Abstract
Early detection is critical for the treatment of colon carcinoma. However, current biomarkers for its diagnosis and prognosis are insufficient and improvement is required. Aberrantly expressed microRNAs (miRNAs/miRs) in colon carcinoma have been identified to function as potential diagnostic and prognostic biomarkers. In the present study, 245 differentially expressed miRNAs between colon carcinoma and normal tissues were identified by a bioinformatics analysis of a dataset from The Cancer Genome Atlas. A six-miRNA (miR-149, miR-3189, miR-3677, miR-3917, miR-4999 and miR-6854) prognostic prediction system was established, which is able to independently and effectively predict the prognosis of colon carcinoma patients [P<0.001, area under the receiver operating characteristic curve (AUC)=0.763]. Furthermore, the six miRNAs were highly correlated with the tumor-nodes-metastasis (TNM) stage and were able to distinguish between different stages (high vs. low TNM stage, P<0.001). Of note, combination of the six-miRNA signature and TNM stage provides an improved prediction of the patient's prognosis (AUC=0.797). Functional enrichment analysis revealed the possible mechanistic involvement of these predictive miRNAs in cancer-associated biological processes and pathways. Taken together, the present study demonstrated the promising potential of the novel six-miRNA model as an independent factor for the prediction of the progression and prognosis of colon carcinoma.
Keywords: microRNA, colon carcinoma, biomarker, progression, prognosis
Introduction
Colon carcinoma is one of the most common types of malignant disease with high mortality and one of the leading causes of cancer-associated death worldwide (1,2). Half of colon carcinoma patients develop recurrences and metastasis, as treatment tends to commence late due to the lack of symptoms at the early stage (3). Given the rapid increases in the incidence and mortality observed in numerous counties, particularly in Eastern Europe, Asia and South America (4), early detection and primary prevention, which may significantly reduce the mortality of colon carcinoma patients, are therefore imperative (5). Unfortunately, the symptoms are frequently not obvious at the earliest stage and there has been little success in the screening for effective molecular markers for colon cancer (6). Carcinoembryonic antigen (CEA), the most utilized colon carcinoma marker, is limited by its low specificity (7). Thus, novel molecular markers with high sensitivity and specificity, which significantly optimize the use of therapies and provide a benefit for patients, are desired.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs, which regulate post-transcription gene expression. Certain miRNAs are aberrantly expressed during colorectal cancer (CRC) development and progression and exert regulatory roles in cancer-associated processes (8). Several studies have reported that miRNAs may function as potential diagnostic and prognostic biomarkers for colon carcinoma (9–11).
In the present study, the differential miRNA expression profiles of 457 colon carcinoma tissues vs. 8 adjacent non-tumorous tissues from The Cancer Genome Atlas (TCGA) database we compared by a bioinformatics analysis. Furthermore, a novel six-miRNA signature was proposed by using the univariate and multivariate Cox's proportional hazard regression model, which has the capacity to effectively predict the prognosis and survival of colon carcinoma patients [P<0.001, area under the receiver operating characteristic (ROC) curve (AUC)=0.763]. In addition, a prognostic prediction model was constructed based on the six miRNAs that were highly correlated with the tumor-nodes-metastasis (TNM) stage. By combining the six-miRNA model and the TNM stage, an improved prediction of the patient's survival status (AUC=0.797) was possible. Taken together, the present results indicated that the six miRNAs may be reliable biomarkers for monitoring the progression of colon carcinoma and predicting the prognosis of affected patients.
Materials and methods
miRNA expression dataset
The miRNA sequence data and clinical characteristics were downloaded from the publicly available TCGA database (https://cancergenome.nih.gov/). Consequently, a total of 465 samples were enrolled in our study, including 457 colon carcinoma tissues and 8 matched normal tissues. Patients without complete information of clinical characteristics or survival time were excluded from the further analysis. Finally, we got 453 colon carcinoma patients and the clinical features recorded included age, sex, TNM stage, tumor (T) stage, extent of spread to the lymph nodes as indicated by the nodal (N) stage, presence of metastasis as indicated by the metastasis (M) stage and residual tumor. The TNM staging system of the American Joint Committee on Cancer (AJCC) is one of the most commonly used tumor staging systems for colon carcinoma. The data processing was in accordance with the publication guidelines provided by TCGA (http://cancerge-nome.nih.gov/publications/publicationguidelines).
Differentially expressed miRNAs (DEmiRNAs)
The package 'edgeR' version 3.22.3 (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) in R version 3.4.2 was used to identify DEmiRNAs between colon carcinoma and normal tissues. The miRNAsb that could not be determined in all samples were excluded. A |log2 fold change (log2FC)| of >2 and an adjusted P-value of <0.01 were considered as the cut-off criteria. The expression levels of the miRNAs were log2 transformed for further analysis.
Construction of the miRNA signature associated with overall survival (OS)
The association between DEmiRNAs and the patients' OS was evaluated by using the univariate Cox's proportional hazard ratio (HR) model using the 'survival' package in R. The miRNAs with P<0.01 were selected for further multivariate Cox's proportional hazard regression to identify the independent prognostic miRNAs. Using the logistic regression method, a prognostic model based on the independent prognostic miRNAs was constructed to evaluate the survival risk of each patient. The risk score was calculated as follows: Risk score = expmiRNA1 × βmiRNA1 + expmiRNA2 × βmiRNA2 + …expmiRNAn × βmiRNAn, where exp is the expression level and β is the regression coefficient derived from the multivariate Cox regression model (12,13).
Utilizing the median risk score as the cut-off point, patients with colon carcinoma were categorized into high-risk and low-risk groups. It was investigated whether the median survival time was significantly different between these two groups using a Kaplan-Meier estimation and log-rank test. The predictability of the model was evaluated by the AUC using the 'survival ROC' package in R (14).
Association between risk score and other clinical characteristics
Next, the association between the risk score of the six miRNAs and the clinical characteristics of patients with colon carcinoma, including age, sex, TNM stage, T stage, N stage, M stage and residual tumor, was assessed. Furthermore, univariate Cox proportional hazard regression analyses were performed to investigate the effects of various clinical features and risk score on the OS of patients with colon adenocarcinoma. The HR and 95% confidence interval were assessed. Multivariate Cox regression analysis was used to verify the independent predictive capacity of the risk score when compared with that of other clinical factors.
Prognostic prediction for colon carcinoma based on risk score
To assess whether the risk score may be used to assess tumor progression, the risk score in different stages, including TNM stage, T stage, N stage and M stage, were compared. Furthermore, the association of integrated risk score with clinical characteristics was assessed to evaluate its predictive value regarding the prognosis in colon carcinoma patients. We investigated if the median survival time was significantly different between patients separated by both risk score and staging (low/high-risk score + high/low stage) using a Kaplan-Meier estimator and log-rank test. The risk score was compared via Multivariate Cox regression analysis as aforementioned and separated by its median. To investigate the performance of the prognostic risk score of the six miRNAs and TNM stage in predicting the outcome for colon carcinoma patients, the AUC of the ROC was calculated and compared.
Functional enrichment analysis
To reveal the possible mechanism of action of the six miRNAs involved in colon carcinoma, the target genes of prognostic miRNAs were predicted using TargetScan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/miRDB/), mirPath (https://mpd.bioinf.uni-sb.de/overview.html) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). Only the overlapping genes were identified to further enhance the reliability of the analysis. A Venn diagram was drawn using the 'VennDiagram' package.
To further elucidate the biological function of the targeted genes of the six miRNAs, a Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway biological enrichment analysis was preformed through the Database for Annotation, Visualization and Integrated Discovery bioinformatics tool (15). P<0.05 was set as the cut-off criterion.
Statistical analysis
Kaplan-Meier survival analysis and univariate/multivariate Cox proportional hazard regression analysis were conducted using the 'survival' packageversion 2.41 (https://github.com/therneau/survival) in R to compare each miRNA (low vs. high level) and prognostic miRNA signatures (low vs. high-risk). Pearson correlation analysis was used to estimate the correlation between the miRNAs and clinical features. An independent-samples t-test was performed to examine the difference in risk score distribution between clinical characteristics (TNM stage), as the risk score was abnormally distributed in those categories. P<0.05 was considered to indicate a statistically significant difference. The statistical analysis was performed using the SPSS 20.0 software package (IBM Corp., Armonk, NY, USA).
Results
Identification of prognostic miRNAs in colon carcinoma
According to the cut-off criteria (P<0.01 and |log2FC|>2.0), a total of 245 DEmiRNAs were obtained, including 174 upregulated and 71 downregulated miRNAs. The result is presented as volcano plot in Fig. 1.
Figure 1.
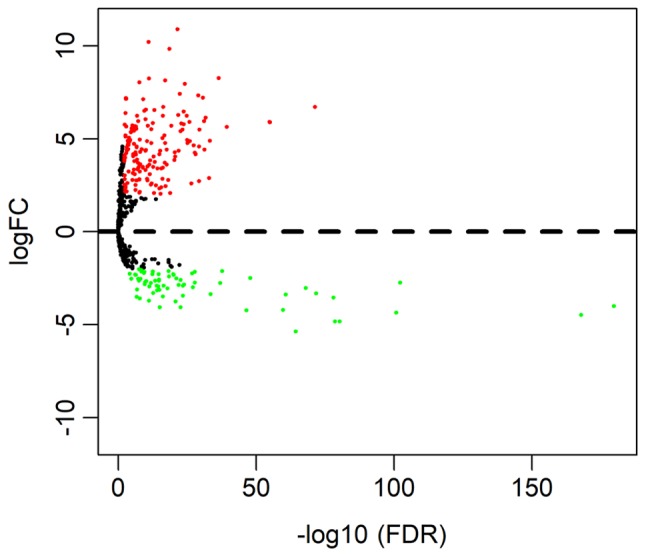
Volcano plot of differentially expressed miRNAs. The red dots represent upregulated miRNA, and green dots represent downregulated miRNA. miRNA, microRNA.
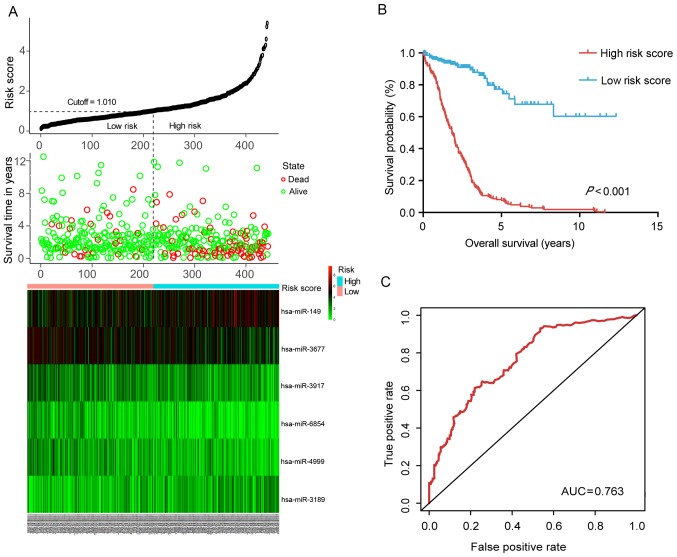
The association between the expression of the 245 DEmiRNAs and patient survival was evaluated by using univariate Cox proportional hazard regression. Based on the criteria (P<0.01), eight miRNAs that were significantly associated with OS were obtained. Furthermore, six of the eight miRNAs were screened out by multivariate Cox regression and a diagnostic prediction model was constructed based on the expression levels of the six miRNAs weighed by their relative coefficient. The miRNA risk score was calculated as follows: miRNA risk score = 0.204 × exphsa-mir-149 − 0.244 × exphsa-mir-3189 − 0.164 × exphsa-mir-3677 − 0.269 × exphsa-mir-3917 + 0.224 × exphsa-mir-4999 − 0.380 × exphsa-mir-6854. The detailed information of these miRNAs is presented in Fig. 2A and Table I. By dividing the risk score according to its median (median=1.010), 453 patients were stratified into the high-risk and low-risk groups. The survival analysis performed using the Kaplan-Meier method revealed that the low-risk group had a significantly better OS (P<0.001) than the high-risk group (Fig. 2B). The ROC curve further demonstrated that the risk score model was able to effectively predict the prognosis of colon cancer patients (AUC=0.763; Fig. 2C).
Figure 2.
Predictive value of the six-miRNA signature for colon carcinoma. (A) The distributions of the risk score, survival status of colon carcinoma patients and heatmap of six miRNA expression profiles which are ranked by risk score. (B) Kaplan-Meier curve of overall survival probability for colon carcinoma patients divided into high- and low-risk score groups based on the risk score of the six-miRNA signature. (C) Receiver operating characteristic curve of the risk score for predicting five-year survival. AUC, area under curve; miR/miRNA, microRNA; hsa, Homo sapiens.
Table I.
Characteristics of the six deregulated miRNAs and their regression coefficient derived from the multivariate Cox regression model in colon carcinoma prognosis prediction.
| miRNA | Ensembl ID | Coefficient | SE | P-value |
|---|---|---|---|---|
| hsa-miR-149 | ENSG00000207611 | 0.204 | 0.093 | 0.029 |
| hsa-miR-3917 | ENSG00000283938 | −0.269 | 0.102 | 0.008 |
| hsa-miR-3677 | ENSG00000266643 | −0.164 | 0.102 | 0.108 |
| hsa-miR-6854 | ENSG00000278412 | −0.380 | 0.111 | <0.001 |
| hsa-miR-4999 | ENSG00000265390 | 0.224 | 0.112 | 0.045 |
| hsa-miR-3189 | ENSG00000264175 | −0.244 | 0.130 | 0.060 |
The coefficient and P-value are derived from the multivariate Cox regression analysis. miR/miRNA, microRNA; hsa, Homo sapiens; SE, standard errors of coefficients.
Correlation between risk score and other clinical characteristics
The correlation between the risk score based on the signature of DEmiRNAs and various clinical features was then assessed. The results indicated that the risk score was significantly correlated with the TNM stage (P=0.007), N stage (P<0.001), M stage (P<0.001) and residual tumor (P=0.021; Table II).
Table II.
Correlation between the risk score and clinical features.
| Variable | Total (n) (%) | Risk score of six-miRNA signature
|
χ2 test P-value |
|
|---|---|---|---|---|
| Low (n) (%) | High (n) (%) | |||
| Age (years) | ||||
| <60 | 125 (27.6) | 65 (52.0) | 60 (48.0) | 0.596 |
| ≥60 | 328 (72.4) | 162 (49.4) | 166 (50.6) | |
| Sex | ||||
| Male | 239 (52.8) | 121 (50.6) | 118 (49.4) | 0.294 |
| Female | 214 (47.2) | 106 (49.5) | 108 (50.5) | |
| TNM stage | ||||
| I+III | 250 (56.6) | 134 (53.6) | 116 (46.4) | 0.007 |
| III+IV | 192 (43.4) | 89 (46.4) | 103 (53.6) | |
| T-stage | ||||
| T1+T2 | 89 (19.6) | 55 (61.8) | 34 (38.2) | 0.070 |
| T3+T4 | 364 (80.4) | 172 (47.3) | 192 (52.7) | |
| N-stage | ||||
| N0 | 266 (58.7) | 140 (52.6) | 126 (47.4) | 0.007 |
| N1+N2 | 187 (41.3) | 87 (46.5) | 100 (53.4) | |
| M-stage | ||||
| M0 <0.001 | 332 (83.4) | 173 (52.2) | 159 (47.8) | |
| M1 | 64 (16.6) | 27 (42.2) | 37 (57.8) | |
| Residual tumor | ||||
| R0 | 327 (92.1) | 175 (53.5) | 152 (46.5) | 0.021 |
| R1+R2 | 28 (7.9) | 13 (46.4) | 15 (53.6) | |
The risk score was based on the 6-miRNA signature and the median was used as a cut-off to distinguish between high and low-risk. Values are expressed as n (%). a TNM, tumor-nodes-metastasis.
In addition, to further verify the independent prognostic value of the six miRNAs, univariate and multivariate Cox regression analysis was used to evaluate the effect of the six miRNA-based risk score (high- vs. low-risk) as well as that of other clinical parameters on OS. As presented in Table III, the univariate analysis revealed that the TNM stage (HR=2.884, P<0.001), T stage (HR=2.895, P=0.007), N stage (HR=2.538, P<0.001), M stage (HR=4.253, P<0.001), residual tumor (HR=4.094, P<0.001) and risk score of the six-miRNA signature (HR=3.504, P<0.001) were significantly associated with the OS of colon carcinoma patients. In the multivariate analysis, TNM stage (HR=5.461, P=0.007), N stage (HR=0.331, P=0.034) and the six-miRNA signature (HR=3.991, P<0.001) were independent prognostic factor.
Table III.
Uni- and multivariate Cox regression analysis for the prognostic value of various parameters in colon carcinoma patients.
| Variable | Univariate analysis
|
Multivariate analysis
|
||
|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Age (≥60 vs. <60) | 1.346 (0.845–2.144) | 0.211 | ||
| Sex (male vs. female) | 1.124 (0.759–1.665) | 0.559 | ||
| TNM stage (III+IV vs. I+II) | 2.884 (1.890–4.402) | <0.001 | 5.461 (1.605–18.585) | 0.007 |
| T stage (T3+T4 vs. T1+T2) | 2.895 (1.340–6.252) | 0.007 | 6.549 (0.877–48.892) | 0.067 |
| N stage (N1+N2 vs. N0) | 2.538 (1.699–3.792) | <0.001 | 0.331 (0.119–0.918) | 0.034 |
| M stage (M1 vs. M0) | 4.253 (2.710–6.673) | <0.001 | 1.687 (0.788–3.612) | 0.178 |
| Residual tumor (R1+R2 vs. R0) | 4.094 (2.249–7.450) | <0.001 | 1.809 (0.867–3.777) | 0.114 |
| Risk score (high vs. low) | 3.504 (2.237–5.489) | <0.001 | 3.991 (2.040–7.809) | <0.001 |
HR, hazard ratio; CI, confidence interval; TNM, tumor-nodes-metastasis.
Prognostication of colon carcinoma patients based on risk score
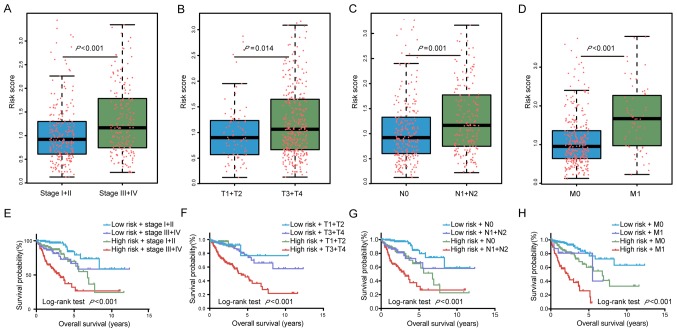
To further assess the capacity of the risk score to predict tumor progression, the risk score was compared different tumor stages. Patients with a high TNM stage (stage III+IV) had significantly higher risk scores than those with a low TNM stage (stage I+II; P<0.001). In addition, the risk score was significantly higher in tissues from patients with high vs. low T (P=0.014), N (P=0.001) and M stages (P<0.001; Fig. 3A–D). Collectively, these results suggest that the risk score of the six miRNAs may have a utility in predicting tumor progression and surveillance for recurrence.
Figure 3.
Prognostic prediction for colon carcinoma patients based on risk score. The risk score in different stages, including (A) TNM stage, (B) T stage, (C) N stage and (D) M stage. The red dots represent the risk score for colon carcinoma patients, the horizontal lines represented median, the error bars represented the upper and lower quartiles of risk score. The integrated risk score and (E) TNM stage, (F) T stage, (G) N stage and (H) M stage for the prognostication of colon carcinoma patients. TNM, tumor-nodes-metastasis.
The AUC of the ROC indicated that the prediction model had higher prognostic accuracy than any other factors. The AJCC staging system is widely applied in various tumor types, including colon carcinoma (16). The data of the present study indicated that the TNM stage was able to predict patient survival (AUC=0.751). However, combination of the TNM stage and risk score by binary logistic regression analysis significantly improved the prognostic value (AUC=0.797; Fig. 4).
Figure 4.
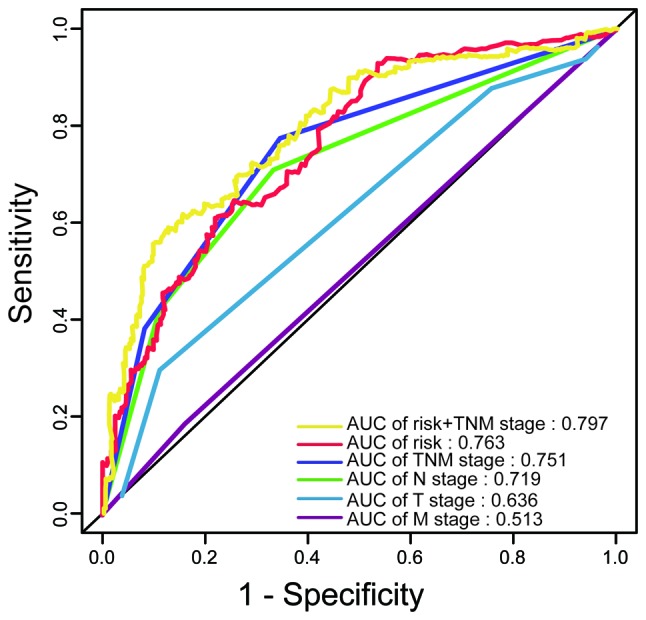
Time-dependent receiver operating characteristic curves comparing the prognostic accuracy of the risk score and the TNM, T, N or M stage, as well as the combination of the risk score and TNM stage. AUC, area under curve; TNM, tumor-nodes-metastasis.
The Kaplan-Meier curves also indicated that patient prognosis for patients stratified by their risk score combined with staging was significantly different (P<0.0001; Fig. 3E–H). All of these results demonstrated that the six miRNAs may be utilized for the prediction of the progression of colon carcinoma and the prognosis of affected patients. Furthermore, a more accurate prognosis was obtained by using a combination of the TNM stage and risk score.
Target prediction and functional analysis
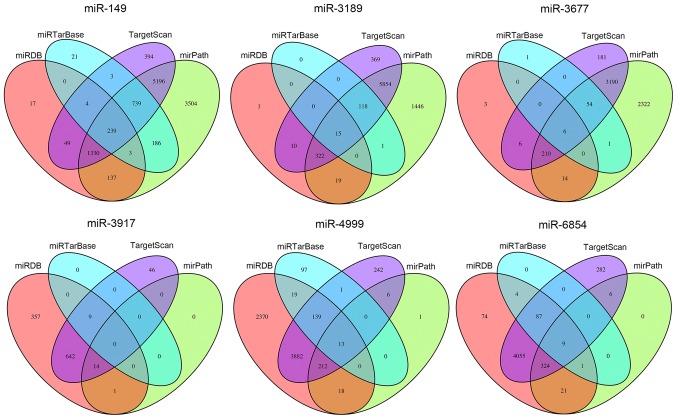
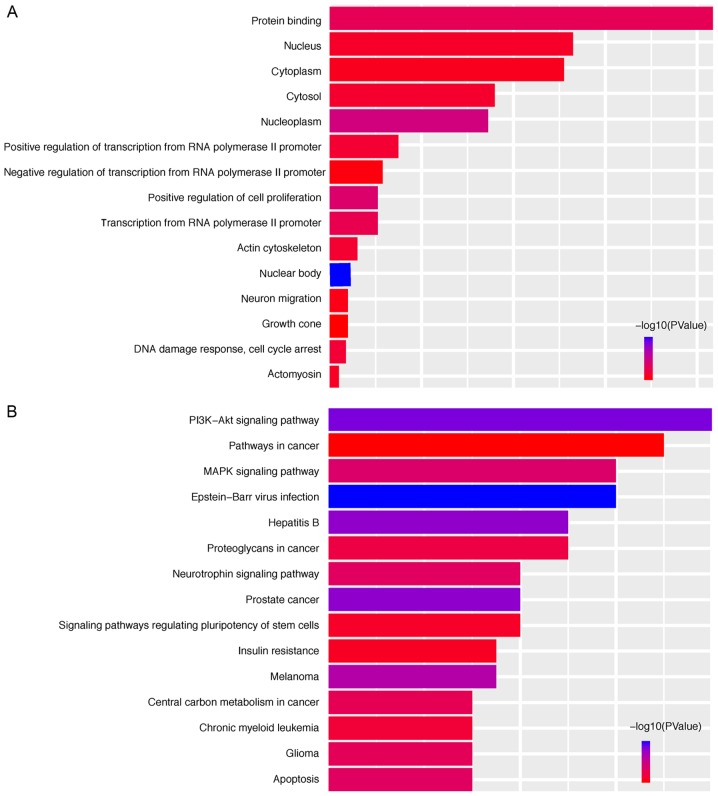
To elucidate the possible mechanisms that the six miRNAs are involved in, their target genes were predicted using the TargetScan, miRDB, mirPath and miRTarBase online analysis tools. A total of 239 overlapping genes for miR-149, 15 overlapping genes for miR-3189, 6 overlapping genes for miR-3677, 0 overlapping genes for miR-3917, 13 overlapping genes for miR-4999 and 9 overlapping genes for miR-6854 were obtained (Fig. 5). Subsequently, a bioinformatics enrichment analysis was performed. The overlapping genes of the DEmiRNAs were significantly enriched in KEGG pathways 'phosphoinositide-3 kinase (PI3K)-Akt signaling pathway', 'prostate cancer', 'mitogen-activated protein kinase (MAPK) signaling pathway', 'apoptosis' and 'neurotrophin signaling pathway'. The top five GO terms were 'nuclear body', 'nucleoplasm', 'positive regulation of cell proliferation', 'protein binding' and 'transcription from RNA polymerase II promoter' (Fig. 6). This functional enrichment analysis revealed the potential mechanisms of the prognostic miRNAs in the genesis/progression of colon cancer.
Figure 5.
Target genes of the six miRNAs were predicted using the TargetScan, miRDB, miRPath and miRTarBase online analysis tools, and the overlapping genes were selected. miRNA/miR, microRNA.
Figure 6.
Functional analysis of the target genes. (A) Top 15 significantly enriched Gene Ontology terms in the category biological process of target genes. (B) Top 15 significantly enriched Kyoto Encyclopedia of Genes and Genomes pathways of the target genes. PI3K, phosphoinositide-3 kinase; MAPK, mitogen-activated protein kinase.
Discussion
Colon carcinoma is one of the leading causes of cancer-associated mortality worldwide (2). Early diagnosis and radical resection may improve patient prognosis (17,18). However, the symptoms are frequently not obvious at the earliest stage, and at present, the detection of colon carcinoma relies on screening and endoscopy (8,19). Although the detection of serum CEA is currently available as a non-invasive method for the diagnosis and surveillance of colon cancer, its clinical utility is limited by its low sensitivity and specificity (20). Therefore, it is necessary to further elucidate the molecular mechanisms of colon cancer development, which may lead to the identification of novel accurate markers.
miRNAs are small non-coding RNAs, whose aberrant expression may be involved in the initiation and progression of colon carcinoma (21,22). Monitoring of changes in the expression of miRNAs may aid in the diagnosis and prediction of the prognosis of colon carcinoma and management of its recurrence (23–25).
In the present study, DEmiRNAs between colon carcinoma tissues and normal tissues were identified. A prognostic model using six miRNAs was then constructed based on univariate (data not shown) and multivariate Cox regression analysis. Furthermore, multivariate Cox regression and stratification analyses suggested that the six-miRNA signature was an independent prognostic factor for survival prediction of colon carcinoma patients. The risk score, based on which it was possible to effectively distinguish colon cancer patients with significantly different prognoses, was validated as an independent prognostic risk factor.
Furthermore, previous studies indicated that miR-149 is downregulated in breast cancer and colonic carcinoma cells and functions as a tumor suppressor by controlling breast epithelial cell migration and invasion, which is in agreement with the present results (26,27). miR-3189 has been previously reported to exert antitumor effects in glioblastoma and gastric cancer (28,29), miR-3677 may be used as a potential molecular marker to predict the prognosis of hepatic carcinoma patients (30,31), and miR-3917 may be useful for the early screening of high-risk populations and early diagnosis of lung cancer (32). Furthermore, miR-4999 and miR-6854, which have not been previously reported in tumors, to the best of our knowledge, may improve the accuracy of colon carcinoma prognostication.
The mRNAs regulated by the six-miRNA signature were enriched in pathways that were involved in cancer progression and prognosis. The six miRNAs were able to regulate several key signaling pathways, including the PI3K-Akt signaling pathway, apoptosis, the MAPK signaling pathway and the neurotrophin signaling pathway, which is in accordance with the results of the present bioinformatics analysis (33,34). The results indicated that the six-miRNA signature was highly correlated with cancer, which suggested the possibility of using it as a prognostic factor for colon carcinoma.
The current management of colon cancer relies on clinical and histopathologic factors, including TNM stage, tumor margin involvement, differentiation and lymphovascular invasion (35). The TNM staging system is the preferred staging system for colon cancer, and it may be beneficial to identify reliable biomarkers that are able to distinguish between different stages, to further assess tumor progression and aid the development of therapeutic strategies. The risk score postulated in the present study exhibited high sensitivity and specificity, and was correlated with the TNM stage and prognosis of colon cancer patients, which further suggests that it may be able to predict tumor occurrence and development. In addition, combining the risk score and TNM stage provided an improved prediction of the patients' survival status.
Of note, the present study was is entirely based on the TCGA dataset and experiments should be performed to further verify the mechanisms of the miRNAs involved in the tumorigenesis of colon cancer. In addition, further clinical investigations should be performed to validate the utility of this model for early diagnosis and evaluation of therapeutic efficacy.
In conclusion, the present study postulated a six-miRNA model that provides effective mortality risk stratification of colon carcinoma patients and may be a potential prognostic indicator. Combination with the TNM stage further improved the capacity of the risk score to predict patient prognosis. It may also be possible to utilize the six miRNAs for early diagnosis, allowing for a timely intervention.
Acknowledgments
Not applicable.
Funding
This study was supported in part by grants from the Jiangsu Provincial Key research development program (grant nos. BE2016794 to JF and BE2016795 to JW).
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding authors on reasonable request.
Authors' contributions
JF and JW contributed to the design of the study. HZ performed the bioinformatic analysis and wrote the manuscript. ZW and RM were responsible for article revision. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Villar-Vázquez R, Padilla G, Fernández-Aceñero MJ, Suárez A, Fuente E, Pastor C, Calero M, Barderas R, Casal JI. Development of a novel multiplex beads-based assay for autoantibody detection for colorectal cancer diagnosis. Proteomics. 2016;16:1280–1290. doi: 10.1002/pmic.201500413. [DOI] [PubMed] [Google Scholar]
- 4.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 5.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 6.Pucci S, Bonanno E, Sesti F, Mazzarelli P, Mauriello A, Ricci F, Zoccai GB, Rulli F, Galatà G, Spagnoli LG. Clusterin in stool: A new biomarker for colon cancer screening? Am J Gastroenterol. 2009;104:2807–2815. doi: 10.1038/ajg.2009.412. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Jiang X, Dong S, Shen J, Yu H, Zhou J, Li J, Ma H, He E, Skog S. Serum TK1 is a more reliable marker than CEA and AFP for cancer screening survey in a study of 56,286 people. Cancer Biomark. 2016;16:529–536. doi: 10.3233/CBM-160594. [DOI] [PubMed] [Google Scholar]
- 8.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng JH, Liang L, He RQ, Tang RX, Cai XY, Chen JQ, Luo DZ, Chen G. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget. 2017;8:16811–16828. doi: 10.18632/oncotarget.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sui J, Xu SY, Han J, Yang SR, Li CY, Yin LH, Pu YP, Liang GY. Integrated analysis of competing endogenous RNA network revealing lncRNAs as potential prognostic biomarkers in human lung squamous cell carcinoma. Oncotarget. 2017;8:65997–66018. doi: 10.18632/oncotarget.19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Hari DM, Bilchik AJ. Clinical decision-making and implementation challenges with the AJCC VII staging system for colorectal carcinoma. J Surg Oncol. 2011;105:221–222. doi: 10.1002/jso.22086. [DOI] [PubMed] [Google Scholar]
- 17.van Rossum LG, van Rijn AF, Laheij RJ, Van Oijen MG, Fockens P, Van Krieken HH, Verbeek AL, Jansen JB, Dekker E. Random comparison of guaiac and immunochemical fecal occult blood tests for colorectal cancer in a screening population. Gastroenterology. 2008;135:82–90. doi: 10.1053/j.gastro.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 18.Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, Jemal A, Schymura MJ, Lansdorp-Vogelaar I, Seeff LC, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116:544–573. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: A potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 20.Spratlin JL, Hui D, Hanson J, Butts C, Au HJ. Community compliance with carcinoembryonic antigen: Follow-up of patients with colorectal cancer. Clin Colorectal Cancer. 2008;7:118–125. doi: 10.3816/CCC.2008.n.016. [DOI] [PubMed] [Google Scholar]
- 21.Ito M, Mitsuhashi K, Igarashi H, Nosho K, Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507–2515. doi: 10.1002/ijc.28920. [DOI] [PubMed] [Google Scholar]
- 22.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng M, Wu D, Yang C, Peng H, Wang G, Wang T, Li X. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2017;181:108–120. doi: 10.1016/j.trsl.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 25.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Bischoff A, Huck B, Keller B, Strotbek M, Schmid S, Boerries M, Busch H, Müller D, Olayioye MA. miR149 functions as a tumor suppressor by controlling breast epithelial cell migration and invasion. Cancer Res. 2014;74:5256–5265. doi: 10.1158/0008-5472.CAN-13-3319. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G, Liu X, Li Y, Wang Y, Liang H, Li K, Li L, Chen C, Sun W, Ren S, et al. EphB3-targeted regulation of miR-149 in the migration and invasion of human colonic carcinoma HCT116 and SW620 cells. Cancer Sci. 2017;108:408–418. doi: 10.1111/cas.13161. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Jeansonne D, DeLuca M, Marrero L, Lassak A, Pacifici M, Wyczechowska D, Wilk A, Reiss K, Peruzzi F. Anti-tumoral effects of miR-3189-3p in glioblastoma. J Biol Chem. 2015;290:8067–8080. doi: 10.1074/jbc.M114.633081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian Y, Guo J, Qiao L, Sun X. miR-3189-3p mimics enhance the effects of S100A4 siRNA on the inhibition of proliferation and migration of gastric cancer cells by targeting CFL2. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19010236. pii: E236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Chong CC, Chen GG, Lai PB. A Seven-microRNA expression signature predicts survival in hepatocellular carcinoma. PLoS One. 2015;10:e0128628. doi: 10.1371/journal.pone.0128628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775–8784. doi: 10.18632/oncotarget.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Sui J, Shen X, Li C, Yao W, Hong W, Peng H, Pu Y, Yin L, Liang G. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of lung cancer. Oncol Rep. 2017;37:3543–3553. doi: 10.3892/or.2017.5612. [DOI] [PubMed] [Google Scholar]
- 33.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 35.Kandimalla R, Gao F, Matsuyama T, Ishikawa T, Uetake H, Takahashi N, Yamada Y, Becerra CR, Kopetz S, Wang X, Goel A. Genome-wide discovery and identification of a novel miRNA signature for recurrence prediction in stage II and III colorectal cancer. Clin Cancer Res. 2018 Mar 7; doi: 10.1158/1078-0432.CCR-17-3236. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding authors on reasonable request.