Abstract
Feed-forward loops (FFLs) are three-gene modules that exert significant effects on a series of biological processes and carcinogenesis development. MicroRNA-associated FFLs (miR-FFLs) represent a new era in disease research. However, analysis of the miR-FFL network motifs has yet to be systematically performed, and their potential role in cardiac hypertrophy and acute myocardial infarction (AMI) requires investigation. The present study used a computational method to establish a comprehensive miR-FFL network for cardiac hypertrophy and AMI, by integrating high-throughput data from different sources and performing multi-aspect analysis of the network features. Several heart disease-associated miR-FFL motifs were identified that were specific or common to the two diseases investigated. Functional analysis further revealed that miR-FFL motifs provided specific drug targets for the clinical treatment of cardiac hypertrophy and AMI. Associations between specific drugs associated with heart disease and dysregulated FFLs were also identified. The present study highlighted the components of FFL motifs in cardiac hypertrophy and AMI, and revealed their possibility as heart disease biomarkers and novel treatment targets.
Keywords: feed-forward loop, cardiac hypertrophy, acute myocardial infarction, topological feature, biomarker, drug target
Introduction
Heart disease threatens the health of humans worldwide; however, it is often difficult to detect at the level of transcriptional regulation. Abnormal transcriptomic regulation is complicated and affected by a number of complex mechanisms at both a transcriptional and post-transcriptional level (1,2). Transcription factors (TFs) determine the level of gene expression by recognizing specific DNA sequences in diverse cell types (3). MicroRNAs (miRNAs) are important gene expression regulators, which bind with their target mRNAs, and repress or degrade them (4). An increasing number of studies have emphasized that miRNAs are essential in numerous types of heart diseases. Therefore, constructing a clear map of the network between miRNAs, their target genes and TFs may be beneficial for identifying dysregulated genes and signaling pathways. This would then increase the understanding on the roles of specific genes in heart disease pathogenesis.
TFs, miRNAs and their shared target genes form miRNA-associated feed-forward loops (miR-FFLs), in which miRNAs and TFs co-ordinate to regulate gene expression (5). The regulatory units within the miR-FFL network consist of an miRNA, a TF and their shared target genes (6,7). FFLs govern a number of biological processes, such as cell differentiation, and cause the development of certain diseases, including cancer (8,9). However, the varied roles of FFLs in certain common heart diseases have not been globally studied in a systematic manner.
Cardiovascular disease is an important cause of mortality worldwide and affects a great number of individuals annually (10). Cardiac hypertrophy is typically an inherited cardiovascular disease caused by abnormal gene mutations (11). Transcriptome reprogramming in the diseased heart can result in the development of abnormal pathological features (12). Several previous studies have demonstrated the association between miRNAs and cardiac hypertrophy (13-15).
Acute myocardial infarction (AMI) is a pathological and life-threatening condition, in which blood is unable to flow into the heart due to the blockage of a coronary artery, leading to the death of a part of the myocardial muscle (16). AMI and its associated risk factors require urgent assessment (17). A number of previous studies have also demonstrated the essential roles of miRNAs in AMI (18-20). Furthermore, recent studies have reported an association between cardiac hypertrophy and AMI. AMI treatment may be influenced by cardiac hypertrophy, as it appears to decrease the efficiency of ischemic preconditioning (21). Left ventricular (LV) hypertrophy and remodeling subsequent to MI are vital predictors of a patient's prognosis (22). However, the association between the two diseases on the miR-FFL level has yet to be globally investigated.
In the present study, a global miR-FFL network was constructed and its topological features were analyzed. In addition, a comprehensive method was designed that collects regulatory interaction and high-throughput expression profiles for miRNAs, TFs and target genes in order to characterize common or specifically dysregulated miR-FFL motifs in cardiac hypertrophy and AMI. The dysregulated miR-FFLs were revealed to be associated with cardiac-associated functions and signaling pathways. The genes and miRNAs in these miR-FFLs were also demonstrated to be vital drug targets. In conclusion, the present study highlighted the effect of dysregulated miR-FFL motifs in cardiac hypertrophy and AMI, which revealed their possibility as novel biomarkers and treatment targets in heart disease.
Materials and methods
Collecting high-throughput data for miRNAs, TFs and genes
The gene expression profiles of cardiac hypertrophy and AMI were downloaded from the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). Two studies in which the samples provided both miRNA and gene expression profiles were extracted. The cardiac hypertrophy expression data included three diseases samples and three control samples (accession no. GSE60291) (23). The AMI expression data included four disease samples and one control sample (accession no. GSE24591; unpublished data).
Establishing a genome-wide miR-FFL network
miR-FFL motifs are three-gene modules consisting of an miRNA, a TF and their shared target gene. The miRNA is regulated by the TF, while the target gene is regulated by both the miRNA and TF. Three types of regulatory associations were used to construct a comprehensive miR-FFL network. Firstly, the TF-miRNA association was obtained from a public database known as TransmiR, which contains experimentally validated information regarding TFs and miRNAs (24). Secondly, TF-gene regulatory pairs were obtained from the TRANSFAC Professional database (release date, February 2014) (24). Finally, experiments supporting the miRNA-gene regulatory association were obtained from TarBase v6.0, which is a high quality and widely used database (25). The data were merged, and the TFs and gene names were mapped to gene symbols, while the miRNA names were mapped to miRBase accession numbers for mature miRNAs (http://www.mirbase.org/).
Dissecting topological features for the miR-FFL network
Four measurements were used to assess the entire miR-FFL network, including degrees, topological coefficients, neighborhood connectivity and the clustering coefficients of nodes. All analyses were performed using Cytoscape 3.0 (http://www.cytoscape.org/).
Identifying dysregulated miR-FFLs in cardiac hypertrophy and AMI
An integrative method was designed to identify dysregulated miR-FFL motifs in cardiac hypertrophy and AMI, which used the miR-FFL regulatory networks and the expression data. Initially, Student's t-test was performed to compare the differences in expression between the TFs, miRNAs and genes in the patients with disease and the corresponding controls, for each miR-FFL motif. Next, for each interacting pair in the miR-FFL motif (TF-miRNA, TF-gene and miRNA-gene), the Pearson correlation coefficients (PCCs) and the statistical difference between them in the disease and control samples were calculated. The association between regulatory interactions was represented using the absolute difference of PCCs between the disease and control samples. In addition, the integrated comprehensive scores (Sdif and Spcc) for the FFLs were determined using the following equations for the differential expression of the P-value and PCCs:
In these equations, Pm, PT and Pg refer to the P-values of the miRNAs, TFs and genes, respectively, in each miR-FFL motif that were derived from the t-test. Sdif is the difference between the expression level of the miR-FFL motif between the patients with disease and the corresponding controls. DTm, DTg and Dmg correspond to the PCCs of the three regulatory pairs, including the TF-miRNA, TF-gene and miRNA-gene interactions for the disease samples, respectively. CTm, CTg and Cmg represent the PCCs of the same three regulatory interaction pairs for the control samples, respectively. Spcc is the absolute distinction of the PCC score between the disease and control samples in the entire miR-FFL motif. An integrative and equally-weighted method was also used to rank all the FFL motifs according to the Sdif and Spcc scores (26). Once two ranked lists were obtained, the ranking positions of the two lists were integrated to calculate the final ranking score list for each miR-FFL motif. A higher-ranking score represented an increased level of the dysregulated motif in the disease compared with the control. To obtain the significant P-value for each miR-FFL motif, each final motif ranking score was compared with the permutation-based final ranking score list, which was generated after randomly disturbing all sample labels in expression profiles 1,000 times. Finally, the significantly dysregulated motifs (P<0.05) for cardiac hypertrophy and AMI were obtained (27). All aforementioned analyses were performed using R software (version 3.2.3; https://www.r-project.org/).
Gene Ontology (GO) enrichment analysis
Enriched GO terms (P<0.01) were obtained from Database for Annotation, Visualization and Integrated Discovery web server using the default parameters (28).
Drug target analysis for miRNAs and genes
SM2miR (29) and DrugBank (30) databases were used to investigate the association between drugs and genes, and drugs and miRNAs, respectively.
Results
Characterizing topological properties of the miR-FFL network
A total of 429 miR-FFL network motifs were obtained using multiple data resources. Each network motif consisted of a TF, an miRNA and their shared target gene. A genome-wide miR-FFL network was established by integrating the network motifs (Fig. 1A), which included 235 nodes (122 genes, 35 TFs and 78 miRNAs) and 578 edges. Similar to the majority of biological networks, the transcription regulatory network obtained in the present study had a scale-free distribution (Fig. 1B). In addition, other topological features of all nodes were identified, including connectivity, topological coefficient and clustering coefficient (Fig. 1C–E), all of which displayed scale-free properties. The scale-free network demonstrated that the miR-FFL network was similar to a small-world network (31). Neighborhood connectivity was measured based on the average connectivity of certain neighbors (neighbors = 0, 1, 2…n). In the current study, a decreased degree distribution of the network was found to be accompanied by a decrease in the topological coefficient, which suggests that there was a hierarchical modularity phenomenon within the network. In addition, the nodes with a high and low degree were linked by the majority of the edges in the network, and these edges followed the decreased distribution, suggesting sub-networks were present within the network (32). The results also revealed that other types of disease networks, which shared similar topological properties, generally exhibited the same features as the miR-FFL network reported in the present study (33).
Figure 1.
Global topological features of the miR-FFL network. (A) Comprehensive miR-FFL network, where different colors represent different transcripts, including miRNAs (yellow), genes (blue) and transcription factors (red). Detailed topological features of nodes in the miR-FFL network, including the (B) degree, (C) connectivity, (D) topological coefficients and (E) clustering coefficients. miR-FFL, microRNA-associated feed-forward loops; Avg, average.
Specific miR-FFL motifs are significantly dysregulated in cardiac hypertrophy and AMI
The significantly dysregulated miR-FFL network motifs were characterized globally to determine the effect of miR-FFL motifs in cardiac hypertrophy and AMI. Next, specific dysregulated miR-FFL sub-networks were established for cardiac hypertrophy and AMI (Fig. 2). Among all background miR-FFLs, there were 22 miR-FFL motifs (9.4%) dysregulated in cardiac hypertrophy that included 13 genes, 6 TFs and 17 miRNAs (Fig. 2A and B). These dysregulated cardiac hypertrophy-associated miR-FFLs were ranked in the top 22 of all the background miR-FFLs following permutation testing (Fig. 2C). For AMI, there were 25 dysregulated miR-FFLs with 20 genes, 6 TFs and 10 miRNAs, which represented 10.59% of all background miR-FFLs (Fig. 2D and E). In total, 96% of these dysregulated AMI-associated miR-FFLs were ranked in the top 25 background miR-FFLs following permutation testing. For both cardiac hypertrophy and AMI, almost all the dysregulated miR-FFLs identified by the method reported in the current study were revealed to be statistically significant following permutation testing (P<0.05).
Figure 2.
Identified dysregulated miR-FFL motifs in the sub-network. (A) Different colors correspond to the different type of nodes in each dysregulated sub-network, including miRNAs (yellow), genes (blue) and TFs (red). (B) The number of miR-FFLs, TFs, genes and miRNAs among the cardiac hypertrophy-associated miR-FFLs are illustrated in the bar graph. (C) Pie chart showing the proportion of cardiac hypertrophy-associated miR-FFLs among all the background miR-FFLs. (D) Significantly dysregulated sub-networks associated with AMI. (E) Bar graph showing the number of miR-FFLs, TFs, genes and miRNAs among the AMI-associated miR-FFLs. (F) Pie chart showing the proportion of AMI-associated miR-FFLs among all the background miR-FFLs. miR-FFL, microRNA-associated feed-forward loops; TFs, transcription factors; AMI, acute myocardial infarction.
In the identified dysregulated miR-FFLs, certain miRNAs have previously been reported to serve essential roles in heart disease. For instance, miR-21 can affect the hypertrophic response process through exosome-associated transmission (34). In addition, miR-34a has a critical effect in heart development, and the inhibition of miR-34a is beneficial for cardiac function (35). Certain genes have also been reported to be strongly associated with heart disease. For instance, TP53 is a prominent tumor suppressor gene, and its downregulation in cardiac fibroblasts is regulated by miR-155 (36).
Common miR-FFLs motifs between cardiac hypertrophy and AMI revealing the disease mechanism
The results of the present study demonstrated that the majority of dysregulated miR-FFLs were specific in cardiac hypertrophy and AMI. A total of 20 miR-FFLs were dysregulated in cardiac hypertrophy alone; 23 miR-FFLs were dysregulated in AMI alone (Fig. 3A and D). For example, miR-FFL, consisting of TP53, mir-34b and CAV1, was only demonstrated to be dysregulated in cardiac hypertrophy (Fig. 3A). In addition, EGR1 was differentially expressed in both cardiac hypertrophy and AMI; however, it also forms miR-FFL with diverse miRNAs and genes (Fig. 3D). The present study analysis revealed two common miR-FFLs between cardiac hypertrophy and AMI (Fig. 3B). The first common miR-FFL consisted of epidermal growth receptor (EGR1), miR-335 and baculoviral IAP repeat-containing protein 5 (BIRC5). This miR-FFL ranked nos. 15 and 16 regarding the top dysregulated miR-FFLs associated with hypertrophy and AMI, respectively. In the present study, miR-335 was differentially expressed between normal and AMI samples. Furthermore, EGR1 and BIRC5 were differentially expressed between normal and cardiac hypertrophy samples (Fig. 3C). In addition, patients suffering from myocardial infarction have lower miR-335 expression, thus, miR-335 may serve as a novel treatment strategy for AMI (37). Furthermore, BIRC5 was downregulated in AMI patients (38). These results suggested that EGR1, miR-335 and BIRC5 were dysregulated and could form a miR-FFL during cardiac hypertrophy and AMI. Another common miR-FFL involved MYC, miR-16 and ABCC1. This miR-FFL ranked no. 20 and 19 regarding the top dysregulated miR-FFLs associated with hypertrophy and AMI, respectively. MYC and miR-16 were differentially expressed between normal and cardiac hypertrophy samples. These findings indicate that a number of the dysregulated FFLs identified in the present analysis were associated with heart disease, particularly the common FFLs, which were almost all associated with cardiac hypertrophy and AMI. Therefore, the authors suggest that numerous miRNAs, TFs and genes may have roles in cardiac hypertrophy and AMI via formation of FFLs.
Figure 3.
Common and specific miR-FFLs in cardiac hypertrophy and acute myocardial infarction. (A) miR-FFLs that are only dysregulated in cardiac hypertrophy, with two miR-FFLs presented as examples. (B) The Venn diagram reveals the intersection between the two diseases. (C) miR-FFLs that are only dysregulated in AMI, (D) with two miR-FFLs presented as examples. miR-FFL, microRNA-associated feed-forward loops.
Functional analysis revealing the roles of miR-FFL motifs in cardiac disease
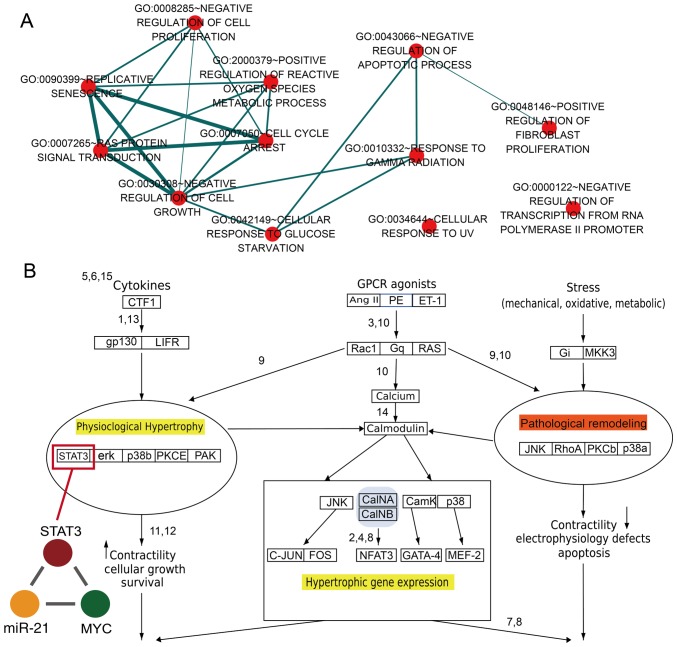
GO analysis was performed based on all the TFs and genes in dysregulated miR-FFLs for cardiac hypertrophy and AMI. It was observed that these dysregulated genes and TFs were enriched in specific GO terms associated with cell proliferation, fibroblast proliferation and reactive oxygen species metabolism (Fig. 4A). A previous study reported that basic fibroblast growth factor promotes human cardiosphere-derived cell engraftment in order to enhance cardiac repair, thus representing a potential treatment target for myocardial infarction (39). In addition, the result of the present study revealed that numerous GO terms shared common genes (Fig. 4A). For example, the 'replicative senescence' and 'negative regulation of cell growth' GO terms shared numerous genes. The results demonstrated that a single gene may participate in diverse functions via formation of diverse miR-FFLs involving different miRNAs and TFs. The current study results also revealed that STAT3 was a key gene for the pathological and physiological processes within the heart (Fig. 4B). Previously, it has been observed that STAT3 may be a novel therapeutic biomarker for cardiac hypertrophy treatment, and downregulation of STAT3 expression resulted in increased collagen synthesis and the limitation of hypertrophy (40). In addition, ANG II type 1 receptor activation mediated STAT3 gene regulation, resulting in the nuclear accumulation of U-STAT3, and this process was notably associated with the development of cardiac hypertrophy (41). In present study, the results revealed that STAT3, miR-21 and MYC formed a miR-FFL. It has also been reported that interferon (IFN) is able to greatly induce miR-21 expression based on the STAT3-associated signaling pathway, and miR-21 was characterized as a new target that exerts negative feedback on the IFN-induced apoptosis effect (42). Therefore, the results of the present study suggest the presence of an association among STAT3, miR-21 and heart disease.
Figure 4.
Functional analysis of transcription factors and genes in dysregulated miR-FFLs. (A) The results of gene enrichment analysis are illustrated as a network. Each red node corresponds to a single GO term result, and the nodes are clustered and annotated based on the similarity between them. Node size corresponds to the quantity of genes for each GO term. Nodes are connected by the common genes between GO terms. (B) Physiological and pathological hypertrophy of the heart signaling pathway. miR-FFL, microRNA-associated feed-forward loops; GO, Gene Ontology.
miR-FFLs contain the drug target miRNAs and genes
Subsequently, the association of the TFs, genes and miRNAs in the dysregulated miR-FFLs with a potential drug target was further explored. Initially, the study searched for miR-FFLs that were common between cardiac hypertrophy and AMI, including EGR1, miR-335 and BIRC5, using the DrugBank and SM2miR databases (Fig. 5A). SM2miR database is an integrated database of the experimentally validated effects of small molecules on miRNA expression (29).
Figure 5.
Drug targets of transcription factors, miRNAs and genes in dysregulated miR-FFLs for cardiac hypertrophy and AMI. (A) Association of drugs with miRNAs and genes in an miR-FFL network that is common between cardiac hypertrophy and AMI. (B) miRNAs as targets for the drugs isoproterenol and (C) propranolol in cardiac hypertrophy and AMI, respectively. miR-FFL, microRNA-associated feed-forward loops; AMI, acute myocardial infarction.
MiR-335 was found to be associated with doxorubicin, a chemotherapy drug used for the treatment of a number of cancer types (Fig. 5A). Furthermore, BIRC5 was observed to be a target of the drug reserpine (Fig. 5A), which is a type of antiadrenergic agent occasionally used to control high blood pressure and treat schizophrenia. However, this drug is rarely used in clinical practice due to its side effects.
All the miRNAs investigated in the current study were found to be associated with isoproterenol or propranolol (Fig. 5B and C). Isoproterenol is generally used to treat heart diseases, such as abnormal heartbeats, heart block and heart failure. It was observed that 7 miRNAs were associated with isoproterenol in cardiac hypertrophy, while 3 miRNAs in AMI were associated with isoproterenol. The miR-FFL that included MYC, STAT3 and miR-21, as well as the miR-FFL network that included MYC, TCF7L2 and miR-21, were observed to form a more complex and larger motif by sharing miR-21 and MYC (Fig. 5B). Furthermore, the miR-FFL network involving MYC, miR-29b and VEGFA, along with the miR-FFL that consisted of MYC, miR-29c and VEGFA, also formed a more complex motif by sharing VEGFA and MYC. Therefore, it may be suggested that miR-FFL motifs dysregulate significant biological pathways and networks, and trigger important biological responses, thereby severely affecting disease mechanisms and influencing the effect of drugs. Additionally, propranolol is a β-blocker that has an effect on human heart function and is widely used to treat high blood pressure and abnormal heart conditions. In the current analysis, the miR-FFL network consisting of TP53, miR-125b and TP53INP1 was found to be associated with propranolol in cardiac hypertrophy. Taken together, the aforementioned findings indicate that dysregulated miR-FFLs may be a novel target for drug treatment in future studies investigating hypertrophy and AMI.
Discussion
Various transcripts, including TFs, miRNAs and genes, can form complex regulatory associations, such as FFLs, which may influence pathogenic mechanisms in certain heart diseases. Therefore, the present study investigated TF-miRNA-gene FFLs motifs and used a comprehensive method to detect their features by integrating multiple regulatory associations and expression profiles from large repositories. Dysregulated miR-FFL motifs were found to be present in cardiac hypertrophy and AMI, which are two diseases with a high prevalence worldwide. In addition, certain dysregulated miR-FFLs that were shared or specific to the two diseases were identified, focusing on tissue-specific analyses conducted in previous studies (43). Disease-specific miR-FFL motifs may be beneficial for drug discovery, and may increase the therapeutic effects of drugs. As in other similar studies, the disease and drug target-associated research conducted in the present study revealed the potential functionality of certain miR-FFL motifs, which may serve as novel biomarkers in the treatment of heart diseases (27). The current study also revealed that miR-FFLs may provide another method for studying heart disease.
In addition, it was observed that certain miR-FFLs share common miRNAs, genes or TFs, forming larger complexes that regulate the biological network. These results indicated that, although miR-FFLs may serve as unitary motifs and participate in disease progression, complex associations within the disease network may also have a specific function. The present study also demonstrated that cardiac hypertrophy and AMI are two complex diseases with numerous miRNAs, genes, TFs and other factors that participate in the process of disease development. These factors may form various different motifs to serve their specific roles. There are also other types of miRNA-mediated FFLs, which differ from the ones focused on in the present study, such as those involving miRNA-regulated TFs (44). These alternative miR-FFLs have also been reported to participate in the development of several types of cancer (8). Therefore, the authors recognize that the effects of certain miR-FFLs may strongly depend on the cell type and context. Future studies should investigate an increased number of cell types in a variety of contexts to validate the accuracy and stability of the method used in the present study. Finally, our method identified novel candidates associated with disease development, which require further investigation and experimental validation.
In conclusion, the present study examined and highlighted the potential functional mechanism of miRNAs, genes and TFs within cardiac hypertrophy and AMI. Dysregulated miR-FFLs were identified, and miR-FFLs common to the two diseases were further investigated. The functional and drug analyses revealed the essential role of miR-FFLs in heart diseases. The present study also provided a systematic and novel approach that revealed the dysregulated cross-talk in cardiac hypertrophy and AMI by identifying specific functional motifs.
Acknowledgments
Not applicable.
Funding
This work was supported by a grant from the National Natural Science Foundation of China Project (grant no. 81670381 awarded to Bingchen Liu).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XC, BL and WQ conceived and designed the present study. FZ, SM and SZ performed the experiments. SS, YZ and LS analyzed the data. WQ, BL and XC wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Matsuoka R. Mutations of transcription factors in human with heart disease for understanding the development and mechanisms of congenital cardiovascular heart disease. Adv Exp Med Biol. 2005;565:349–415. doi: 10.1007/0-387-24990-7_27. [DOI] [PubMed] [Google Scholar]
- 2.Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. doi: 10.1016/j.lfs.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Latchman DS. Transcription factors: An overview. Int J Biochem Cell Biol. 1997;29:1305–1312. doi: 10.1016/S1357-2725(97)00085-X. [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Z, Shah PK, Amin SB, Samur MK, Huang N, Wang X, Misra V, Ji H, Gabuzda D, Li C. Integrative analysis of gene and miRNA expression profiles with transcription factor-miRNA feed-forward loops identifies regulators in human cancers. Nucleic Acids Res. 2012;40:e135. doi: 10.1093/nar/gks395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HM, Kuang S, Xiong X, Gao T, Liu C, Guo AY. Transcription factor and microRNA co-regulatory loops: Important regulatory motifs in biological processes and diseases. Brief Bioinform. 2015;16:45–58. doi: 10.1093/bib/bbt085. [DOI] [PubMed] [Google Scholar]
- 10.Mendis S, Davis S, Norrving B. Organizational update: The world health organization global status report on noncommunicable diseases 2014 one more landmark step in the combat against stroke and vascular disease. Stroke. 2015;46:e121–122. doi: 10.1161/STROKEAHA.115.008097. [DOI] [PubMed] [Google Scholar]
- 11.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–255. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- 12.Sala V, Gallo S, Leo C, Gatti S, Gelb BD, Crepaldi T. Signaling to cardiac hypertrophy: Insights from human and mouse RASopathies. Mol Med. 2012;18:938–947. doi: 10.2119/molmed.2011.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diniz GP, Huang ZP, Liu J, Chen J, Ding J, Fonseca RI, Barreto-Chaves ML, Donato J, Jr, Hu X, Wang DZ. Loss of microRNA-22 prevents high-fat diet induced dyslipidemia and increases energy expenditure without affecting cardiac hypertrophy. Clin Sci. 2017;131:2885–2900. doi: 10.1042/CS20171368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heggermont WA, Papageorgiou AP, Quaegebeur A, Deckx S, Carai P, Verhesen W, Eelen G, Schoors S, van Leeuwen R, Alekseev S, et al. Inhibition of MicroRNA-146a and overexpression of its target dihydrolipoyl succinyltransferase protect against pressure overload-induced cardiac hypertrophy and dysfunction. Circulation. 2017;136:747–761. doi: 10.1161/CIRCULATIONAHA.116.024171. [DOI] [PubMed] [Google Scholar]
- 15.Nagalingam RS, Sundaresan NR, Gupta MP, Geenen DL, Solaro RJ, Gupta M. A cardiac-enriched microRNA, miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J Biol Chem. 2017;292:5123. doi: 10.1074/jbc.A112.442384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shafei AE, Ali MA, Ghanem HG, Shehata AI, Abdelgawad AA, Handal HR, Talaat KA, Ashaal AE, El-Shal AS. Mesenchymal stem cells therapy: A promising cell based therapy for treatment of myocardial infraction. J Gene Med. 2017 Dec; doi: 10.1002/jgm.2995. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 17.Liang H, Qiu H, Tian L. Short-term effects of fine particulate matter on acute myocardial infraction mortality and years of life lost: A time series study in Hong Kong. Sci Total Environ. 2018;615:558–563. doi: 10.1016/j.scitotenv.2017.09.266. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Zhao Y, Chen W, Xie L, Zhao ZA, Yang J, Chen Y, Lei W, Shen Z. MicroRNA-133 overexpression promotes the therapeutic efficacy of mesenchymal stem cells on acute myocardial infarction. Stem Cell Res Ther. 2017;8:268. doi: 10.1186/s13287-017-0722-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayoumi AS, Park KM, Wang Y, Teoh JP, Aonuma T, Tang Y, Su H, Weintraub NL, Kim IM. A carvedilol-responsive microRNA, miR-125b-5p protects the heart from acute myocardial infarction by repressing pro-apoptotic bak1 and klf13 in cardiomyocytes. J Mol Cell Cardiol. 2018;114:72–82. doi: 10.1016/j.yjmcc.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zhang Y, Du W, Liang H, He H, Zhang L, Pan Z, Li X, Xu C, Zhou Y, et al. MiR-223-3p as a novel MicroRNA regulator of expression of voltage-gated K+ channel Kv4.2 in acute myocardial infarction. Cell Physiol Biochem. 2016;39:102–114. doi: 10.1159/000445609. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi T, Ishii Y, Kikuchi K, Hasebe N. Ischemic preconditioning effect of prodromal angina is attenuated in acute myocardial infarction patients with hypertensive left ventricular hypertrophy. Circ J. 2011;75:1192–1199. doi: 10.1253/circj.CJ-10-0906. [DOI] [PubMed] [Google Scholar]
- 22.Esen O, Agus HZ, Guler GB, Açar G, Avci A, Güler E, Karaca O, Geçmen C, Bulut M, Emiroğlu Y, et al. Relationship between circulating soluble Fas ligand and preexisting left ventricular hypertrophy in the setting of left ventricular remodeling after acute myocardial infarction. Coron Artery Dis. 2011;22:294–298. doi: 10.1097/MCA.0b013e328345c216. [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal P, Turner A, Matter A, Kattman SJ, Stoddard A, Lorier R, Swanson BJ, Arnett DK, Broeckel U. RNA expression profiling of human iPSC-derived cardiomyocytes in a cardiac hypertrophy model. PLoS One. 2014;9:e108051. doi: 10.1371/journal.pone.0108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Lu M, Qiu C, Cui Q. TransmiR: A transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. TarBase 6.0: Capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, et al. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 27.Jiang W, Mitra R, Lin CC, Wang Q, Cheng F, Zhao Z. Systematic dissection of dysregulated transcription factor-miRNA feed-forward loops across tumor types. Brief Bioinform. 2016;17:996–1008. doi: 10.1093/bib/bbv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Wang S, Meng F, Wang J, Zhang Y, Dai E, Yu X, Li X, Jiang W. SM2miR: A database of the experimentally validated small molecules' effects on microRNA expression. Bioinformatics. 2013;29:409–411. doi: 10.1093/bioinformatics/bts698. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amaral LA, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci USA. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simos T, Georgopoulou U, Thyphronitis G, Koskinas J, Papaloukas C. Analysis of protein interaction networks for the detection of candidate hepatitis B and C biomarkers. IEEE J Biomed Health Inform. 2015;19:181–189. doi: 10.1109/JBHI.2014.2344732. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z, Shao T, Zhang J, Wang L, Li X. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015;43:8169–8182. doi: 10.1093/nar/gkv853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Indolfi C, Curcio A. Stargazing microRNA maps a new miR-21 star for cardiac hypertrophy. J Clin Invest. 2014;124:1896–1898. doi: 10.1172/JCI75801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, Fisch S, Unno K, Sereti KI, Liao R. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res. 2015;117:450–459. doi: 10.1161/CIRCRESAHA.117.305962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He W, Huang H, Xie Q, Wang Z, Fan Y, Kong B, Huang D, Xiao Y. MiR-155 knockout in fibroblasts improves cardiac remodeling by targeting tumor protein p53-inducible nuclear protein 1. J Cardiovasc Pharmacol Ther. 2016;21:423–435. doi: 10.1177/1074248415616188. [DOI] [PubMed] [Google Scholar]
- 37.Liang J, Bai S, Su L, Li C, Wu J, Xia Z, Xu D. A subset of circulating microRNAs is expressed differently in patients with myocardial infarction. Mol Med Rep. 2015;12:243–247. doi: 10.3892/mmr.2015.3422. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Li Z, Liu T, Xue X, Jiang H, Huang J, Wang H. Impaired cardioprotective function of transplantation of mesenchymal stem cells from patients with diabetes mellitus to rats with experimentally induced myocardial infarction. Cardiovasc Diabetol. 2013;12:40. doi: 10.1186/1475-2840-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takehara N, Tsutsumi Y, Tateishi K, Ogata T, Tanaka H, Ueyama T, Takahashi T, Takamatsu T, Fukushima M, Komeda M, et al. Controlled delivery of basic fibroblast growth factor promotes human cardiosphere-derived cell engraftment to enhance cardiac repair for chronic myocardial infarction. J Am Coll Cardiol. 2008;52:1858–1865. doi: 10.1016/j.jacc.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 40.Mir SA, Chatterjee A, Mitra A, Pathak K, Mahata SK, Sarkar S. Inhibition of signal transducer and activator of transcription 3 (STAT3) attenuates interleukin-6 (IL-6)-induced collagen synthesis and resultant hypertrophy in rat heart. J Biol Chem. 2012;287:2666–2677. doi: 10.1074/jbc.M111.246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue H, Li W, Desnoyer R, Karnik SS. Role of nuclear unphosphorylated STAT3 in angiotensin II type 1 receptor-induced cardiac hypertrophy. Cardiovasc Res. 2010;85:90–99. doi: 10.1093/cvr/cvp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Fang Z, Xu C, Li Y, Cai X, Ren S, Liu H, Wang Y, Wang F, Chen R, Qu M, et al. A feed-forward regulatory loop between androgen receptor and PlncRNA-1 promotes prostate cancer progression. Cancer Lett. 2016;374:62–74. doi: 10.1016/j.canlet.2016.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.