Abstract
The present study aimed to develop a pathway-based prognosis prediction model for glioblastoma (GBM). Univariate and multivariate Cox regression analysis were used to identify prognosis-related genes and clinical factors using mRNA-seq data of GBM samples from The Cancer Genome Atlas (TCGA) database. The expression matrix of prognosis-related genes was transformed into pathway deregulation score (PDS) based on the Gene Set Enrichment Analysis (GSEA) repository using Pathifier software. With PDS scores as input, L1-penalized estimation-based Cox-proportional hazards (PH) model was used to identify prognostic pathways. Consequently, a prognosis prediction model based on these prognostic pathways was constructed for classifying patients in the TCGA set or each of the three validation sets into two risk groups. The survival difference between these risk groups was then analyzed using Kaplan-Meier survival analysis and log-rank test. In addition, a gene-based prognostic model was constructed using the Cox-PH model. The model of prognostic pathway combined with clinical factors was also evaluated. In total, 148 genes were discovered to be associated with prognosis. The Cox-PH model identified 13 prognostic pathways. Subsequently, a prognostic model based on the 13 pathways was constructed, and was demonstrated to successfully differentiate overall survival in the TCGA set and in three independent sets. However, the gene-based prognosis model was validated in only two of the three independent sets. Furthermore, the pathway+clinic factor-based model exhibited better predictive results compared with the pathway-based model. In conclusion, the present study suggests a promising prognosis prediction model of 13 pathways for GBM, which may be superior to the gene-level information-based prognostic model.
Keywords: pathway, gene, clinical variable, prognosis index, pathway dysregulation score
Introduction
Glioblastoma, namely glioblastoma multiforme (GBM), is the most fatal cancer developed within the brain, which is characterized by rapid progression, common therapeutic resistance and a high probability of recurrence (1,2). The mean survival time of patients is 12 to 15 months following diagnosis, with a small portion of patients surviving longer than five years (3). Disappointingly, small improvement has been made in GBM patients' prognosis over the years (4). Therefore, powerful prognostic model based on molecular biomarkers are needed to facilitate accurate prediction of GBM prognosis.
Identifying prognostic biomarkers for GBM has attracted increasing attention. For instance, gene signature-based survival models for prognosis prediction with high-throughput data have been investigated in multiple studies (5,6). Sana et al (7) have suggested a six-microRNA (miRNA) signature-based risk score model as an independent prognostic predictor of GBM. Based on the observation that closely correlated genes are involved in the same biological processes, incorporating higher-order representative features, such as pathways, is thought to yield more stable and robust prognosis prediction (8). In addition, alterations in multiple pathways have important roles in cancer initiation and progression (9,10). Hence, characterization of pathway-level information is crucial for improving patient survival and developing individualized cancer therapies. Pathifier is an algorithm for pathway analysis of high-throughput data, which could quantify deviation of each pathway from normal behavior in a context-specific manner by using pathway deregulation score (PDS) (11). Pathway-based transcriptomic information of breast cancer has been used for prognosis prediction (12). For GBM, the pathways significantly associated with survival have been explored with Pathifier (11). However, prognosis stratification models based on pathway-level information have not been studied in GBM.
In the present study, based on The Cancer Genome Atlas (TCGA) data of GBM patients, a pathway-based prognosis prediction model was constructed using a combination of univariate and multivariate Cox regression analysis, PDS calculation by Pathifier, and L1-penalized estimation-based Cox-proportional hazards (Cox-PH) model. Additionally, the risk differentiating power of pathway-based model was successfully validated in three independent sets. The pathway-based model was compared to a gene-based model for predictive robustness. Furthermore, the pathway-based information was integrated with clinical features to build a pathway+clinic factor-based model, with the aim of improving the prognostic performance of the pathway-based model. These findings may have important implications for GBM prognosis and may hold promising potential for personalized therapeutic intervention.
Materials and methods
Data source and preprocessing
The mRNA-seq data of 154 GBM tissue samples which were acquired from the TCGA repository (https://gdc-portal.nci.nih.gov/, platform: Illumina HiSeq 2000 RNA Sequencing) were considered as a training set in the current study. An additional three validation sets were used in the study: The gene expression profiles of 128 GBM samples numbered 'Part A' (13,14), which were downloaded from the Chinese Glioma Genome Atlas database (CGGA, http://cgga.org.cn/); the GSE13041 dataset (platform, GPL96; Affymetrix Human Genome U133 Array) including gene expression data of 191 GBM samples downloaded from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/); and the GSE74187 dataset (platform, GPL6480; Agilent-014850 Whole Human Genome Microarray 4×44K) of 60 GBM samples downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/). The clinical characteristics of all these four sets are listed in Table I.
Table I.
Clinical features of TCGA set and three validation sets.
| Clinical factor | TCGA (n=154) | GSE13041 (n=191) | CGGA (n=128) | GSE74187 (n=60) |
|---|---|---|---|---|
| Age (years, mean ± SD) | 59.84±13.54 | 53.83±13.65 | 47.41±11.83 | – |
| Sex (male/female/−) | 99/54/1 | 116/74/1 | 62/39/27 | – |
| Chemotherapy (yes/no/−) | 44/91/19 | – | – | – |
| Drug therapy (yes/no/−) | 19/115/20 | – | – | – |
| Pharmaceutical therapy (yes/no/−) | 55/84/15 | – | – | – |
| Radiation therapy (yes/no/−) | 19/120/15 | – | – | – |
| Targeted molecular therapy (yes/no/−) | 18/116/20 | – | – | – |
| Progression free survival (yes/no) | – | – | – | 51/9 |
| Progression free survival months (mean ± SD) | – | – | – | 14.94±10.56 |
| Death (dead/alive) | 102/50 | 176/15 | 68/33 | 46/14 |
| Overall survival (months, mean ± SD) | 12.06±10.41 | 19.37±19.41 | 14.24±7.85 | 19.15±10.58 |
TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas; SD, standard deviation; −, information unavailable.
Raw data (CEL files) in GSE13041 (platform, GPL96) were processed for background correction and normalization by oligo package (15) (http://www.bioconductor.org/packages/release/bioc/html/oligo.html) in R language (version 3.4.1). With regard to the CGGA and GSE74187 datasets downloaded in the GPL6480 platform, probes were annotated to genes according to platform annotation profiles. By using the limma package (16) (https://bioconductor.org/packages/release/bioc/html/limma.html) in R language (version 3.4.1), data was log2 transformed to achieve normal distribution, and standardized using median normalization.
Identification of differentially expressed genes (DEGs)
In the TCGA set, the patients that died within 6 months following diagnosis were classified as bad prognosis, and the patients with survival time >12 months were considered as good prognosis. The DEGs between the bad prognosis and good prognosis patients were screened using edgeR package in R language (version 3.4.1) with the thresholds of |log fold change (FC)|>0.585 and false discovery rate (FDR) <0.05.
Screening for prognosis-related genes and clinical features
Using the survival package in R language (version 3.4.1; http://bioconductor.org/packages/survivalr/), univariate Cox regression analysis (17) was performed to reveal the DEGs and clinical features that are significantly associated with survival. The genes and clinical characteristics with log-rank P-value <0.05 were further subjected to multivariate Cox regression analysis to identify the prognosis-related genes (17). According to the expression levels of the prognosis-related genes, two-way hierarchical clustering analysis based on centered Pearson correlation algorithm (18) was conducted with the pheatmap package (19) in R language (https://bioconductor.org/packages/release/bioc/html/pheatmap.html; version 3.4.1).
Constructing a pathway-based prognosis prediction model
The Gene Set Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/) software (20) is a freely available tool for analysis of microarray data at the gene-level, including 217 Biocarta pathways and 186 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. In order to evaluate pathway deregulation associated with GBM, the Pathifier package (8) (http://bioconductor.org/packages/pathifier/) in R language (version 3.4.1) was applied to calculate a PDS for each pathway in each sample based on the expressions of the prognosis-related genes in the TCGA set. The PDS score was indicative of the degree of deviation in the activity of a pathway in GBM compared to the activity in normal tissue.
The PSD matrix was inputted, and least absolute shrinkage and selection operator (LASSO) estimation-based Cox-PH model was then used to identify the specific predictive pathways of prognosis by penalized package in R language (version 3.4.1). The optimal parameter 'lambda' was determined by running 1,000 simulations through cross-validation likelihood. Consequently, a pathway-based prognostic model was constructed with the predictive pathways and their Cox-PH coefficients and PDS scores. The prognosis index was calculated as follows: ; where CoefPi stands for the Cox-PH coefficient of pathway i; and PDSPi stands for the PDS score of pathway i.
All samples in the training set were divided into high-risk group (above median PI) or low-risk group (below median PI). The overall survival time of the two groups was compared using Kaplan-Meier survival analysis (21) and log-rank test. Similarly, all samples in each validation set (CGGA set, GSE13041 and GSE74187) were classified by PI into two risk groups, followed by comparison of overall survival time between the two groups. Receiver operating characteristic (ROC) curve analysis was conducted to compare the sensitivity and specificity of the prognosis prediction model. The area under the curve (AUC) was calculated as well. For log-rank test and ROC analysis, significance level was set at P<0.05.
Constructing a gene-based prognosis prediction model
In order to establish a prognosis prediction model based on gene expression, the prognostic genes for GBM were identified by LASSO estimation-based Cox-PH model, with expression matrix of the prognosis-related genes as input. Expression data of these prognostic genes and their Cox-PH coefficients were used to develop a gene-based prognosis prediction model as follows: ; where CoefPi denotes the Cox-PH coefficient of gene i; and expgenei denotes expression level of gene i.
By using this model, all samples in the TCGA set, CGGA set, GSE13041, or GSE74187 were classified by PI into a high-risk group and a low-risk group, separately. The overall survival time of the two risk groups was compared by Kaplan-Meier survival analysis and log-rank test for each set, separately, followed by ROC analysis.
Developing a pathway+clinic factor-based model
In order to investigate whether clinical features could improve the predictive performance of the pathway-based model, the significant clinical features extracted from multivariate Cox regression analysis were combined with prognostic pathways into the LASSO penalized step to construct a pathway+clinic factor-based model. Similarly, samples were separated into high-risk group and low-risk group by PI for TCGA set, CGGA set, GSE13041, and GSE74187, separately. The model performance was evaluated similarly to that of the pathway-based model.
Results
DEGs in TCGA set
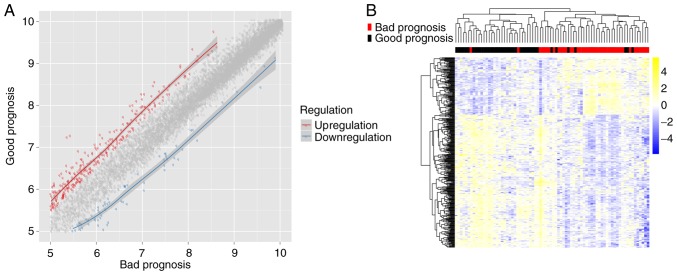
There were 38 bad prognosis samples and 38 good prognosis samples in TCGA set. A total of 402 DEGs were identified between the good and bad prognosis samples, including 84 downregulated DEGs and 318 upregulated DEGs (Fig. 1A). Two-way hierarchical clustering analysis of these DEGs revealed that the good prognosis samples were distinguished from the bad prognosis samples based on expression pattern of these DEGs (Fig. 1B).
Figure 1.
Analysis of DEGs between good and bad prognosis groups. (A) Scatter plot of DEGs. Red spots represent upregulated genes in the good prognosis group; blue spots represent downregulated genes; grey spots represent the non-differentially expressed genes between good and bad prognosis groups. (B) Heatmap showing the hierarchical clustering of DEGs. DEGs, differentially expressed genes.
Selection of prognosis-related genes and clinical characteristics
The 402 DEGs and clinical characteristics were subjected to univariate Cox regression analysis. Three clinical features, including age, chemotherapy and pharmaceutical therapy were significantly associated with overall survival by univariate Cox regression analysis (P<0.05, Table II). In addition, multivariate Cox regression analysis revealed that 148 genes and pharmaceutical therapy were independent biomarkers for prognosis in GBM (Table II).
Table II.
Clinical factors significantly associated with prognosis.
| Clinical factor | Univariable Cox regression
|
Multivariable Cox regression
|
||
|---|---|---|---|---|
| P-value | HR (95% CI) | P-value | HR (95% CI) | |
| Age (≤60/>60 years) | 0.015 | 1.631 (1.096-2.427) | 0.132 | 1.409 (0.902-2.201) |
| Sex (male/female) | 0.289 | 0.806 (0.540-1.202) | 0.210 | 0.746 (0.472-1.179) |
| Chemo therapy (yes/no) | 0.003 | 0.503 (0.319-0.793) | 0.375 | 1.491 (0.618-3.598) |
| Drug therapy (yes/no) | 0.104 | 0.622 (0.349-1.108) | 0.676 | 1.203 (0.506-2.864) |
| Pharmaceutical therapy (yes/no) | <0.001 | 0.351 (0.221-0.558) | 0.003 | 0.249 (0.010-0.626) |
| Radiation therapy (yes/no) | 0.111 | 0.587 (0.303-1.138) | 0.168 | 0.545 (0.230-1.291) |
| Targeted molecular therapy (yes/no) | 0.466 | 0.803 (0.444-1.451) | 0.949 | 0.975 (0.449-2.114) |
HR, hazard ratio; CI, confidence interval.
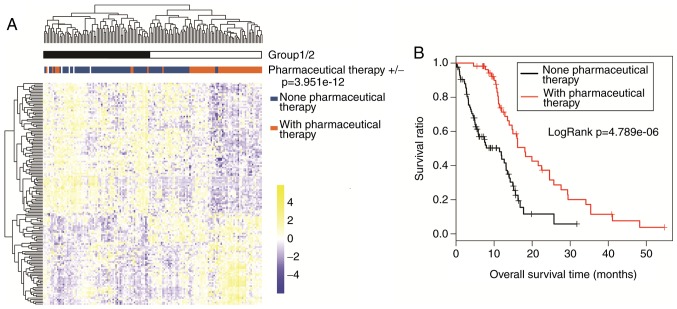
As illustrated in Fig. 2A, two-way hierarchical clustering analysis demonstrated that TCGA samples were classified into two groups based on expression data of the 148 prognosis-related genes. Group 1 included 8 patients that received pharmaceutical therapy and 64 patients that did not receive pharmaceutical therapy. Group 2 was comprised of 47 patients that received pharmaceutical therapy and 20 patients that did not receive pharmaceutical therapy. Chi-square test revealed that the two groups clustered by the 148 prognosis-related genes had a significant correlation to pharmaceutical therapy (χ2=48.149, P=3.951×10−12). In addition, Kaplan-Meier survival analysis revealed that group 2 had significantly improved survival compared with group 1 (P=1.663×10−04; data not shown). According to Kaplan-Meier survival analysis, the patients with pharmaceutical therapy had significantly improved survival than the patients without pharmaceutical therapy in the TCGA set (P=4.789×10−06; Fig. 2B). These results indicate that the pharmaceutical therapy is closely associated with prognosis.
Figure 2.
Association of pharmaceutical therapy with prognosis. (A) Two-way hierarchical clustering analysis of the 148 prognosis-related genes. TCGA samples were categorized into group 1 and group 2, based in the hierarchical clustering. The two groups were compared for pharmaceutical therapy using chi-square test (P=3.951×10−12). (B) Kaplan-Meier survival analysis of patients with or without pharmaceutical therapy in TCGA set. TCGA, The Cancer Genome Atlas.
Identification of prognostic pathways
The expression matrix of the 148 prognosis-related genes was transformed into PDS matrix, which was then used as initial input for LASSO estimation-based Cox-PH model. When the maximal cross-validation likelihood was −490.999, the optimal lambda value reached 19.700. Consequently, 13 pathways were selected to be prognostic pathways (Table III). These pathways involved 19 prognosis-related genes (Table IV).
Table III.
Thirteen prognostic pathways identified by LASSO estimation-based univariate Cox-proportional hazards model.
| Pathways | Coefficient | HR | P-value |
|---|---|---|---|
| BIOCARTA_HIVNEF_PATHWAY | 0.113 | 1.002 | <0.001 |
| KEGG_ARACHIDONIC_ACID_METABOLISM | 3.310 | 3.168 | <0.001 |
| KEGG_AXON_GUIDANCE | 2.784 | 2.124 | 0.001 |
| KEGG_CYTOKINE-CYTOKINE_RECEPTOR_INTERACTION | 0.609 | 1.021 | 0.002 |
| KEGG_ENDOCYTOSIS | 0.647 | 1.118 | 0.003 |
| KEGG_FOCAL_ADHESION | 1.666 | 1.460 | 0.001 |
| KEGG_INSULIN_SIGNALING_PATHWAY | −1.653 | 0.322 | 0.001 |
| KEGG_NITROGEN_METABOLISM | 2.046 | 1.769 | 0.001 |
| KEGG_O-GLYCAN_BIOSYNTHESIS | 1.133 | 1.391 | 0.001 |
| KEGG_PATHWAYS_IN_CANCER | 2.272 | 1.889 | 0.001 |
| KEGG_TYPE_II_DIABETES_MELLITUS | 2.394 | 2.081 | 0.001 |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 1.875 | 1.682 | 0.002 |
| KEGG_CELL_ADHESION_MOLECULES | 3.288 | 2.963 | 0.002 |
LASSO, least absolute shrinkage and selection operator; HR, hazard ratio.
Table IV.
List of genes involved in the 13 prognostic pathways.
| Gene name | Count of involved pathways | HR | P-value |
|---|---|---|---|
| MET | 12 | 0.591 | 0.049 |
| PLA2G5 | 11 | 0.944 | 0.022 |
| BIRC3 | 10 | 1.024 | 0.002 |
| HK3 | 7 | 0.662 | 0.013 |
| SOCS1 | 6 | 1.098 | 0.006 |
| NGFR | 5 | 0.906 | 0.033 |
| L1CAM | 3 | 2.716 | 0.016 |
| NKX3-1 | 2 | 3.109 | <0.001 |
| NRXN3 | 2 | 1.953 | 0.001 |
| PTGES | 2 | 0.909 | 0.010 |
| ACAP1 | 1 | 0.326 | 0.002 |
| ALOX15B | 1 | 1.176 | 0.036 |
| CA14 | 1 | 1.236 | 0.005 |
| CA9 | 1 | 1.875 | 0.013 |
| CDH4 | 1 | 1.780 | 0.010 |
| EPHA5 | 1 | 1.336 | 0.043 |
| EPHB6 | 1 | 0.341 | 0.007 |
| GALNT12 | 1 | 0.607 | 0.003 |
| GALNT6 | 1 | 0.537 | 0.046 |
HR, hazard ratio.
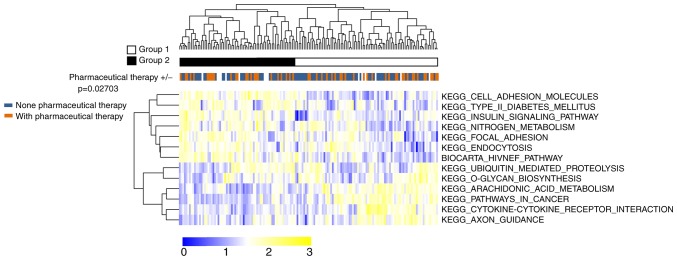
As illustrated in Fig. 3, two-way hierarchical clustering analysis based on the 13 prognostic pathways PDS scores categorized all samples of TCGA set into two groups: Group I (n=69) and group II (n=85). There were 35 samples with pharmaceutical therapy and 25 samples without pharmaceutical therapy in Group I. Group II had 30 samples with pharmaceutical therapy and 49 samples without pharmaceutical therapy. The proportion of pharmaceutical therapy was significantly different between the two groups (χ2=4.899, P=0.027), indicating that this grouping approach had a significant association with pharmaceutical therapy. These results indicate that pharmaceutical therapy is an important prognostic clinical feature for GBM.
Figure 3.
Two-way hierarchical clustering analysis of TCGA samples based on pathway dysregulation score of the 13 prognostic pathways. TCGA samples were clustered into two groups (group 1 and group 2). Chi-square test revealed that the two groups were significantly different in the proportion of pharmaceutical therapy (P=0.02703). TCGA, The Cancer Genome Atlas.
Prognostic performance of the pathway-based model
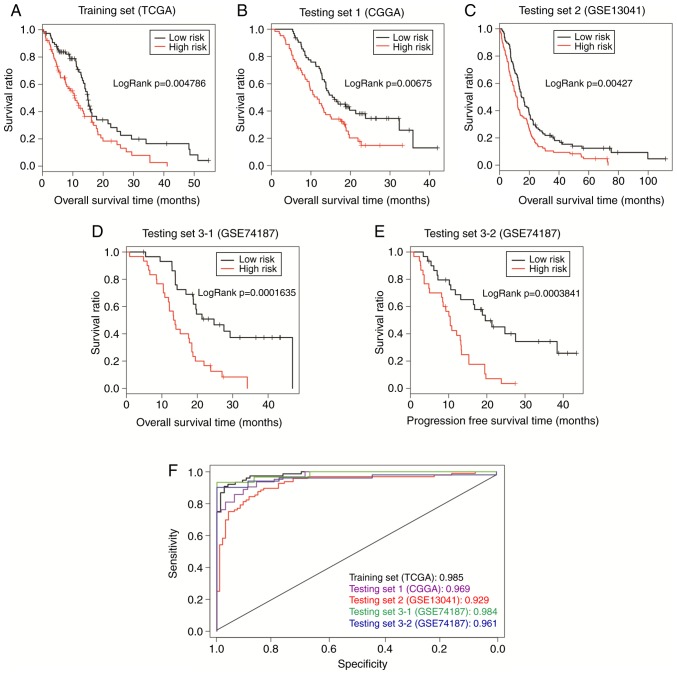
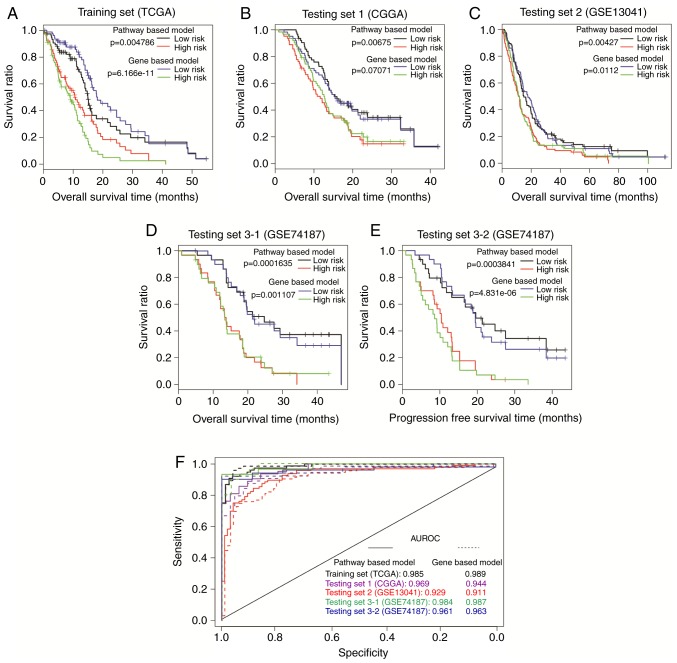
The pathway-based model calculated a PI for each sample with Cox-PH coefficients of the 13 prognostic pathways, and the TCGA samples were then separated by PI into the high-risk group (n=77) and the low-risk group (n=77). According to Kaplan-Meier survival curves (Fig. 4A), the high-risk group had significantly shorter overall survival time compared with the low-risk group (10.38±8.68 vs. 13.74±11.71 months, respectively; P= 0.005). The AUC was 0.987 (Fig. 4F), suggesting that the pathway-based model could predict the survival outcome of GBM patients.
Figure 4.
Prognosis performance of the pathway-based model. Samples in (A) TCGA set, (B) CGGA set, (C) GSE13041, and (D and E) GSE74187 were dichotomized by PI into a high-risk group and a low-risk group. The two groups were compared by Kaplan-Meier curves, and the P-value was calculated by log-rank test. (F) Comparison of ROC curves between different sets. ROC curves were generated with PI values as predictions compared to overall survival or progression-free survival. TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas; PI, prognosis index; ROC, receiver operating characteristic.
The robustness of this pathway-based model was validated in the CGGA, GSE13041 and GSE74187 sets. All samples in each set were classified into high-risk group or low-risk group with the threshold of median PI. For CGGA set, improved survival was observed in the low-risk group compared with the high-risk group (16.69±8.35 vs. 12.28±7.16 months, respectively; P=0.007; Fig. 4B), with an AUC of 0.969 (Fig. 4F). The pathway-based model also exhibited good predictive power with P=0.004 (22.56±21.47 vs. 16.21±16.65 months, respectively; Fig. 4C) and AUC of 0.929 (Fig. 4F) in the GSE13041 set. In the GSE74187 set, the low-risk patients (n=30) had markedly longer overall survival time (23.40±11.56 vs. 14.89±7.54 months; P=1.635×10−04; Fig. 4D) and progress-free survival (PFS) time (19.23±12.01 month vs. 10.66±6.68; P=0.0003841; Fig. 4E), compared with the high-risk patients (n=30). Furthermore, the ROC curve demonstrated an AUC value of 0.984 for overall survival, and 0.961 for PFS (Fig. 4F). These results indicate that the prognostic power of the pathway-based model is successfully validated in all the three independent sets.
Prognostic performance of the gene-based model
Based on expression data of the 148 prognosis-related genes, LASSO estimation-based Cox-PH model uncovered 22 genes that were significantly associated with survival (Table V). The 22-gene signature-based model calculated a PI for each sample as described above. All patients of the TCGA set were divided by PI into high-risk group (n=77) or low-risk group (n=77). According to Kaplan-Meier survival analysis, overall survival time was significantly different between the high-risk group and the low-risk group (8.007±6.43 vs. 16.12±11.98 months, respectively; P=6.166×10−11; Fig. 5A). The ROC value was 0.989 (Fig. 5F), consistent with the results of the Kaplan-Meier curve analysis.
Table V.
Twenty-two prognostic genes identified by LASSO-based univariate Cox-proportional hazards model.
| Gene name | Coefficient | HR | P-value |
|---|---|---|---|
| AZGP1 | 0.138 | 1.375 | 0.049 |
| CA9 | 0.019 | 1.875 | 0.013 |
| COL22A1 | 0.844 | 1.592 | 0.002 |
| CPNE6 | 0.249 | 1.006 | 0.030 |
| EN2 | 0.063 | 0.838 | 0.016 |
| FERMT1 | −0.649 | 0.355 | 0.003 |
| GPC5 | 0.162 | 2.420 | 0.002 |
| HES5 | −0.098 | 0.776 | 0.019 |
| HIST3H2A | −0.865 | 0.997 | 0.002 |
| HOXB2 | 0.365 | 1.519 | 0.012 |
| HOXC10 | 0.229 | 1.250 | 0.024 |
| IGFBP6 | 0.216 | 1.415 | 0.000 |
| L1CAM | 0.336 | 2.716 | 0.016 |
| LRRC61 | 0.429 | 2.325 | <0.001 |
| MSTN | −0.659 | 1.389 | 0.001 |
| NEUROD1 | −0.254 | 1.066 | 0.011 |
| NRXN3 | 0.236 | 1.953 | 0.001 |
| OLFM1 | 0.842 | 2.143 | 0.005 |
| PTPRN | 0.594 | 0.723 | <0.001 |
| PYROXD2 | 0.226 | 0.920 | 0.002 |
| RGS7 | 0.012 | 0.915 | 0.011 |
| RPL39L | 1.051 | 2.100 | 0.000 |
LASSO, least absolute shrinkage and selection operator; HR, hazard ratio.
Figure 5.
Prognosis performance of the gene-based model. Samples in the (A) TCGA set, (B) CGGA set, (C) GSE13041, and (D and E) GSE74187 were divided by PI into a high-risk group and a low-risk group. Differences in overall survival or progression-free survival between the two groups were analyzed by Kaplan-Meier curves. P-values were produced by log-rank test. (F) ROC curves and AUC analysis. ROC curves were generated for PI-based classification for each set. TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas; PI, prognosis index; ROC, receiver operating characteristic; AUC, area under the curve.
The CGGA, GSE13041 and GSE74187 sets were then used to validate the pathway-based model. The gene-based model gave a P-value of 0.071 (13.49±7.41 vs. 15.48±8.59 months; Fig. 5B) and an AUC value of 0.94 (Fig. 5F) for the CGGA set, and a P-value of 0.011 (16.59±18.08 vs. 22.18±20.38 months; Fig. 5C) and an AUC value of 0.911 (Fig. 5F) for GSE13041. Additionally, there were obvious differences in both overall survival time (14.94±9.08 vs. 23.36±10.42 months; P=0.001; Fig. 5D) and PFS time (9.41±7.16 vs. 20.06±11.90 months; P=4.831×10−06; Fig. 5E) between the different risk groups in GSE74187, with an AUC value of 0.987 for overall survival and 0.963 for PFS time (Fig. 5F). These results demonstrated that the gene-based model could successfully classify patients into two risk groups with significantly different overall survival time in GSE13041 and GSE74187. However, poor risk differentiation power was observed for the gene-based model in the CGGA set (P=0.071; Fig. 5B). Thus, the gene-based model was inferior to the pathway-based model for prognosis prediction in GBM.
Prognostic performance of the pathway+clinic factor-based model
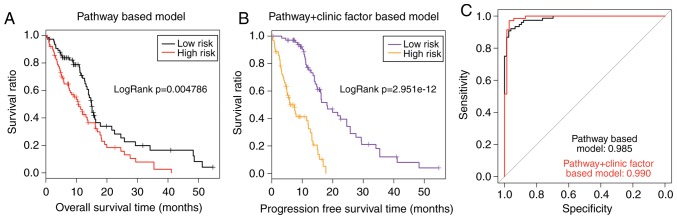
In order to improve the prognostic performance of the pathway-based model, a prognosis prediction model of the 13 prognostic pathways combined with pharmaceutical therapy was constructed in the present study. Based on the PI calculated by this pathway+clinic factor-based model, all patients in the TCGA set were classified into high-risk group (n=77) and low-risk group (n=77). Significantly different overall survival time was observed between the high-risk and low-risk group (6.54±4.62 months vs. 15.79±10.89 months; P=2.951×10−12; Fig. 6B). The AUC value was 0.990 for the TCGA set (Fig. 6C). As presented in Fig. 6A and B, the P-value of the pathway+clinic factor-based model was markedly more significant compared with the pathway-based model (2.951×10−12 vs. 0.004786). In addition, the AUC value of the pathway+clinic factor-based model was larger compared with the pathway-based model (0.990 vs. 0.985; Fig. 6C). These results demonstrated that the pathway+clinic factor-based model had better performance than the pathway-based model for risk assessment of GBM.
Figure 6.
Comparison of the pathway-based model and the pathway+clinic factor-based model in TCGA set. (A) Kaplan-Meier curve of the two risk groups dichotomized by the pathway-based model. (B) Kaplan-Meier curve of the two risk groups dichotomized by the pathway+clinic factor-based model. (C) Comparison of the two prognostic models for AUC. TCGA, The Cancer Genome Atlas; AUC, area under the curve.
Discussion
GBM, the most common malignancy in brain, which is characterized by inevitable recurrence and dismal prognosis results in decreased health-related quality of life (22). It has been hypothesized that the prognosis models based on higher-order representative relationships with genes, such as pathways and network modules, have more stable and accurate results (23–25). Therefore, the present study focused on exploring prognosis models based on the degree of pathway dysregulation caused by GBM, in combination with Cox-PH model and LASSO estimation. A total of 148 genes were identified to be significantly associated with prognosis by Cox regression analysis. Based on the expression matrix of the 148 prognosis-related genes, LASSO estimation-based Cox-PH model identified 13 prognostic pathways. A pathway-based model was constructed with the Cox-PH coefficients and the PDS scores of these pathways. The pathway-based model was trained on the TCGA set, and tested on three independent sets (CGGA, GSE13041, and GSE74187) of different sample sizes, which were downloaded from different platforms. The results demonstrated that the pathway-based model could successfully classify patients in each set into two risk groups with significantly different survival outcome. In addition, the present study also constructed a gene-based prognosis prediction model with the expression matrix and the Cox-PH coefficients of the 22 prognostic genes. The prognostic power of this gene-based model was validated in only two of the three validation sets, suggesting that the pathway-based model performed better than the gene-based model in terms of outcome prediction. Therefore, the prognostic performance of the pathway+clinic factor-based model was evaluated next, since the pathway-based model was superior to the gene-based model for prognosis prediction in GBM.
It has been demonstrated that models using genomic data combined with clinical data exhibit more accurate prognosis prediction compared with models using genomic data or clinical data alone (26). Cheng et al (27) have demonstrated that combined clinical and genomic model is superior over models based on either data type in terms of prognostic accuracy. Additionally, Huang et al (12) have provided strong evidence that integration of clinical and genomic information could greatly improve prognosis prediction compared with using only one type of information. In order to further improve the prediction power of this pathway-based model, a pathway+clinic factor-based prognostic model was developed with the 13 prognostic pathways and a significant clinical factor (pharmaceutical therapy) identified by Cox regression analysis. The pathway+clinic factor-based model exhibited improved performance compared with the pathway-based model and the gene-based model in the TCGA set, with improved P-values (2.951e-12 vs. 0.004786 vs. 6.166e-11) and AUC values (0.999 vs. 0.985 vs. 0.989). In addition, this combined model displayed considerably better prognostic performance compared with the prognostic three-gene signature for patients with MGMT promoter-methylated GBM, as described in the study of Wang et al (P=0.0033) (28).
In the present study, the pathway-based predictive model was based on 13 prognostic pathways, such as endocytosis pathway, insulin signaling pathway, ubiquitin-mediated proteolysis pathway, focal adhesion and cell adhesion pathways. These pathways might have great biological relevance to GBM prognosis. The endocytosis pathway is an active transport form, which is strengthened in cancer (29). The insulin signaling pathway is critical for glucose metabolism. Abnormal glucose metabolism has an important role in GBM growth and chemoresistance, suggesting glucose metabolism might be a promising target for developing GBM therapies (30). There is indeed evidence that the total lesion glycolysis in hypoxia (hTLG) representing hypoxic glucose metabolism is a signifi-cant prognostic factor for GBM (31). Ubiquitin-mediated proteolysis is a complex protein degradation process. Deregulation of the ubiquitin system in the ubiquitin-mediated proteolysis pathway has been demonstrated to be a causative factor of several types of cancer (32). Focal adhesion and cell adhesion molecules are critical determinants in cancer cell resistance to therapy (33).
There are several potential limitations in the present study. Firstly, the CGGA, GSE13041 and GSE74187 data-sets do not have information concerning pharmaceutical therapy. Hence, the pathway+clinic factor-based prognostic model was not validated in these sets. Therefore, further analysis with more datasets is necessary to fully test the robustness of the pathway+clinic factor-based model. Additionally, the study only analyzed gene-level information of 403 pathways in the GSEA repository, and some gene information may be inevitably lost. The pathway-based prognostic model will be applied to larger groups of GBM patients in future studies, in order to further validate its prognostic significance.
In conclusion, the present in silico study presents a promising prognosis prediction model based on 13 pathways, which is constructed by a combination of PDS-based Pathifier method and LASSO estimation-based Cox-PH model. The pathway-based model exhibited stronger prognostic power compared with the gene-based model. Furthermore, incorporating the clinical information of pharmaceutical therapy to the pathway-based model resulted in improved prognostic performance. Application of these pathway-based prognostic models might improve stratification of GBM patients and offer considerable potential for individualized GBM management.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
RL and MW performed the data analysis and wrote the manuscript. GZ, HZ and YZ contributed to the data analysis and manuscript revision. ZS conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kesari S. Understanding glioblastoma tumor biology: The potential to improve current diagnosis and treatments. Semin Oncol. 2011;38(Suppl 4):S2–S10. doi: 10.1053/j.seminoncol.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Aldape K, Zadeh G, Mansouri S, Reifenberger G, Deimling AV. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 3.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108:11–27. doi: 10.1007/s11060-011-0793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ, Yan W, Liu YW, Zhang W, Chen L, Jiang T. Prognostic value of a nine-gene signature in glioma patients based on mRNA expression profiling. CNS Neurosci Ther. 2014;20:112–118. doi: 10.1111/cns.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YW, Koul D, Kim SH, Lucioeterovic AK, Freire PR, Yao J, Wang J, Almeida JS, Aldape K, Yung WK. Identification of prognostic gene signatures of glioblastoma: A study based on TCGA data analysis. Neuro Oncol. 2013;15:829–839. doi: 10.1093/neuonc/not024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sana J, Radova L, Lakomy R, Kren L, Fadrus P, Smrcka M, Besse A, Nekvindova J, Hermanova M, Jancalek R, et al. Risk Score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis. 2014;35:2756–2762. doi: 10.1093/carcin/bgu212. [DOI] [PubMed] [Google Scholar]
- 8.Ma S, Kosorok MR, Huang J, Dai Y. Incorporating higher-order representative features improves prediction in network-based cancer prognosis analysis. BMC Med Genomics. 2011;4:5. doi: 10.1186/1755-8794-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 10.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drier Y, Sheffer M, Domany E. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci USA. 2013;110:6388–6393. doi: 10.1073/pnas.1219651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Yee C, Ching T, Yu H, Garmire LX. A novel model to combine clinical and pathway-based transcriptomic information for the prognosis prediction of breast cancer. PLoS Comput Biol. 2014;10:e1003851. doi: 10.1371/journal.pcbi.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan W, Zhang W, You G, Zhang J, Han L, Bao Z, Wang Y, Liu Y, Jiang C, Kang C, et al. Molecular classification of gliomas based on whole genome gene expression: A systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro Oncol. 2012;14:1432–1440. doi: 10.1093/neuonc/nos263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Zhang W, Chen D, Lv Y, Zheng J, Lilljebjörn H, Ran L, Bao Z, Soneson C, Sjögren HO, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc Natl Acad Sci USA. 2014;111:3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrish RS, Spencer HJ., III Effect of normalization on significance testing for oligonucleotide microarrays. J Biopharm Stat. 2004;14:575–589. doi: 10.1081/BIP-200025650. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P, Wang Y, Hang B, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–55351. doi: 10.18632/oncotarget.10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, Wang Y. RNA-seq analyses of multiple meri-stems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14:169. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilford CA, Siemers NO. Gene set enrichment analysis. Methods Mol Biol. 2009;563:99–121. doi: 10.1007/978-1-60761-175-2_6. [DOI] [PubMed] [Google Scholar]
- 21.Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–278. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. doi: 10.1007/978-1-60327-492-0_14. [DOI] [PubMed] [Google Scholar]
- 23.Abraham G, Kowalczyk A, Loi S, Haviv I, Zobel J. Prediction of breast cancer prognosis using gene set statistics provides signature stability and biological context. BMC Bioinformatics. 2010;11:277. doi: 10.1186/1471-2105-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Akker EB, Passtoors WM, Jansen R, van Zwet EW, Goeman JJ, Hulsman M, Emilsson V, Perola M, Willemsen G, Penninx BW, et al. Meta-analysis on blood transcriptomic studies identifies consistently coexpressed protein-protein interaction modules as robust markers of human aging. Aging Cell. 2014;13:216–225. doi: 10.1111/acel.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4:e1000217. doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pittman J, Huang E, Dressman H, Horng CF, Cheng SH, Tsou MH, Chen CM, Bild A, Iversen ES, Huang AT, et al. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proc Natl Acad Sci USA. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng F, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, Perou CM. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Zhang L, Wang Z, Yang F, Wang H, Liang T, Wu F, Lan Q, Wang J, Zhao J. A three-gene signature for prognosis in patients with MGMT promoter-methylated glioblastoma. Oncotarget. 2016;7:69991–69999. doi: 10.18632/oncotarget.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellman I, Yarden Y. Endocytosis and cancer. Cold Spring Harb Perspect Biol. 2013;5:a016949. doi: 10.1101/cshperspect.a016949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen H, Decollogne S, Dilda PJ, Hau E, Chung SA, Luk PP, Hogg PJ, McDonald KL. Dual-targeting of aberrant glucose metabolism in glioblastoma. J Exp Clin Cancer Res. 2015;34:14. doi: 10.1186/s13046-015-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyonaga T, Yamaguchi S, Hirata K, Kobayashi K, Manabe O, Watanabe S, Terasaka S, Kobayashi H, Hattori N, Shiga T, et al. Hypoxic glucose metabolism in glioblastoma as a potential prognostic factor. Eur J Nucl Med Mol Imaging. 2017;44:611–619. doi: 10.1007/s00259-016-3541-z. [DOI] [PubMed] [Google Scholar]
- 32.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 33.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.