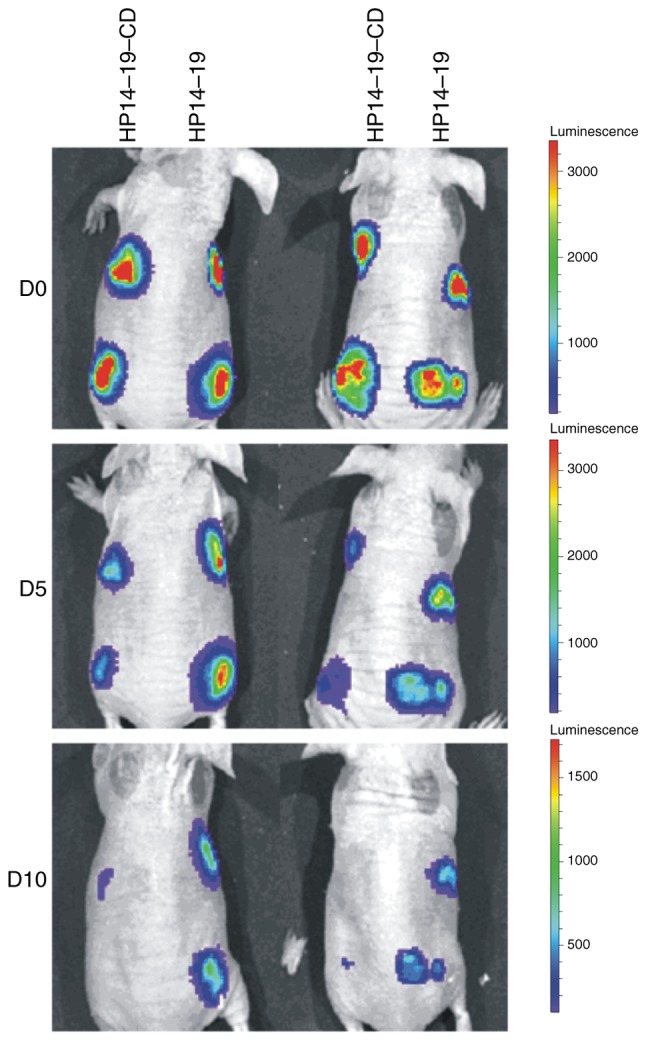
Effect of CD gene expression on cell suicide in vivo. HP14-19 and HP14-19-CD cells were labelled with adenovirus-firefly luciferase and then subcutaneously inoculated into the front and rear notum on the left and/or right side of 6 nude mice (1×106 cells/injection). 5-FC at 300 mg/kg per day was intragastrically administered to the nude mice in vivo every 2 days following cell transplantation. On days 0, 5 and 10 days following implantation, optical in vivo imaging was performed to dynamically observe luciferase signaling in the surviving cells. Representative results from 2 nude mice are presented. Ad, adenovirus; CD, cytosine deaminase; D, day.
HP14-19-CD cells repair the hepatic function of mice with acute liver failure (ALF)
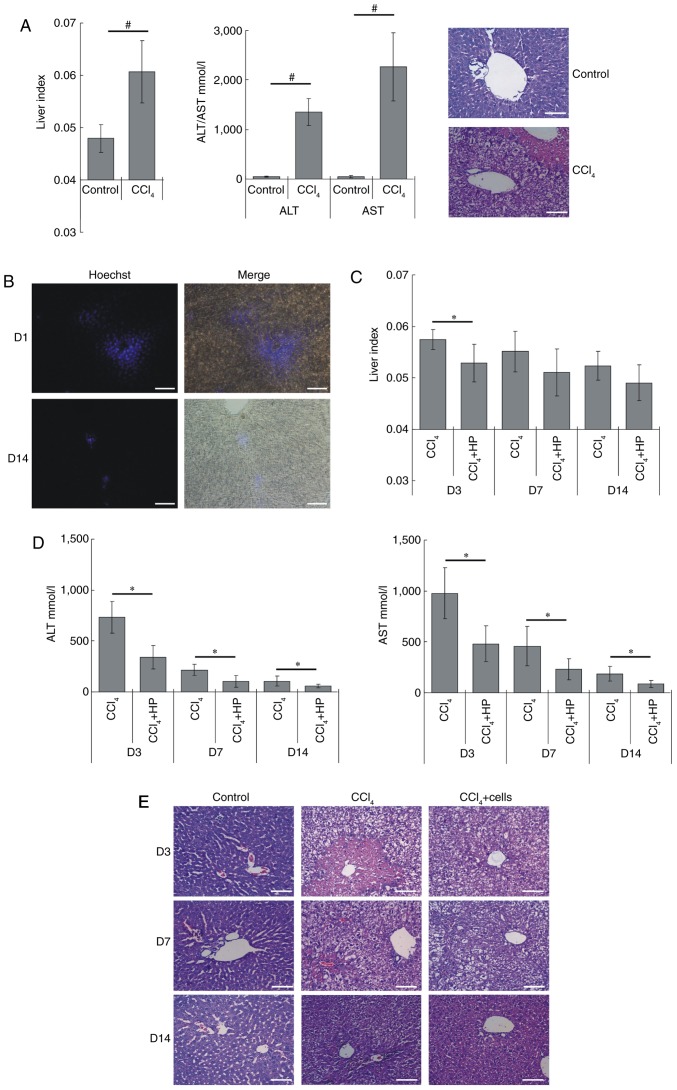
The recovery efficiency of HP14-19-CD cells in vivo was next investigated. First, an ALF model was successfully established using 2% CCl4 gavage. Compared with normal nude mice, the liver index of the ALF model was significantly increased (P<0.05), and the levels of ALT and AST also increased >10-fold (P<0.05). H&E staining revealed that there was hepatocyte degeneration in the ALF liver tissues, as well as hepatocyte steatosis, vacuolar degeneration, focal necrosis, hepatic cord disorder, and nuclear condensation or disappearance (Fig. 7A). Hoechst 33342-pre-labelled cells were injected into the splenic vein, and the majority of cells were distributed around the portal vein in the hepatic portal area on day 1 post-transplantation. After 14 days, the majority of the cells had survived and a proportion of cells had migrated into the liver parenchyma (Fig. 7B). The liver has a good ability to self-repair, thus, when the model mice no longer received CCl4 stimulation, gradually the liver index, ALT/AST levels and hepatic pathological changes recovered to a certain extent. The liver index of the cell+CCl4 group was consistently lower compared with that of the CCl4 group, with a significant difference observed on day 3; however, no significant differences were identified following day 7 (Fig. 7C). The ALT and AST levels of the cell+CCl4 group were significantly decreased compared with the CCl4 group at each time point (Fig. 7D). The H&E staining results presented in Fig. 7E revealed that, when compared with the CCl4 model group, the transplanted cells markedly repaired the liver pathological structure. Damaged hepatocytes were markedly repaired on the third day following cell injections, whereas the structure of the hepatic cords had almost completely recovered after 14 days. Therefore, these results indicated that HP14-19-CD cell transplantation may improve hepatic recovery.
Figure 7.
HP14-19-CD cell transplantation improves liver functional recovery in mice with acute liver injury. (A) Acute liver failure model was established by using 2% CCl4 gavage. After 24 h, the liver index (liver wet weight/body weight ×100%), serum levels of ALT and AST, and the pathological changes in liver were determined to evaluate liver structure and function. Scale bar, 200 µm. (B) Hoechst 33342-pre-labelled cells were injected into the splenic vein. The survival and distribution of exogenous cells were detected on day 1 and at 14 days after transplantation. Scale bar, 200 µm. (C) The liver index of each group was determined at the indicated time points. (D) The serum levels ALT and AST in each group were detected at the indicated time points. (E) The specimens were fixed in 4% paraformaldehyde, embedded in paraffin following dehydration and serially cut into 5-µm-thick sections. Serial sections were stained with hematoxylin and eosin. Scale bar, 200 µm. #P<0.05 vs. control group; *P<0.05 vs. CCl4 model group. ALT, alanine aminotransferase; AST, aspartate amino-transferase; CD, cytosine deaminase; D, day.
Discussion
At present, identifying a reliable cell source is the most important issue to be addressed in BAL technology (16). To date, cell sources for liver cell-based therapies have included autologous, allogeneic or xenogeneic mature hepatic cells, differentiated hepatic cells from bone marrow hematopoietic stem cells, embryonic liver cells and hepatic oval cells (4,17,18). For BAL application, primary hepatic cells must be amplified to several orders of magnitude in vitro, and must also be capable of protein synthesis and metabolic detoxification functioning. However, primary hepatocytes lack the sufficient number of cells required to be a reliable cell source and exhibit a poor proliferative ability in vitro (19,20). Furthermore, stem cell differentiated hepatic-like cells have a limited ability to exert specific liver functions. To this end, previous studies have focused on how to obtain hepatocytes with good proliferation abilities and normal liver functions in vitro in order to identify a stable and safe source of cells for BAL therapy (21,22).
SV40 is a double-stranded DNA virus (23), in which SV40T is a key early protein that is essential for driving viral replication and inducing cellular transformation (24). SV40T stimulates the entry of quiescent cells into the cell cycle by binding to the host key cell cycle regulators retinoblastoma protein 1 and tumor protein 53, then regulating cell proliferation in order to establish immortalized cells. A previous study demonstrated that the delivery of SV40T into primary adult hepatocytes is able to establish immortalized liver cell lines with high levels of differentiation, and typical characteristics and similar biological functions of primary hepatocytes (25). The immortalized cells may be cultured long-term in vitro, and secrete albumin, ALT, AST and lactate dehydrogenase; they also express various liver-specific markers (including apolipoprotein B, tyrosine aminotransferase and cytoker-atin 18). Therefore, a sufficient number of hepatocytes may be obtained using this method and thus this strategy provides a sufficient source of cells for BAL (7,8,25,26). However, these long-term immortalized hepatocytes possess certain disadvantages. As an oncogene, SV40T may induce further malignant transformation, thereby increasing the tumorigenic risk in vivo to unacceptable levels due to continued proliferation and cell transformation (27). Therefore, previous studies have investigated the safety of the clinical application of SV40T-immortalized hepatocytes (8,28–30).
The Cre-LoxP recombination system is the most commonly used method to produce reversible immortalized hepatocytes. The SV40T gene is flanked by two homodromous LoxP sites (5′-ATA ACT TCG TAT AAT GTA TGC TAT ACG AAG TTAT-3′); by using Cre-LoxP site-specific recombination, the temporal expression of the SV40T gene is removed, thereby reverting the levels of immortalized proliferation to those observed prior to cell transplantation, such as that reported previously in the NKNT-3 cell line (31). Another approach is to integrate suicide genes into immortalized liver cells, then induce cell death using prodrugs once malignant transformation has occurred. Kobayashi et al (32) transduced SV40T into fetal liver cells, then inserted the HSV-tk gene in order to establish the tightly regulated immortalized hepatocyte line OUMS-29/HSV-tk. The cells exhibited a normal liver cell phenotype and physiological function; they were also markedly sensitive to GCV, thus serving as a safeguard for BAL clinical application.
However, the strategies currently available still have disadvantages. The activity of Cre and the removal efficiency of LoxP recombination cannot be controlled, and so the removal of the immortalized gene cannot be guaranteed. In the present study, cells with Cre-LoxP recombination still exhibited low levels of SV40T expression. Therefore, the levels of tumori-genesis, the risk of cell transformation and other biosafety issues in immortalized cells require further verification (8,33). Although the suicide gene scheme increases the safety of cell application in vivo, the SV40T gene has been identified to inhibit the terminal differentiation of certain cell types, including skeletal muscle cells, myoblasts and adipocytes; it has also been identified to affect the differentiation and functional maturation of cells in vivo (34–36).
In the present study, reversible immortalized hepatocytes with double suicide genes were generated. The HSV-tk suicide and SV40T genes were both flanked with LoxP sites, and HSV-tk mediated a negative selection of reversed immortalization. Following selection in medium containing GCV, cells did not express the SV40T and HSV-tk genes. The reversed hepatocytes exhibited a good recovery function in ALF; however, they still had the potential biohazard of malignant transformation and could not be easily removed following transplantation in vivo. In addition, CD suicide gene expression could control cell survival in the presence of 5-FC.
In conclusion, the procedure of the present study established a novel reversible immortalized hepatocyte cell line containing double suicide genes, which exhibited the cellular phenotype and recovery function of normal liver cells. These cells were able to proliferate markedly in order to obtain numerous cell sources. In addition, prior to in vivo application, the removal of the immortalized SV40T gene was guaranteed through HSV-tk/GCV selection, thereby avoiding the risk of negatively influencing cell function as well as the risk of cell malignant transformation. Following transplantation into the body, cells with potential risk could be selectively killed via the action of the CD/5-FC suicide system. This method achieved controllable regulation of immortalized cell safety through recombination and drug screening; thus, the maximal biological safety of immortalized hepatocytes for in vivo application was guaranteed, providing a reliable, safe and ideal cell material for the BAL technique.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Chongqing City (grant no. csct2016jcyjA0228) and the National Natural Science Foundation of China (grant no. 81100309).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
S-YF, C-QH and M-NL carried out the experiments; YB wrote the main manuscript text; YW and M-JG participated in cell culture; LT executed statistical analyses; YB and YH designed the research; T-CH led the whole project. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The use and care of animals was approved by the Institutional Animal Care and Use Committee of The Children's Hospital of Chongqing Medical University (Chongqing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bunchorntavakul C, Reddy KR. Acute Liver Failure. Clin Liver Dis. 2017;21:769–792. doi: 10.1016/j.cld.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Jiang W, Li J, Guo Q, Sun J, Chen C, Shen Z. Liver transplantation for hepatocellular carcinoma. Drug Discov Ther. 2015;9:331–334. doi: 10.5582/ddt.2015.01048. [DOI] [PubMed] [Google Scholar]
- 3.Al MI, Abaalkhail FA, Bahili HA, Abdo AH, Elsiesy HA, Al MS, El Sheikh YM, Hegab BS, Kamel YM, AlGoufi TT, et al. Liver transplantation at KFSHRC: Achievement and challenges. Ann Saudi Med. 2014;34:103–106. doi: 10.5144/0256-4947.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan XP, Li LJ. Advances in cell sources of hepatocytes for bioartificial liver. Hepatobiliary Pancreat Dis Int. 2012;11:594–605. doi: 10.1016/S1499-3872(12)60230-6. [DOI] [PubMed] [Google Scholar]
- 5.Nibourg GA, Chamuleau RA, van der Hoeven TV, Maas MA, Ruiter AF, Lamers WH, Oude Elferink RP, van Gulik TM, Hoekstra R. Liver progenitor cell line HepaRG differentiated in a bioartificial liver effectively supplies liver support to rats with acute liver failure. PLoS One. 2012;7:e38778. doi: 10.1371/journal.pone.0038778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakiyama R, Blau BJ, Miki T. Clinical translation of bioartificial liver support systems with human pluripotent stem cell-derived hepatic cells. World J Gastroenterol. 2017;23:1974–1979. doi: 10.3748/wjg.v23.i11.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen TH, Mai G, Villiger P, Oberholzer J, Salmon P, Morel P, Bühler L, Trono D. Treatment of acetaminophen-induced acute liver failure in the mouse with conditionally immortalized human hepatocytes. J Hepatol. 2005;43:1031–1037. doi: 10.1016/j.jhep.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Meng FY, Liu L, Yang FH, Li CY, Liu J, Zhou P. Reversible immortalization of human hepatocytes mediated by retroviral transfer and site-specific recombination. World J Gastroenterol. 2014;20:13119–13126. doi: 10.3748/wjg.v20.i36.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chendeb M, Schneider R, Davidson I, Fadloun A. Selective elimination of long Interspersed element-1 expressing tumour cells by targeted expression of the HSV-TK suicide gene. Oncotarget. 2017;8:38239–38250. doi: 10.18632/oncotarget.16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai LP, Wang ZF, Liang WY, Chen L, Chen D, Wang AX, Zhang ZQ. In vitro and in vivo effect of 5-FC combined gene therapy with TNF-α and CD suicide gene on human laryngeal carcinoma cell line Hep-2. PLoS One. 2013;8:e61136. doi: 10.1371/journal.pone.0061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi Y, He Y, Huang J, Su Y, Zhu GH, Wang Y, Qiao M, Zhang BQ, Zhang H, Wang Z, et al. Functional characteristics of reversibly immortalized hepatic progenitor cells derived from mouse embryonic liver. Cell Physiol Biochem. 2014;34:1318–1338. doi: 10.1159/000366340. [DOI] [PubMed] [Google Scholar]
- 12.Westerman KA, Leboulch P. Reversible immortalization of mammalian cells mediated by retroviral transfer and site-specific recombination. Proc Natl Acad Sci U S A. 1996;93:8971–8976. doi: 10.1073/pnas.93.17.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezzat T, Dhar DK, Malago M, Olde Damink SW. Dynamic tracking of stem cells in an acute liver failure model. World J Gastroenterol. 2012;18:507–516. doi: 10.3748/wjg.v18.i6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu G, Xu JJ, Deng ZH, Feng J, Jin Y. Supernatant of bone marrow mesenchymal stromal cells induces peripheral blood mononuclear cells possessing mesenchymal features. Int J Biol Sci. 2011;7:364–375. doi: 10.7150/ijbs.7.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei L, Fraser JL, Lu ZY, Hu X, Yu SP. Transplantation of hypoxia preconditioned bone marrow mesenchymal stem cells enhances angiogenesis and neurogenesis after cerebral ischemia in rats. Neurobiol Dis. 2012;46:635–645. doi: 10.1016/j.nbd.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao LF, Pan XP, Li LJ. Key challenges to the development of extracorporeal bioartificial liver support systems. Hepatobiliary Pancreat Dis Int. 2012;11:243–249. doi: 10.1016/S1499-3872(12)60155-6. [DOI] [PubMed] [Google Scholar]
- 17.van Wenum M, Adam AA, Hakvoort TB, Hendriks EJ, Shevchenko V, van Gulik TM, Chamuleau RA, Hoekstra R. Selecting cells for bioartificial liver devices and the importance of a 3D culture environment: A functional comparison between the HepaRG and C3A cell lines. Int J Biol Sci. 2016;12:964–978. doi: 10.7150/ijbs.15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, You S, Rong Y, Wu Y, Zhu B, Wan Z, Liu W, Mao P, Xin S. Newly established human liver cell line: A potential cell source for the bioartificial liver in the future. Hum Cell. 2013;26:155–161. doi: 10.1007/s13577-013-0068-5. [DOI] [PubMed] [Google Scholar]
- 19.Yu Y, Wang X, Nyberg SL. Potential and challenges of induced pluripotent stem cells in liver diseases treatment. J Clin Med. 2014;3:997–1017. doi: 10.3390/jcm3030997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nibourg GA, Chamuleau RA, van Gulik TM, Hoekstra R. Proliferative human cell sources applied as biocomponent in bioartificial livers: A review. Expert Opin Biol Ther. 2012;12:905–921. doi: 10.1517/14712598.2012.685714. [DOI] [PubMed] [Google Scholar]
- 21.van Wenum M, Chamuleau RA, van Gulik TM, Siliakus A, Seppen J, Hoekstra R. Bioartificial livers in vitro and in vivo: Tailoring biocomponents to the expanding variety of applications. Expert Opin Biol Ther. 2014;14:1745–1760. doi: 10.1517/14712598.2014.950651. [DOI] [PubMed] [Google Scholar]
- 22.Ordovás L, Park Y, Verfaillie CM. Stem cells and liver engineering. Biotechnol Adv. 2013;31:1094–1107. doi: 10.1016/j.biotechadv.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Fiers W, Contreras R, Haegemann G, Rogiers R, Van de Voorde A, Van Heuverswyn H, Van Herreweghe J, Volckaert G, Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978;273:113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- 24.Tevethia MJ, Ozer HL. SV40-mediated immortalization. Methods Mol Biol. 2001;165:185–199. doi: 10.1385/1-59259-117-5:185. [DOI] [PubMed] [Google Scholar]
- 25.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 26.Shahid JM, Iwamuro M, Sasamoto H, Kubota Y, Seita M, Kawamoto H, Nakaji S, Noguchi H, Yamamoto K, Kobayashi N. Establishment of an immortalized porcine liver cell line JSNK-1 with retroviral transduction of SV40T. Cell Transplant. 2010;19:849–856. doi: 10.3727/096368910X508979. [DOI] [PubMed] [Google Scholar]
- 27.Prives C. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell. 1990;61:735–738. doi: 10.1016/0092-8674(90)90179-I. [DOI] [PubMed] [Google Scholar]
- 28.Matsumura T, Takesue M, Westerman KA, Okitsu T, Sakaguchi M, Fukazawa T, Totsugawa T, Noguchi H, Yamamoto S, Stolz DB, et al. Establishment of an immortalized human-liver endothelial cell line with SV40T and hTERT. Transplantation. 2004;77:1357–1365. doi: 10.1097/01.TP.0000124286.82961.7E. [DOI] [PubMed] [Google Scholar]
- 29.Ramboer E, De Craene B, De Kock J, Vanhaecke T, Berx G, Rogiers V, Vinken M. Strategies for immortalization of primary hepatocytes. J Hepatol. 2014;61:925–943. doi: 10.1016/j.jhep.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan X, Wang Y, Yu X, Li J, Zhou N, Du W, Zhang Y, Cao H, Zhu D, Chen Y, Li L. Establishment and characterization of an immortalized human hepatic stellate cell line for applications in co-culturing with immortalized human hepatocytes. Int J Med Sci. 2015;12:248–255. doi: 10.7150/ijms.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi N, Noguchi H, Westerman KA, Watanabe T, Matsumura T, Totsugawa T, Fujiwara T, Leboulch P, Tanaka N. Cre/loxP-based reversible immortalization of human hepatocytes. Cell Transplant. 2001;10:383–386. [PubMed] [Google Scholar]
- 32.Kobayashi N, Noguchi H, Totsugawa T, Watanabe T, Matsumura T, Fujiwara T, Miyazaki M, Fukaya K, Namba M, Tanaka N. Insertion of a suicide gene into an immortalized human hepatocyte cell line. Cell Transplant. 2001;10:373–376. [PubMed] [Google Scholar]
- 33.Wang Z, Zhang J, Zeng Y, Sun S, Zhang J, Zhang B, Zhu M, Ouyang R, Ma B, Ye M, et al. Knockout of 4.1B triggers malignant transformation in SV40T-immortalized mouse embryo fibroblast cells. Mol Carcinog. 2017;56:538–549. doi: 10.1002/mc.22515. [DOI] [PubMed] [Google Scholar]
- 34.Tátrai P, Szepesi Á, Matula Z, Szigeti A, Buchan G, Mádi A, Uher F, Német K. Combined introduction of Bmi-1 and hTERT immortalizes human adipose tissue-derived stromal cells with low risk of transformation. Biochem Biophys Res Commun. 2012;422:28–35. doi: 10.1016/j.bbrc.2012.04.088. [DOI] [PubMed] [Google Scholar]
- 35.Van De Klundert FA, Bloemendal H. SV40 large T antigen-induced inhibition of terminal differentiation of primary skeletal muscle cells is associated with a block in the expression of MyoD and myogenin. Mol Biol Rep. 1994–1995;20:143–148. doi: 10.1007/BF00990546. [DOI] [PubMed] [Google Scholar]
- 36.Li M, Chen Y, Bi Y, Jiang W, Luo Q, He Y, Su Y, Liu X, Cui J, Zhang W, et al. Establishment and characterization of the reversibly immortalized mouse fetal heart progenitors. Int J Med Sci. 2013;10:1035–1046. doi: 10.7150/ijms.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.