Abstract
Hepatocellular carcinoma (HCC) is the 5th most common cancer, but the 3rd leading cause of cancer death globally with approximately 700,000 fatalities annually. The severity of this cancer arises from its difficulty to detect and treat. The major etiologies of HCC are liver fibrosis or cirrhosis from chronic viral infections, as well as metabolic conditions. Since most cases arise from prior pathologies, biomarker surveillance in high-risk individuals is an essential approach for early detection and improved patient outcome. While many molecular biomarkers have been associated with HCC, there are few that have made clinical impact for this disease. Here we review some major approaches used for HCC biomarker discovery – proteomics and glycomics – and describe new methodologies being tested for biomarker development.
Introduction
Hepatocellular carcinoma (HCC) is a malignancy of hepatocytes that arises within the liver. This cancer occurs in the background of patients with underlying liver disease such as liver fibrosis and cirrhosis often associated with chronic viral infections. Additionally, obesity-associated nonalcoholic fatty liver disease/nonalcoholic steatohepatitis has been recently considered a major etiology of HCC[1]. The survival rate of people with primary liver cancers is very low, with a 0.95 ratio of mortality to incidence[1]. The low survival rates have been attributed to late diagnosis and limited treatment options[2]. Although liver transplantation is the preferred option for surgical treatment of HCC, the paucity of organ donors means that partial hepatic resection is a common treatment[3]. Unfortunately, even with advances in surgery and patient care, reported 5-year survival rates are around 50%[4]. HCC is consequently responsible for approximately 700,000 deaths annually and ranks as the 3rd leading cause of cancer death worldwide[3,5]. The incidence of HCC has shown a drastic increase in the United States over the last 35 years[6], mainly attributed to hepatitis C virus infection and rising obesity/metabolic challenges[1].
Treatment of HCC
As a highly lethal cancer, successful treatment options for HCC are few. According to the American Association for the Study of Liver Diseases treatment guidelines for HCC, surgical resection or ablative strategies can be therapeutically valuable options for patients with small lesions and well-managed liver disease[5]. Candidates for resection are those without severe cirrhosis and who have 1–3 unilobar lesions (<3cm for multiple lesions or <5cm for one lesion), and this therapy is recommended over radiofrequency ablation[5]. Unfortunately, only about 10% of HCC patients are acceptable for resection[3], and there is significant risk of recurrence or de-novo tumor development following the resection or ablation of HCC lesions[4]. The most effective treatment option for HCC patients is liver transplantation, as it rids the patient of both the cancer and the underlying liver disease. Transplantation thus provides the best outcomes for patients, with 5-year survival rates of 70% and low risk of recurrence[4]. However, the major limits to liver transplantation are the shortage of organ donors as well as the stringent criteria for transplantation[3]. Even though liver transplantation is often viewed as a cure for HCC, intra-hepatic tumor recurrence can occur and is especially a risk for those patients with large initial tumors[7]. Chemotherapeutic options for HCC are limited and the frontline agent for those with non-ablatable tumors is the multi-kinase inhibitor sorafenib, sold under the brand name Nexavar. Sorafenib is a general tyrosine and serine/threonine protein kinase inhibitor with activity against vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF) receptors as well as intracellular kinases B-Raf and Raf-1[8]. Agents that specifically target one growth receptor, such as enhanced VEGFR inhibitors have failed to show activity against HCC[8]. It is noted, however, that sorafenib’s activity against HCC is limited, with improved survival times of only a few months[9]. These bleak treatment options –both in their availability and efficacy – highlight the necessity for early detection of HCC.
Clinical detection of HCC
The current clinical gold standards for detection of HCC are magnetic resonance imaging (MRI), ultrasound (US), and computed tomography (CT) scans to detect lesions. However, a retrospective analysis performed in 2011 indicates shockingly low sensitivity of US to detect small lesions of HCC, with sensitivity being improved upon the addition of MRI and or CT scans[10]. The proposed sensitivity levels of US, CT, and MRI were 46%, 65%, and 72%[10], which are far below commonly desired values for a clinical biomarker. This highlights a disconnect between current clinical practice and the expectations for biomarker performance in clinical trials. A prognostic biomarker is a biological molecule that can predict the occurrence of a disease state – often before any noticeable lesion or physical abnormality may arise, creating significant pressure on biomarkers to indicate what is to come. Thus, the commonly-held view of biomarkers as stand-alone clinical tests for early detection may be unrealistic. However, combining current clinical modalities with prognostic biomarkers could have significant benefit for detection, and a surveillance program study found that US screening combined with the glycoprotein biomarker alpha-fetoprotein (AFP) significantly increased the sensitivity of US screens from 43.9% to 90.2%[11]. The combination of US and AFP is now one of the most widely-used screening methods for HCC[10,11]. Along with prognostic biomarkers for detection, predictive biomarkers for HCC are also needed to suggest an individual’s response to treatment. These predictive biomarkers could serve to assist clinicians in selecting appropriate candidates for liver resection/transplantation as well as predicting disease recurrence[4]. Described below are two current techniques for identifying biomarkers of HCC: proteomics and glycomics. Multiple markers have been observed via each method, yet their clinical impact is little to none at present.
Proteomic identification of biomarkers of HCC
The liver secretes many proteins into the blood, allowing for non-invasive collection of proteins for analysis. Various proteomic methodologies have been proposed to identify proteins that are altered in the serum of those with HCC, and most have involved the comparative analysis of several patient groups: healthy subjects, those infected with hepatitis B or C, those with liver cirrhosis, and those with both liver cirrhosis and HCC. By utilizing sensitive machines and methods, often some form of mass spectrometry or liquid chromatography, low abundance proteins that change with cancer development can be found and related to the cancer. Using such methods, proteins such as peroxiredoxin 3, osteopontin, and AFP have been identified as potential markers of HCC, with upregulation of these proteins observed in HCC patient samples compared to healthy individuals or those with liver disease[12–15]. As mentioned previously, AFP is currently used as a biomarker in the clinic alongside ultrasound, yet it lacks the specificity and sensitivity to stand alone as a powerful biomarker. Another serum protein that has shown potential as a biomarker is des-gamma carboxyprothrombin (DCP), and studies suggest it to be a more powerful biomarker than AFP for larger tumors as well as those arising from viral etiology[16,17]. Recent experiments have begun to utilize combinations of protein markers to create more sensitive biomarker panels, for example combining AFP with another serum protein, fibronectin 1[18]. This multi-marker panel approach illustrates that detection performance can be improved by integrating separately-characterized protein biomarkers.
Glycomic identification of biomarkers of HCC
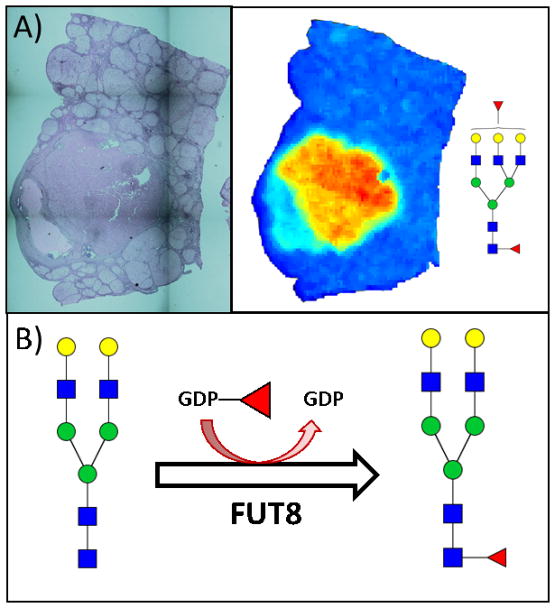
Glycomics is the profiling of glycans (sugar structures) attached to larger molecules such as proteins. The variety of glycan structures that may be attached to a protein creates a post-translational diversity often ignored. There is significant evidence illustrating that glycan structures are altered in the presence of cancer[19,20], and thus there is great potential for glycomic biomarkers as cancer-specific alterations attached to normal serum proteins. In regard to liver cancer, glycomic methodologies have long been used to either improve or discover biomarkers of liver cancer. Initial work showed that AFP with an attached α 1,6 core fucosylated glycan was a better marker of HCC than AFP alone, and it became a USFDA approved biomarker known as AFP-L3[21,22]. There is now substantial evidence to suggest that increased fucosylation occurs directly in the tumor and also that it plays a role in cancer development[23], Figure 1. One major issue with AFP-L3 is the protein to which the glycans are attached. That is, total AFP has a sensitivity of ~40–60%, a value which is not improved by the examination of the fucosylated glycoform[16]. Glycoforms are just a subset of the total AFP protein level, thus the sensitivity is not necessarily improved. However, as the results with AFP-L3 indicate that fucosylation is a highly specific HCC modification, groups have combined this glycomic information with proteomics to identify other proteins with glycan changes that could be used as biomarkers of liver cancer[24–30].
Figure 1.
N-linked glycan changes observed in HCC tissue. A) Recent experiments in our laboratory by MALDI Imaging Mass Spectrometry have shown an increase in core fucosylated glycans in HCC tumor tissue compared to the surrounding tissue. The images shown here are from a formalin fixed paraffin embedded HCC tissue: H&E stain (left) and MALDI image heatmap visualization of a core fucosylated glycan (right, mass value 2320.754 Da) illustrating its increased abundance in the tumor region. B) A recent genomic characterization of HCC noted that the FUT8 gene is overexpressed in response to the decrease of miR-122, a microRNA involved in the differentiation of hepatocytes that is downregulated in HCC[31]. Elevated levels of FUT8 would lead to a subsequent increase in core fucosylated N-glycans via the reaction shown here.
The importance of glycosylation in HCC progression has been observed with α 1,6-fucosyltransferase (FUT8), the enzyme responsible for catalyzing core fucosylation of N-glycans. Experiments with FUT8 knockouts showed significant reduction in growth factor signaling via the epidermal growth factor (EGF) and hepatocyte growth factor (HGF) receptors, as well as inhibited tumor formation in mice[23]. Additionally, a recent genomic analysis of HCC showed overexpression of the FUT8 gene, highlighting the likelihood for increased core fucosylation to be found on glycoproteins of HCC patients[31]. These data suggest that specific glycan alterations are key drivers for HCC carcinogenesis, making them important biomarkers as well as potential therapeutic targets.
Novel techniques to identify biomarkers of HCC
In the desperate search for biomarkers of HCC progression, there is also a need for new biomarker discovery techniques as well as the development of biomarker algorithms. Recent work has highlighted diagnostic algorithms that either utilize AFP alone[32,33] or AFP along with other novel biomarkers such as fucosylated kininogen[30,34] to greatly improve detection of early stage HCC. Other attempts to improve on AFP’s performance as a biomarker include immunochips as microfluidic[35] or Raman spectroscopy devices[36].
Another biomarker discovery technique, known as liquid biopsies, has recently grown in popularity with the isolation of cell-free DNA. A pilot study reported using cell-free DNA in urine as a marker for recurrence of HCC, with results illustrating the detection of recurrence up to 9 months before detectable by MRI[37]. While this study looked at recurrence, the method may have relevance for initial HCC detection as well. Another group studied the size profiles of cell-free DNA in circulation, and they observed a difference in the length of DNA molecules from those patients with HCC[38]. Most recently, a test referred to as CancerSEEK was created using both cell-free DNA and proteins as biomarkers to look at various cancers, one of them being liver cancer[39]. This test shows the benefit of combining various forms of biomarkers; however, its design highlights a key issue in HCC biomarker discovery and development: biomarkers aren’t needed to differentiate between healthy individuals and those with HCC. Instead, biomarkers are needed for screening and surveillance of those patients who already have underlying liver disease.
Conclusion
Hepatocellular carcinoma is a highly lethal cancer that is difficult to treat. The most effective treatment for HCC remains early detection, with surveillance programs being needed for patients with liver disease. When the cancer is caught early, patients have a significantly increased survival outcome. Therefore, biomarkers for early detection of HCC are paramount for the goal of reducing disease mortality. While there exist many potential biomarkers for differentiating between HCC and non-cancerous patients, few of these markers are currently being used in the clinic for screening and detection purposes. Current identified biomarkers include genomic, proteomic, and glycomic signatures, however they lack diagnostic power to perform as clinical markers due to poor sensitivity and/or specificity. Even the USFDA approved marker AFP-L3 suffers from low sensitivity. Yet a shift in mentality is needed for researchers and clinicians to recognize the place of biomarkers as indicators of future disease development. The predictive nature of biomarkers suggests that there is a low probability of discovering one catch-all biomarker to detect HCC with extremely high sensitivity and specificity. This is also limited by the heterogeneity of tumors and individuals.
Instead, combining biomarkers with clinical modalities such as ultrasound or creating panels of various combined biomarkers appear the most effective routes for increased sensitivity and specificity of detection. In order to continue identifying biomarkers for HCC to add to these panels, high-throughput methods for analysis of human samples such as tissue and serum are needed. Advances in mass spectrometry, genomics, and on-chip methodologies may be the future for discovering more effective biomarkers for HCC.
Highlights.
Early detection is an essential effective treatment for hepatocellular carcinoma
Current biomarkers for early detection of hepatocellular carcinoma are lacking
Proteomics and glycomics may provide key sources of cancer biomarkers
Acknowledgments
This work was supported by grants R01 CA120206 (ASM) and U01 CA168856 (ASM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petrick Braunlin, Laversanne Valery, Bray McGlynn. International trends in liver cancer incidence, overall and by histologic subtype, 1978–2007. International Journal of Cancer. 2016;139:1534–1545. doi: 10.1002/ijc.30211. International statistics on increased rates of HCC are discussed here along with projections of future incidence rates. * [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuchiya Biomarkers for the early diagnosis of hepatocellular carcinoma. World Journal of Gastroenterology. 2015;21:10573. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pang Surgical management of hepatocellular carcinoma. World Journal of Hepatology. 2014;7:245. doi: 10.4254/wjh.v7.i2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet J, Schwartz M, Mazzaferro V. Resection and Liver Transplantation for Hepatocellular Carcinoma. 2005;25:181–200. doi: 10.1055/s-2005-871198. [DOI] [PubMed] [Google Scholar]
- 5.Heimbach, Kulik, Finn, Sirlin, Abecassis, Roberts, Zhu, Murad, Marrero AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 6.Petrick, Kelly, Altekruse, McGlynn, Rosenberg Future of Hepatocellular Carcinoma Incidence in the United States Forecast Through 2030. Journal of Clinical Oncology. 2016;34:1787–1794. doi: 10.1200/JCO.2015.64.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welker, Bechstein, Zeuzem, Trojan Recurrent hepatocellular carcinoma after liver transplantation – an emerging clinical challenge. Transplant International. 2013;26:109–118. doi: 10.1111/j.1432-2277.2012.01562.x. [DOI] [PubMed] [Google Scholar]
- 8.Keating Sorafenib: A Review in Hepatocellular Carcinoma. Targeted Oncology. 2017;12:243–253. doi: 10.1007/s11523-017-0484-7. [DOI] [PubMed] [Google Scholar]
- 9.Llovet, Ricci, Mazzaferro, Hilgard, Gane, Blanc, ACDO, Santoro, Raoul, Forner, et al. Sorafenib in Advanced Hepatocellular Carcinoma. The New England Journal of Medicine. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 10.Yu, Chaudhari, Raman, Lassman, Tong, Busuttil, Lu CT and MRI Improve Detection of Hepatocellular Carcinoma, Compared With Ultrasound Alone, in Patients With Cirrhosis. Clinical Gastroenterology and Hepatology. 2011;9:161–167. doi: 10.1016/j.cgh.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Singal, Conjeevaram, Volk, Fu, Fontana, Askari, Su, Lok, Marrero Effectiveness of Hepatocellular Carcinoma Surveillance in Patients with Cirrhosis. Cancer Epidemiology and Prevention Biomarkers. 2012;21:793–799. doi: 10.1158/1055-9965.EPI-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Detection and identification of peroxiredoxin 3 as a biomarker in hepatocellular carcinoma by a proteomic approach. nternational Journal of Molecular Medicine. 2012 doi: 10.3892/ijmm.2012.916. [DOI] [PubMed] [Google Scholar]
- 13.Song, Bao, Yu, Xue, Yun, Zhang, He, Liu, Liu, Lu, et al. Comprehensive profiling of metastasis-related proteins in paired hepatocellular carcinoma cells with different metastasis potentials. PROTEOMICS - Clinical Applications. 2009;3:841–852. doi: 10.1002/prca.200780131. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Ki, Lee, Han, Kim, Park, Cho, Hong, Park, Lee, et al. Elevated Plasma Osteopontin Levels in Patients with Hepatocellular Carcinoma. The American Journal of Gastroenterology. 2006;101:ajg2006379. doi: 10.1111/j.1572-0241.2006.00679.x. [DOI] [PubMed] [Google Scholar]
- 15.MAAEM, Abdel-Aleem, Ali, Saber, Shatat, Rahem, Sayed Diagnostic significance of plasma osteopontin in hepatitis C virus-related hepatocellular carcinoma. Annals of hepatology. 2011;10:296–305. [PubMed] [Google Scholar]
- 16.Marrero, Feng, Wang, Nguyen, Befeler, Roberts, Reddy, Harnois, Llovet, Normolle, et al. α-Fetoprotein, Des- Carboxyprothrombin, and Lectin-Bound γ-Fetoprotein in Early Hepatocellular Carcinoma. Gastroenterology. 2009;137:110–118. doi: 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura, Nouso, Sakaguchi, Ito, Ohashi, Kobayashi, Toshikuni, Tanaka, Miyake, Matsumoto, et al. Sensitivity and Specificity of Des-Gamma-Carboxy Prothrombin for Diagnosis of Patients with Hepatocellular Carcinomas Varies According to Tumor Size. The American Journal of Gastroenterology. 2006;101:ajg2006377. doi: 10.1111/j.1572-0241.2006.00681.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim, Park, Kim, Sohn, Yeo, Yu, Yoon, Park, Kim Serum fibronectin distinguishes the early stages of hepatocellular carcinoma. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-09691-3. s41598-017-09691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kailemia, Park, Lebrilla Glycans and glycoproteins as specific biomarkers for cancer. Analytical and Bioanalytical Chemistry. 2017;409:395–410. doi: 10.1007/s00216-016-9880-6. This review highlights glycomic biomarkers for various cancers as well as discusses various mass spectrometry techniques used to identify them. * [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamczyk, Tharmalingam, Rudd Glycans as cancer biomarkers. Biochimica et Biophysica Acta (BBA) - General Subjects. 2012;1820:1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Buamah P, Cornell C, Cassells-Smith A, Harris A. Fucosylation of alpha-fetoprotein in hepatocellular carcinomas. 1986;327:922–923. doi: 10.1016/s0140-6736(86)91032-9. [DOI] [PubMed] [Google Scholar]
- 22.Aoyagi, Isokawa, Suda, Watanabe, Suzuki, Asakura The fucosylation index of α-fetoprotein as a possible prognostic indicator for patients with hepatocellular carcinoma. Cancer. 1998;83:2076–2082. [PubMed] [Google Scholar]
- 23.Wang, Fukuda, Isaji, Lu, Im, Hang, Gu, Hou, Ohtsubo, Gu Loss of α1,6-fucosyltransferase1,6 inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. The FASEB Journal. 2015;29:3217–3227. doi: 10.1096/fj.15-270710. [DOI] [PubMed] [Google Scholar]
- 24.Comunale, Lowman, Long, Krakover, Philip, Seeholzer, Evans, Hann, Block, Mehta Proteomic Analysis of Serum Associated Fucosylated Glycoproteins in the Development of Primary Hepatocellular Carcinoma. Journal of Proteome Research. 2006;5:308–315. doi: 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- 25.Morelle, Flahaut, Michalski, Louvet, Mathurin, Klein Mass spectrometric approach for screening modifications of total serum N-glycome in human diseases: application to cirrhosis. Glycobiology. 2006;16:281–293. doi: 10.1093/glycob/cwj067. [DOI] [PubMed] [Google Scholar]
- 26.Marrero, Romano, Nikolaeva, Steel, Mehta, Fimmel, Comunale, D’Amelio, Lok, Block GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. Journal of Hepatology. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 27.Drake, Schwegler, Malik, Diaz, Block, Mehta, Semmes Lectin Capture Strategies Combined with Mass Spectrometry for the Discovery of Serum Glycoprotein Biomarkers. Molecular & Cellular Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Morota, Nakagawa, Sekiya, Hemken, Sokoll, Elliott, Chan, Dowell A comparative evaluation of Golgi protein-73, fucosylated hemopexin, α-fetoprotein, and PIVKA-II in the serum of patients with chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Clinical Chemistry and Laboratory Medicine. 2011;49:711–718. doi: 10.1515/CCLM.2011.097. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Desmyter, Gao, Laroy, Dewaele, Vanhooren, Wang, Zhuang, Callewaert, Libert, et al. N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus. Hepatology. 2007;46:1426–1435. doi: 10.1002/hep.21855. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Sanda, Comunale, Herrera, Swindell, Kono, Singal, Marrero, Block, Goldman, et al. Changes in the Glycosylation of Kininogen and the Development of a Kininogen-Based Algorithm for the Early Detection of HCC. Cancer Epidemiology and Prevention Biomarkers. 2017;26:795–803. doi: 10.1158/1055-9965.EPI-16-0974. This article describes a newly-developed algorithm combining AFP, fucosylated kininogen, and clinical values for highly accurate early HCC detection. ** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ally, Balasundaram, Carlsen, Chuah, Clarke, Dhalla, Holt, Jones, Lee, Ma, et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. This paper provides rich genomic information characterizing HCC. Of special interest to this discussion of biomarkers is the description of several overexpressed genes such as those encoding for serum proteins albumin and apolipoprotein B, and the enzyme FUT8 which catalyzes core fucosylation of N-glycans. ** [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Devarajan, Singal, Marrero, Dai, Feng, JASR, vastava, Evans, Hann, et al. The Doylestown Algorithm: A Test to Improve the Performance of AFP in the Detection of Hepatocellular Carcinoma. Cancer Prevention Research. 2016;9:172–179. doi: 10.1158/1940-6207.CAPR-15-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Serag, Kanwal, Davila, Kramer, Richardson A New Laboratory-Based Algorithm to Predict Development of Hepatocellular Carcinoma in Patients With Hepatitis C and Cirrhosis. Gastroenterology. 2014;146:1249–1255.e1. doi: 10.1053/j.gastro.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, Pirrie, Cox, Berhane, Teng, Palmer, Morse, Hull, Patman, Kagebayashi, et al. The Detection of Hepatocellular Carcinoma Using a Prospectively Developed and Validated Model Based on Serological Biomarkers. Cancer Epidemiology and Prevention Biomarkers. 2014;23:144–153. doi: 10.1158/1055-9965.EPI-13-0870. [DOI] [PubMed] [Google Scholar]
- 35.Kagebayashi, Yamaguchi, Akinaga, Kitano, Yokoyama, Satomura, Kurosawa, Watanabe, Kawabata, Chang, et al. Automated immunoassay system for AFP–L3% using on-chip electrokinetic reaction and separation by affinity electrophoresis. Analytical Biochemistry. 2009;388:306–311. doi: 10.1016/j.ab.2009.02.030. [DOI] [PubMed] [Google Scholar]
- 36.Ma, Sun, Chen, Cheng, Han, Zhao, He Multiplex Immunochips for High-Accuracy Detection of AFP-L3% Based on Surface-Enhanced Raman Scattering: Implications for Early Liver Cancer Diagnosis. Analytical Chemistry. 2017 doi: 10.1021/acs.analchem.7b01349. [DOI] [PubMed] [Google Scholar]
- 37.Hann, Jain, Park, Steffen, Song, Su Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Research. 2017;3:105–111. doi: 10.20517/2394-5079.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang, CWMC, KCAC, Cheng, Wong, Wong, GLHW, Chan, TSKM, HLYC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proceedings of the National Academy of Sciences. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen Li, Wang Thoburn, Afsari Danilova, Douville Javed, Wong Mattox, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018 doi: 10.1126/science.aar3247. This paper describes the recent development of a multi-cancer biomarker test that has gained a lot of publicity in its detection power through genomic and proteomic biomarker combinations. ** [DOI] [PMC free article] [PubMed] [Google Scholar]