Abstract
The mitogen-activated protein kinases (MAPKs) are integral to the mechanisms by which cells respond to physiological stimuli, such as growth factors, hormones, and cytokines, and to a wide variety of environmental stresses. The MAPKs, which are stimulated by phosphorylation of a TXY motif in their activation loop, are components of signal transduction cascades in which sequential activation of protein kinases culminates in their activation and their subsequent phosphorylation of various effector proteins that mediate the physiological response. MAPKs are also subject to dephosphorylation and inactivation, both by enzymes that recognize the residues of the TXY motif independently and by dual specificity phosphatases, which dephosphroylate both Tyr and Ser/Thr residues. We report the identification and characterization of a novel dual specificity phosphatase. Contrary to expectation, this broadly expressed enzyme did not inactivate MAPKs in transient cotransfection assays but instead displayed the capacity to function as a selective activator of the MAPK Jnk, hence the name, Jnk Stimulatory Phosphatase-1 (JSP-1). This study illustrates a new aspect of the regulation of MAPK-dependent signal transduction and raises the possibility that JSP-1 may offer a different perspective to the study of various inflammatory and proliferative disorders associated with dysfunctional Jnk signaling.
Cells are capable of responding to various environmental cues, including physiological stimuli, such as growth factors, hormones, and cytokines, and stresses, such as osmotic shock, radiation, and ischemic injury. One of the most important aspects of such cellular responses is the stimulation of mitogen-activated protein kinases (MAPKs) (1–4). MAPKs are the terminal component of a core “signaling module” that comprises a series of three protein kinases. In this signaling module, MAPK kinase kinases (MAPKKK or MEKK) phosphorylate MAPK kinases (MAPKK or MEK) on Ser/Thr residues with concomitant activation. The MAPKKs are dual specificity kinases that, on activation, phosphorylate both the tyrosine and the threonine residue of a conserved TXY motif in the activation loop of MAPKs, again with concomitant activation. These signaling modules are themselves activated in response to various stimuli, via small GTP-binding proteins, such as those of the Ras superfamily, and STE20-like protein kinases, such as HPK1, MST1, NIK, and GLK, acting at the level of membrane recruitment, oligomerization, and phosphorylation of MAPKKKs (1–4). The effects of activation of these signaling modules are manifested by the phosphorylation of a wide variety of substrates by MAPKs, including effector protein kinases, such as MAPK-activated protein kinases (MAPKAPs) and transcription factors, such as the AP-1 complex (1–4).
Approximately 20 MAPKs have been identified to date in mammalian systems, and it is thought likely that more will be discovered (1–3). Although four major subgroups (Erk 1 and 2, Erk 5, p38 α, β, γ, and δ, and Jnk 1, 2, and 3) have been delineated, on the basis of their sequence homology and their abilities to respond to different stimuli, additional less well characterized MAPKs (such as MAK, MRK, and NLK) have also been identified (3). The first major subgroup is represented by the classical MAPKs, Erks1 and 2, in which the TXY motif in the activation loop is TEY. Erks1 and 2 are broadly expressed and activated in response to a wide variety of stimuli associated with both proliferation and differentiation (1–3). The p38 group of MAPKs, which are characterized by a TGY motif, comprise four distinct gene products, with additional diversity introduced through alternative mRNA splicing. These enzymes are activated by a variety of cytokines and hormones as well as in response to environmental stress. Of particular importance to this study is the Jnk family, characterized by a TPY motif. There are three distinct genes, encoding Jnk1, Jnk2, and Jnk3, with further structural diversity manifested at the level of alternative mRNA splicing. Members of this group are predominantly activated after exposure of cells to proinflammatory cytokines and a variety of environmental stresses, such as radiation and osmotic stress (1–4).
Each of the MAPK subgroups responds to distinct MAPKKs. A combination of studies involving gene knockouts and the use of dominant-negative mutants have implicated both MKK4 and MKK7 in the phosphorylation and activation of Jnk (5–7). However, differences have been ascribed to these enzymes. For example, whereas MKK7 is primarily activated in response to cytokines, MKK4 primarily responds to extracellular stress (4, 8). In addition, at least in vitro, MKK4 preferentially phosphorylates the tyrosine residue of the TPY motif of the activation loop of the Jnks, whereas MKK7 preferentially recognizes the threonine (9). Thus, it is possible that these enzymes may function in cooperation to integrate signals at the level of JNK activation. The MAPKKs are themselves activated by dual phosphorylation of their activation loops, but unlike for the MAPKs, this occurs on Ser and Thr residues (1–3). The MAPKKKs encompass a broad group of kinases from several divergent families, with the heterogeneity in these enzymes consistent with the diversity of stimuli that lead to activation of MAPK signaling modules (1–3). With the exception of Raf and the activation of Erks1 and 2 via MEK1/2, the contribution of the various MAPKKKs to the activation of distinct MAPK signaling modules remains unclear (1–4, 10).
The duration and extent of MAPK activation are governed by the balance between the activity of the MAPKKs and that of the protein phosphatases that dephosphorylate either or both of the Thr and Tyr residues in the TXY motif. Thus, protein phosphatases are also critical regulators of MAPK-dependent signaling events. Recently, several members of the protein tyrosine phosphatase (PTP) family have been implicated in the dephosphorylation of MAPKs (11, 12). In addition, a subgroup within the PTP family, termed dual specificity phosphatases (DSPs), has been identified (13). Although DSPs display limited sequence identity to the classical PTPs, they are characterized by the presence of the PTP signature motif, HCXXGXXR [S/T], and share striking similarity with the PTPs in their secondary and tertiary structures (11–15). Several of the DSPs have now been shown to recognize MAPKs as substrate and to dephosphorylate both the Thr and Tyr residues of the TXY activation loop motif (12, 16, 17). Interestingly, it has been noted that coexpression of one such DSP, MKP-1, with the MAPKKs MKK1 and MKK2, or with Raf, lead, by an unknown mechanism, to activation of these kinases (18). Nevertheless, even under these conditions, MKP-1 functioned as an inhibitor of the activation of Erk and Erk-dependent signaling (18). To date, 10 DSPs have been identified that have the capacity to dephosphorylate and inactivate MAPKs, including MKPs 1–5, VHR, hVH3/B23, hVH5/M3/6, Pac1 and Pyst2, and which display differences in expression, tissue and subcellular distribution, and specificity for MAPK family members (16, 17). Therefore, these MAPK phosphatases (MKPs) represent a complex response network for regulation of specific MAPK-dependent signaling pathways in particular tissues and subcellular compartments after defined stimuli.
In an effort to identify additional DSPs that may function as regulators of MAPK-dependent signaling pathways, we searched expressed sequence tag (EST) databases for sequences with homology to the signature motif of members of the DSP subfamily of PTPs. We identified and cloned several novel DSPs, including a broadly expressed enzyme, which we have termed JSP-1. In contrast to the established principle that DSPs are negative regulators of MAPKs, we made the striking observation that JSP-1 has the capacity to activate the Jnk signaling pathway specifically (hence, Jnk Stimulatory Phosphatase-1), illustrating a new aspect of the regulation of MAPK-dependent signal transduction.
Material and Methods
Molecular Cloning of DSP3.
We searched the National Center for Biotechnology Information EST database (dbEST) with a tblastn query of the conserved MAPK phosphatase (MKP) active-site domain. Three ESTs were identified as containing sequences of a novel, but incomplete, DSP. To identify the full length open reading frame, a 3′ and 5′ RACE analysis was performed on human skeletal muscle total RNA (CLONTECH) by using the 5′/3′ RACE kit (Boehringer Mannheim) and the manufacturer's instructions. To obtain full length cDNA of this DSP, now termed JSP-1, human skeletal muscle RNA (CLONTECH) was reverse transcribed by using Superscript II reverse transcriptase (GIBCO/BRL). The resultant DNA was used as a template to amplify JSP-1 cDNA, encoding a protein of 184 amino acids, and the PCR product was ligated into the bacterial expression vector pGEX-6P (Amersham Pharmacia Biotech) to generate pGEX-6P-JSP-1.
PCR Analysis of JSP-1 Expression in Human Tissues.
The human multiple tissue cDNA panels (MTC I and MTC II) were used as a source of cDNA (CLONTECH). The expression of JSP-1 was analyzed by PCR by using the primer pairs JSP-1, ATG: 5′-ATGGGGAATGGGATGAACAAGATC-3′ and JSP-1, 3′-ENDr: 5′-CAGTCTTCTGAGAAAGGCCCAGAA-3′. The cycle condition for PCR was 94°C, 30 seconds, and 68°C, 1 minute, with 23 cycles of PCR performed.
Mammalian Expression Constructs.
Flag-p38, and Flag-MKK4 mammalian expression constructs were kindly provided by Roger Davis (Howard Hughes Medical Institute, University of Massachusetts Medical School). Myc-JNK was a gift from Jon Cooper (Fred Hutchinson Cancer Research Center, Seattle). To generate the N-terminally hemagglutinin-tagged JSP-1 expression construct pCGN-JSP-1-WT, pGEX-6P-JSP-1 was used as a template to amplify the full length JSP-1 sequence engineered to contain a 5′ XbaI site and a 3′ KpnI site. The PCR product was ligated into the mammalian expression vector pCGN.
Site-Directed Mutagenesis.
Site-directed mutagenesis was carried out by using the Quick Change protocol (Stratagene). pCGN-JSP-1-WT was used as a template to generate the expression constructs pCGN-JSP-1-CS, in which Cys-88 of JSP-1 was mutated to Ser. The kinase-dead mutant of MKK4, MKK4-KR, was generated by using Flag-MKK4 as template and mutating Lys-131 to Arg.
Protein Expression and Purification.
The Escherichia coli expression constructs GST-JUN and GST-ATF2 were gifts from Roger Davis and were expressed and purified following standard protocols (19). GST-JSP-1 was expressed in E. coli–DH5, the cells were lysed by sonication on ice. Triton X-100 (1% v/v) was added to the sonicate, which was then clarified and added to glutathione-sepharose beads. JSP-1 was released from the beads by treatment with PreScission protease (Amersham Pharmacia).
Assay of Protein Phosphatase Activity.
The substrates polyGlu:Tyr and RCML were phosphorylated with [γ-32P]ATP by using the protein tyrosine kinase GST-FER (20). The stock substrate preparations were 200 μM pTyr-reduced carboxamidamethylated and maleylated lysozyme (RCML) and 80 μM pTyr-polyGlu:Tyr. Casein (Sigma) was phosphorylated with [γ-32P]ATP and protein kinase A, according to the manufacturer's instructions, to yield a stock substrate of 50 μM pSer/pThr. Protein phosphatase assays were performed in a total volume of 100 μl, in an assay buffer of 50 mM Hepes, pH 7.4/4 mM DTT/2 mg/ml of BSA containing 10 μl of the labeled substrate. Assays with polyGlu:Tyr or RCML as substrate contained 100 ng of JSP-1, whereas those using casein as substrate contained 250 ng of JSP-1. Reactions were initiated by addition of JSP-1 and terminated by addition of 800 μl of charcoal mix, followed by centrifugation and counting of 500 μl of the supernatant in scintillant.
Cell Culture and Transfection.
COS-1 cells were maintained at 37°C and 5% CO2 in DMEM + 10% fetal bovine serum. Cells were transfected with various expression plasmids by using the LipofectAMINE reagent, according to the manufacturer's instructions. If no treatment was required, cells were harvested 36–48 hours after transfection. For specific treatments, cells were switched to serum-free DMEM medium 36 hours after transfection, incubated in serum-free medium overnight, then subjected to specific stimuli and harvested. To induce activation of Jnk, cells were treated for 30 minutes with 10 ng/ml of anisomycin.
Immunoprecipitation and Immunoblotting.
Cells were lysed in Nonidet P-40-containing buffer supplemented with protease and phosphatase inhibitors. For immunoprecipitation, cell lysates were incubated with the appropriate primary antibody for 1 hour on ice, followed by 1-hour rotation with protein A sephorose beads at 4°C. The beads were then washed three times in lysis buffer and bound proteins solubilized in sample buffer. For immunoblotting, the nitrocellulose membrane was blocked in 5% milk (or 1% BSA for anti-ACTIVE-JNK antibody) for 1 hour, then incubated with primary antibody for 1 hour at room temperature (or overnight at 4°C for anti-ACTIVE-JNK antibody). After washing, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody for 1 hour and bound proteins detected by enhanced chemiluminescence. A working concentration of 0.2 μg/ml was used for the anti-HA tag antibody 12CA5, the anti-Myc tag antibody 9E10, and the anti-Flag tag antibody (Sigma). The stocks provided by the manufacturers were diluted 1:5,000 for anti-ACTIVE-JNK antibody (Promega) and 1:1,000 for the phospho-SEK1(Thr-261) antibody (Cell Signaling Technology, Beverly, MA).
Immune Complex MAPK Assays.
Cells were lysed in Nonidet P-40-containing buffer supplemented with protease and phosphatase inhibitors. The Flag-tagged p38 was immunoprecipitated with 0.75 μg of anti-Flag antibody. Myc-tagged Erk1 or JNK was precipitated with 1 μg of 9E10 antibody. The immune complex was washed twice in lysis buffer and twice in kinase assay buffer (20 mM Pipes, pH 7.2/10 mM MgCl2/1 mM DTT/0.1% Triton X-100/1 mM sodium vanadate). Reactions were initiated by mixing the immune complex with 80 μl of kinase buffer supplemented with 20 μM ATP/10 μCi [γ-32P]ATP and the appropriate substrate. GST-JUN (2 μg), GST-ATF2 (2 μg), and MBP (20 μg) were used as substrates for JNK, p38, and Erk, respectively. After 10–15 minutes at 30°C, assays were terminated by addition of 80 μl of 2× SDS/PAGE sample buffer. Samples were resolved by SDS/PAGE, transferred to a nitrocellulose membrane, and the incorporation of 32P-phosphate visualized by exposure to X-ray film and quantified by PhosphorImage.
Results
Molecular Cloning of JSP-1.
To identify novel DSPs involved in the regulation of MAPK family members, we searched the National Center for Biotechnology Information dbEST with a tblastn query of the conserved MAPK phoshatase active site domain. We identified three ESTs, AA374753, AA 411671, and H82446, as containing sequences of an incomplete DSP. The full length open reading frame, identified by 3′ and 5′ RACE analysis on human skeletal muscle total RNA as a template, encoded 184 amino acids (Fig. 1) and was designated JSP-1 (defined below). JSP-1 displayed 33–40% identity (58% similarity) to the catalytic domains of the known MKPs, MKP4, MKP5, Pyst1, and hVH5, and contained both the conserved signature motif of the PTP family, HC(X)5R, and AYLM, a conserved motif of MKPs.
Figure 1.
Nucleotide and predicted peptide sequence of JSP-1.
JSP-1 Is Broadly Expressed in Human Tissues.
We examined the tissue distribution of JSP-1 by Northern blot analysis and found two major transcripts, of 4.0 and 1.35 kb, that were broadly expressed. In addition, we performed PCR analysis by using the Multiple Tissue cDNA panels I and II (CLONTECH) as templates. PCR products were detected in all tissues examined, with highest expression observed in heart, placenta, lung, liver, kidney, and pancreas (data not shown), indicating that in humans, JSP-1 is broadly expressed in a variety of tissues.
JSP-1 Is a Dual Specificity Phosphatase.
To demonstrate that JSP-1 exhibits intrinsic DSP activity, we expressed it as a GST-fusion protein in E. coli. The fusion protein was purified over glutathione sepharose and the GST tag removed by PreSission protease cleavage. The activity of purified JSP-1 was then determined by using a variety of substrates, including both proteins and peptides phosphorylated on Tyr residues, such as polyGlu:Tyr and RCML, and proteins phosphorylated on Ser/Thr residues, such as casein and histone. JSP-1 readily dephosphorylated tyrosyl-phosphorylated substrates. The specific activities against polyGlu:Tyr (80 nmol/min/mg) and RCML (144 nmol/min/mg) were similar to those observed for other PTPs (21). However, although JSP-1 dephosphorylated Ser/Thr residues in proteins, it did so with a much lower specific activity, for example ≈1 nmol/min/mg for protein kinase A-labeled casein. Similar preference for tyrosine-phosphorylated substrates has been reported for the DSP VHR (22). Nevertheless, it remains to be determined whether this reflects weaker intrinsic Ser/Thr phosphatase activity of JSP-1 or simply that the substrates tested to date are suboptimal for this phosphatase.
JSP-1 Specifically Activates the Jnk Signaling Pathway.
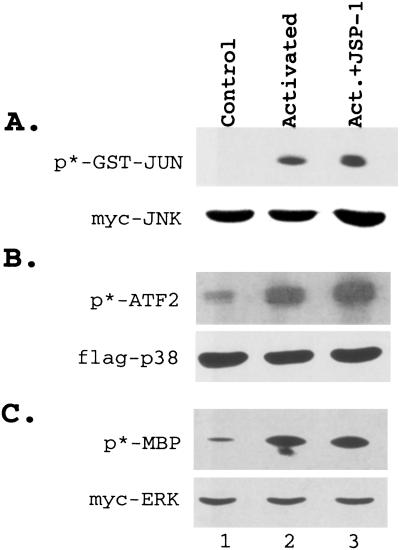
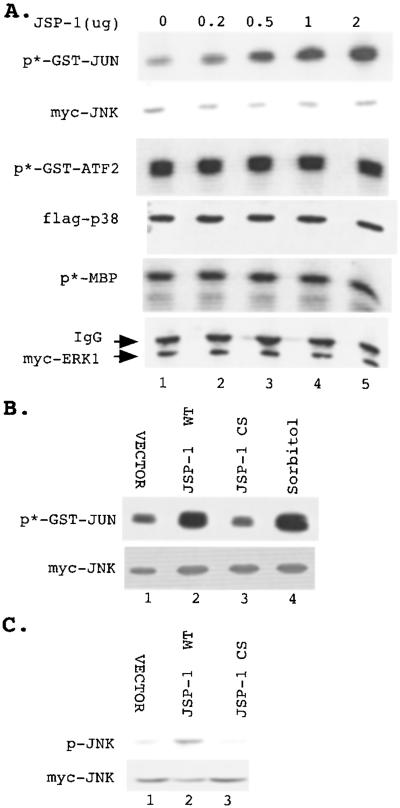
Because many DSPs have been shown to dephosphorylate and inactivate MAPKs, we examined the effects of JSP-1 on different members of the MAPK family. Plasmids encoding hemagglutinin-tagged JSP-1 constructs and epitope-tagged Jnk, epitope p38, or Erk1 MAPKs were cotransfected in COS cells. Each kinase was immunoprecipitated, and its activity was measured in an immune-complex kinase assay. To our surprise, we did not detect inhibition of epidermal growth factor-induced Erk or inhibition of Cdc42 (V12)-induced Jnk or p38 in these assays (Fig. 2). In contrast, we observed that ectopic expression of JSP-1 led to a dose-dependent increase in the activity of coexpressed Jnk, whereas the activities of Erk1 and p38 were not significantly affected (Fig. 3). Furthermore, the extent of activation of Jnk by JSP-1 was similar to that observed in response to the physiological stimulus sorbitol (Fig. 3B). Expression of an inactive mutant form of JSP-1, in which the active site cysteine (Cys-88) was changed to serine, did not lead to activation of Jnk, indicating that the phosphatase activity of JSP-1 is required for this effect (Fig. 3B). It has been shown that dual phosphorylation of both the Thr and Tyr residues in the TPY motif of the activation loop of Jnk is required for activation. To test whether JSP-1-induced activation of Jnk correlated with increased phosphorylation of these sites, we coexpressed Flag-tagged Jnk with either a control vector, wild-type JSP-1 or the C→S mutant form of JSP-1 in COS cells. The phosphorylation status of JNK was then examined by immunoblotting with the anti-ACTIVE-JNK antibody (Promega), which specifically recognizes the dual phosphorylated TPY motif. We observed that overexpression of wild-type JSP-1 increased the phosphorylation of coexpressed JNK, whereas the inactive C→S mutant form of the phosphatase did not. Immunoblotting with an antibody to the Myc tag illustrated that equivalent quantities of JNK were present in each sample (Fig. 3C). On the basis of these data, we refer to this phosphatase as Jnk-Stimulatory Phosphatase-1 (JSP-1).
Figure 2.
JSP-1 did not inactivate MAP kinases. Flag-tagged Jnk (A) or Flag-tagged p38 (B) was coexpressed with a control vector (lane 1) or an active mutant form of Cdc42 (V12) in the absence (lane 2) or presence (lane 3) of JSP-1. Cells were serum starved overnight before harvesting. (C) Myc-tagged Erk was coexpressed with a control vector (lanes 1 and 2) or with JSP-1 (lane 3). Cells were serum starved overnight and harvested without stimulation (lane 1) or following stimulation with epidermal growth factor for 25 minutes before harvesting (lanes 2 and 3). The kinase activity of Jnk, p38, and Erk was determined by immunoprecipitation of the kinases and immune-complex assays with GST-Jun, GST-ATF2, or myelin basic protein (A Upper, B Upper, and C Upper, respectively) as substrate. The level of Jnk, p38, and Erk in each reaction was determined by immunoblotting with the anti-Flag antibody (A Lower and B Lower) or the anti-Myc antibody, 9E10 (C Lower).
Figure 3.
JSP-1 specifically activated Jnk. (A) A control vector (lane 1) or 0.2, 0.5, 1, or 2 μg (lanes 2–5) of plasmid encoding JSP-1 were each cotransfected with 0.5 μg of plasmid encoding Myc-tagged Jnk (Upper), Flag-tagged p38 (Middle), or Myc-tagged Erk (Lower), respectively. The total amount of DNA in each transfection was adjusted to 2.5 μg with control vector. The kinase activities of Jnk, p38, or Erk were determined by immunoprecipitation of the kinase and immune-complex assays with GST-Jun (Upper), GST-ATF2 (Middle), or myelin basic protein (Lower) as substrate. The level of Jnk, p38, or Erk in each kinase reaction was determined by immunoblotting by using anti-Myc antibody 9E10 (Upper and Lower) or the anti-Flag antibody (Middle). (B and C) Myc-tagged Jnk was coexpressed with control vector (lane 1), wild-type JSP-1 (lane 2), or the inactive C→S mutant form of JSP-1 (lane 3). In addition, cells expressing Myc-tagged Jnk were stimulated with 500 mM sorbitol (B, lane 4). Jnk activity was determined by the immune-complex kinase assay (B) or by immunoblotting with the anti-ACTIVE-JNK antibody (C). The amount of Jnk in each sample was determined by immunoblotting using the anti-Myc antibody 9E10.
Activation of Jnk by JSP-1 Is Associated with Activation of MKK4.
The activity of Jnk is controlled by the coordinated actions of positive regulators, such as the MAPKKs, and negative regulators, including MKP-type protein phosphatases. Stimuli such as osmotic stress and various cytokines activate Jnk primarily via activation of MAPKKs, such as MKK4, whereas heat shock and oxidative stress have been shown to prevent inactivation of Jnk by phosphatases (23). Similarly, the stimulatory effects of JSP-1 on Jnk activation could be manifested either through activation of a MAPKK or by prevention of the inactivation of Jnk. To distinguish between these possibilities, we compared the inactivation rate of ectopically expressed Jnk in cells overexpressing either JSP-1 or a vector control. Jnk was activated by pretreating cells with anisomycin for 30 minutes, then the cells were treated with rotenone, an inhibitor of the respiratory chain, and deoxyglucose, an inhibitor of glycolysis, to block ATP synthesis. Under these conditions, the high intracellular ATPase activity leads to a rapid decrease in the levels of cellular ATP, blocking the de novo activation of Jnk by other kinases (23). We observed that Jnk activity, determined by immune-complex kinase assay, was reduced by >70% within 5 minutes of ATP depletion and was essentially completely inactivated within 30 minutes. This rate of inactivation of Jnk was the same in the presence or absence of JSP-1, indicating that JSP-1 is unlikely to exert its effects by preventing inactivation of Jnk.
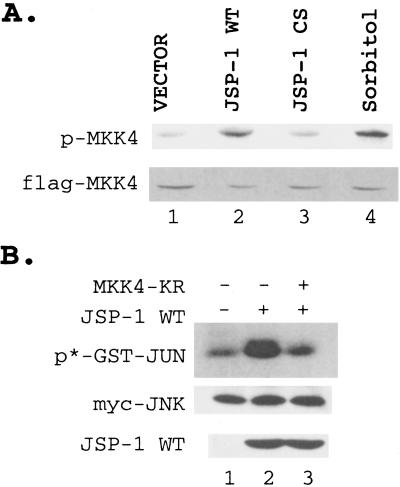
We then tested whether overexpression of JSP-1 led to activation of kinases upstream of Jnk. MKK4 phosphorylates and activates Jnk both in vitro and in vivo (9, 12, 24) and is itself activated through phosphorylation by an upstream MAPKKK (5). Because activation of MKK4 depends on phosphorylation on its activation loop, immunoblot analysis was performed with an antibody that specifically recognizes MKK4 phosphorylated on Thr-261 (antiphopho-SEK), as an indirect measurement of MKK4 activity. MKK4 was coexpressed with either wild-type JSP-1, the C→S inactive mutant form of JSP-1, or a vector control and the phosphorylation level of MKK4 determined by immunoblotting with antiphopho-SEK antibody. Under conditions used in this study, the antiphospho-SEK antibody detected ectopically expressed MKK4, but not the endogenous enzyme, even after anisomycin treatment (data not shown). Enhanced phosphorylation of MKK4 was detected in cells coexpressing MKK4 and wild-type JSP-1, but not in cells coexpressing MKK4 and either the C→S mutant form of JSP-1 or the vector control (Fig. 4A). Furthermore, JSP-1 activated MKK4 to an extent similar to that observed in response to the physiological stimulus sorbitol (Fig. 4A). Thus, overexpression of JSP-1 led to activation of MKK4, and this activation depends on the phosphatase activity of JSP-1.
Figure 4.
JSP-1 activated JNK through activation of MKK4. (A) Flag-tagged MKK4 was coexpressed with a control vector (lane 1), plasmid encoding wild-type JSP-1 (lane 2), or the inactive C→S mutant form of JSP-1 (lane 3). In addition, cells expressing Flag-tagged MKK4 were stimulated with 500 mM sorbitol (lane 4). Cell lysate was resolved by SDS/PAGE, and the MKK4 activity was measured by immunoblotting with antiphospho-SEK antibody (Upper). The amount of MKK4 in each lane was determined by immunoblotting with anti-Flag antibody (Lower). (B) Myc-tagged Jnk was coexpressed with control vector (lane 1) or hemagglutinin-tagged wild-type JSP-1 in the absence (lane 2) or presence of the inactive K→R mutant form of MKK4 (lane 3). Jnk activity was determined by immune-complex kinase assay (Top). The amount of Jnk in each reaction was determined by immunoblotting with the anti-Myc antibody 9E10 (Middle), and the expression of JSP-1 was determined by immunoblotting with the 12CA5 antibody (Bottom).
To investigate further whether MKK4 is required for JSP-1-induced activation of Jnk, we coexpressed myc-tagged Jnk with a control vector, wild-type JSP-1, or JSP-1, together with a dominant-negative form of MKK4 (MKK4-KR) in COS-1 cells. Jnk activity was measured by immune-complex kinase assays in cell lysates. We observed that overexpression of JSP-1 caused a 2.5-fold increase in Jnk activity, which was almost completely abolished by coexpression of the dominant-negative MKK4. Immunoblots using antibodies to Myc-Jnk or hemagglutinin-JSP-1 demonstrated that the differences we observed in Jnk activity did not result from variations in the levels of Jnk or JSP-1 protein (Fig. 4B).
Discussion
The MAPKs are critical regulators of signaling pathways underlying cellular responses to environmental cues that alter proliferation, differentiation, and survival. In addition, the MAPKs are of fundamental importance in embryonic development. Great progress has been made in defining the various three-tiered protein kinase cascades that culminate in activation of MAPKs and in characterizing both the mechanisms by which they are triggered in response to defined stimuli and those by which they effect a physiological response (1–4). Recently, attention has also focused on the protein phosphatases as regulators of these signaling pathways (11–13). The MAPKs are activated by phosphorylation of the Thr and Tyr residues of the TXY motif in their activation loop. Inactivation is mediated in vivo both by single-specificity phosphatases, acting independently on the residues of the TXY motif, and members of a subfamily of the PTPs, termed DSPs, which have the capacity to dephosphorylate both Tyr and Ser/Thr residues in their target substrates (16, 17). In short, it now appears that the regulation of MAPKs that is exerted at the level of the protein phosphatases is as sophisticated as that mediated by the protein kinases. In this study, we reveal a further level of complexity to the regulation of MAPK-dependent signaling pathways. In a search for additional DSPs with the ability to down-regulate MAPK function, we identified an enzyme now termed JSP-1. To our surprise, rather than catalyzing the dephosphorylation and inactivation of MAPKs, we have observed that this DSP has the capacity to function as a specific activator of the Jnk signaling pathway.
Utilizing the full length peptide sequence of JSP-1 and an advanced tblastn query for Mus musculus dbEST sequences in the National Center for Biotechnology Information database, we identified the EST AA103595 as containing sequences homologous to the N-terminal half of human JSP-1. A blast analysis of the Mus musculus dbEST database searching with AA103595 allowed us to identify another overlapping EST, AW413206, which contains the carboxyl-terminal portion of murine JSP-1 (mJSP-1). The combined sequence from both ESTs AA103595 and AW413206 encodes a protein that is closely related to human JSP-1 in the N-terminal 169 residues (95% similarity), suggesting that this combined sequence may represent the murine homolog of human JSP-1. In searching the GenBank nonredundant database with a tblastn query by using the full-length peptide sequence of human JSP-1, we also identified a database entry AK000383, which is a partial human cDNA encoding a peptide displaying 100% identity to residues 65–169 of human JSP-1. The C-terminal 36 residues of sequence translated from AK000383 were different from the C terminus of human JSP-1 but highly related to that of murine JSP-1 (78% identity). Therefore, AK000383 may represent an alternatively spliced variant of human JSP-1. We also searched the dbEST database with JSP-1 peptide sequence looking for matches with ESTs from other species. Several were found, including Rat [BF549803, 92% identity (128/139) and 98% similarity (137/139)], Xenopus [AW646139, 74% identity (128/171) and 86% similarity (149/171)], Drosophila [BF495653, 50% identity (83/163) and 72% similarity (119/163)], and zebrafish [BE557484, 66% identity (99/148) and 83% similarity (125/148)]. In light of the extensive similarity between JSP-1 and these ESTs, it is possible that they may represent homologs of JSP-1, consistent with an important biological function for the enzyme that may be conserved across species.
JSP-1 is a small DSP that is reminiscent of one of the initially characterized members of the family, VHR (22). It lacks the cdc25-homology domains found in the noncatalytic segments of previously characterized MKPs, such as MKP-1. These domains have been implicated in substrate docking and specificity determination in the MKPs (16, 17, 25). Nonetheless, JSP-1 does display specificity in function. Characterization of its enzymatic activity revealed preferential dephosphorylation of tyrosyl residues in proteins, as previously noted for VHR (22). In fact, the activity of JSP-1 toward artificial pTyr-containing substrates in vitro is similar to that observed for other members of the family (21). However, although the activity of JSP-1 was low toward the pSer/pThr-containing proteins that have been tested as substrates to date in vitro, one should not conclude that the phosphatase will recognize pTyr residues preferentially in vivo. Many examples have now been documented in which members of the PTP family display specificity in their recognition of target substrates (12). Therefore, a complete characterization of the activity of JSP-1 and a definition of its preference for phosphorylated residues in proteins will require the identification of its physiological substrates.
Our data illustrate that JSP-1 has the potential to display specificity in a cellular context, in that its expression led to activation of Jnk without altering the activity of Erk or p38 MAPKs. Overexpression of JSP-1 also led to the activation of MKK4, a MAPKK that functions upstream of Jnk. Furthermore, a dominant-negative mutant form of MKK4 abolished JSP-1-induced activation of Jnk. These data suggest that JSP-1 does not exert its effects directly on Jnk but support a site of action of the phosphatase upstream of MKK4. Although overexpression of MKK4 has been reported to activate both JNK and p38 (4), we observed that only JNK was activated after overexpression of JSP-1. Whether this reflects a limited activation of MKK4 by JSP-1 that is sufficient for activation of Jnk but not sufficient for activation of p38 remains to be determined. Interestingly, a similar situation was reported for MLK3, a MAPKKK that functions upstream of MKK4 (26). Although MLK3 is an activator of MKK4, its expression results only in the activation of Jnk and not p38 (27). Many different phosphorylation events have the potential to regulate the Jnk signaling pathway upstream of MKK4, including a large number of MAPKKKs such as MEKK1, MEKK4, and the mixed lineage kinases, as well as Ste20-like kinases that function in linking MAPK cascades to external stimuli. Further insights into how JSP-1 stimulates Jnk signaling will be gained after identification of the physiological substrates and regulators of this phosphatase.
The activation of Jnk has been implicated in a number of important physiological processes, from embryonic morphogenesis to cell survival and apoptosis. Jnk signaling has also been linked to human disease conditions, including tumor development, cardiac hypertrophy, ischemia/reperfusion injury, diabetes, hyperglycemia-induced apoptosis, and several neurodegenerative disorders (1–4). Thus, the Jnks are viewed as exciting candidates for therapeutic intervention. Our studies reveal a novel aspect of the regulation of Jnk-signaling, a DSP that may function in the activation of Jnk without effect on Erk and p38 MAPKs. Therefore, it will be exciting to see whether, with continued investigation, JSP-1 may also be considered as a target for therapeutic intervention in human disease.
Acknowledgments
We thank our colleagues Drs. Roger Davis and Jon Cooper for generously sharing expression constructs with us. This work was supported by National Institutes of Health Grants CA 53840 and GM 55989.
Abbreviations
- MAPK
mitogen-activated protein kinase
- MAPKK or MEK
MAPK kinase
- MAPKKK or MEKK
MAPK kinase kinase
- MKP
MAPK phosphatase
- PTP
protein tyrosine phosphatase
- DSP
dual specificity phosphatase
- EST
expressed sequence tag
- dbEST
database EST
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF424702).
References
- 1.Kyriakis J M, Avruch J. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 2.Chang L, Karin M. Nature (London) 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 3.Pearson G, Robinson F, Beers Gibson T, Xu B, Karandikar M, Berman K, Cobb M H. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 4.Davis R J. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 5.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Nature (London) 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 6.Tournier C, Whitmarsh A J, Cavanagh J, Barrett T, Davis R J. Proc Natl Acad Sci USA. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland P M, Suzanne M, Campbell J S, Noselli S, Cooper J A. J Biol Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 8.Tournier C, Dong C, Turner T K, Jones S N, Flavell R A, Davis R J. Genes Dev. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawler S, Fleming Y, Goedert M, Cohen P. Curr Biol. 1998;8:1387–1390. doi: 10.1016/s0960-9822(98)00019-0. [DOI] [PubMed] [Google Scholar]
- 10.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 11.Neel B G, Tonks N K. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 12.Tonks N K, Neel B G. Curr Opin Cell Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 13.Martell K J, Angelotti T, Ullrich A. Mol Cell. 1998;8:2–11. [PubMed] [Google Scholar]
- 14.Barford D, Flint A J, Tonks N K. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 15.Yuvaniyama J, Denu J M, Dixon J E, Saper M A. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 16.Keyse S M. Curr Opin Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 17.Camps M, Nichols A, Arkinstall S. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- 18.Shapiro P S, Ahn N G. J Biol Chem. 1998;273:1788–1793. doi: 10.1074/jbc.273.3.1788. [DOI] [PubMed] [Google Scholar]
- 19.Smith D B, Corcoran L M. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1994. , Suppl. 28, pp. 16.7.1–16.7.7. [Google Scholar]
- 20.Shen Y, Schneider G, Cloutier J F, Veillette A, Schaller M D. J Biol Chem. 1998;273:6474–6481. doi: 10.1074/jbc.273.11.6474. [DOI] [PubMed] [Google Scholar]
- 21.Yang Q, Co D, Sommercorn J, Tonks N K. J Biol Chem. 1993;268:6622–6628. [PubMed] [Google Scholar]
- 22.Denu J M, Zhou G, Wu L, Zhao R, Yuvaniyama J, Saper M A, Dixon J E. J Biol Chem. 1995;270:3796–3803. doi: 10.1074/jbc.270.8.3796. [DOI] [PubMed] [Google Scholar]
- 23.Meriin A B, Yaglom J A, Gabai V L, Zon L, Ganiatsas S, Mosser D D, Sherman M Y. Mol Cell Biol. 1999;19:2547–2555. doi: 10.1128/mcb.19.4.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D, Tournier C, Wysk M, Lu H T, Xu J, Davis R J, Flavell R A. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanoue T, Adachi M, Moriguchi T, Nishida E. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 26.Rana A, Gallo K, Godowski P, Hirai S, Ohno S, Zon L, Kyriakis J M, Avruch J. J Biol Chem. 1996;271:19025–19028. doi: 10.1074/jbc.271.32.19025. [DOI] [PubMed] [Google Scholar]
- 27.Teramoto H, Coso O A, Miyata H, Igishi T, Miki T, Gutkind J S. J Biol Chem. 1996;271:27225–27228. doi: 10.1074/jbc.271.44.27225. [DOI] [PubMed] [Google Scholar]