Summary
We evaluated the association of Human Pegivirus (HPgV) viraemia with risk of developing lymphoma, overall and by major subtypes. Because this virus has also been associated with better prognosis in the setting of co-infection with human immunodeficiency virus, we further assessed the association of HPgV with prognosis. We used risk factor data and banked plasma samples from 2094 lymphoma cases newly diagnosed between 2002 and 2009 and 1572 frequency-matched controls. Plasma samples were tested for HPgV RNA by reverse transcription polymerase chain reaction (RT-PCR), and those with RNA concentrations <5,000 genome equivalents/ml were confirmed using nested RT-PCR methods. To assess the role of HPgV in lymphoma prognosis, we used 2948 cases from a cohort study of newly diagnosed lymphoma patients (included all cases from the case-control study). There was a positive association of HPgV viraemia with risk of lymphoma overall (Odds ratio=2.14; 95% confidence interval [CI] 1.63–2.80; P<0.0001), and for all major subtypes except Hodgkin lymphoma and chronic lymphocytic leukaemia/small lymphocytic lymphoma, and this was not confounded by other lymphoma risk factors. In contrast, there was no association of HPgV viraemia with event-free survival (Hazard ratio [HR]=1.00; 95% CI 0.85–1.18) or overall survival (HR=0.97; 95% CI 0.79–1.20) for lymphoma overall, or any of the subtypes. These data support the hypothesis for a role of HPgV in the aetiology of multiple lymphoma subtypes.
Keywords: HPgV, infection, lymphoma, risk, prognosis
Lymphoid neoplasms, including Hodgkin lymphoma (HL), non-Hodgkin lymphoma (NHL), chronic lymphocytic leukaemia (CLL)/small lymphocytic lymphoma (SLL) and plasma cell neoplasms, are a heterogeneous group of malignancies, and overall represent the sixth most common cancer in the United States (US). In 2016, there were an estimated 136,690 new cases of lymphoid neoplasms, the vast majority represented by NHL (Teras, et al 2016). Although our understanding of the aetiology of NHL remains incomplete, an increased risk is associated with immunodeficiency, autoimmune disorders and exposure to physical, chemical and infectious agents (Cerhan, et al 2018). Among the latter, several recent studies suggest an association between a common, otherwise non-pathogenic virus, called human pegivirus (HPgV), and NHL (Chang, et al 2014, Krajden, et al 2010).
HPgV was initially called by two names when it was discovered in 1995, GB virus type C (GBV-C) (Simons, et al 1995) and hepatitis G virus (HGV) (Linnen, et al 1996). As it was shown not to cause hepatitis or any other known illness, nor was infection identified in the presumed hepatitis patient with the initials “GB”, neither name accurately described the virus, and it was re-classified as a member of the Pegivirus genus within the Flaviviridae (Adams, et al 2013, Simmonds, et al 2017, Stapleton, et al 2011). HPgV is a single-stranded RNA virus that is transmitted through parenteral, sexual and perinatal routes (Heuft, et al 1998, Mohr and Stapleton 2009). Approximately 2% of healthy US blood donors are viraemic with HPgV at the time of blood donation, while individuals with sexually transmitted infections and human immunodeficiency virus (HIV), and intravenous drug users have a much higher rate of exposure and viraemia, with a cross-sectional viraemia prevalence of approximately 20% (Mohr and Stapleton 2009).
Phylogenetically, HPgV is most closely related human virus to Hepatitis C virus (HCV), which is also associated with risk of NHL (Engels, et al 2004, Giordano, et al 2007, Matsuo, et al 2004, Simmonds, et al 2017). Active HPgV infection may persist for decades, although the majority of infections clear within 2 years in immune competent hosts (Tacke, et al 1997, Thomas, et al 1997, Tillmann, et al 1998). HPgV infection is detected by nucleic acid amplification of viral RNA from plasma or serum (Linnen, et al 1996, Simons, et al 1995). The reason for the lack of hepatic disease presumably relates to its tissue tropism, as HPgV does not appear to replicate in the liver, and based on both ex vivo and in vitro studies it replicates in B and T lymphocytes, including CD4+ and CD8+ lymphocytes, spleen, and bone marrow (Bailey, et al 2015, Berg, et al 1999, Chivero, et al 2014, Fan, et al 1999, George, et al 2006, George, et al 2003, Jablonska, et al 2013, Kisiel, et al 2013, Laskus, et al 1998, Tucker, et al 2000, Xiang, et al 2000). Persistent infection among HIV-infected individuals is highly associated with impaired T-cell receptor and interleukin 2 receptor (IL2R) signalling, resulting in inhibition of T cell apoptosis, and impaired T cell activation and proliferation that may contribute to lymphomagenesis (Bhattarai, et al 2012, Bhattarai, et al 2013, Lanteri, et al 2015, Moenkemeyer, et al 2008, Stapleton, et al 2012, Stapleton, et al 2009).
Because HPgV may cause persistent infection, alters immune function, and is related to HCV, a number of epidemiological studies have been conducted to determine if there is a relationship between HPgV infection and risk of NHL. Several case-control studies initially reported an association of HPgV with NHL, but conclusions were limited due to small sample sizes, use of convenience sample control groups, and combining HPgV viraemia and prior infection (De Renzo, et al 2002, Giannoulis, et al 2004, Kaya, et al 2002, Minton, et al 1998, Zignego, et al 2007). One carefully designed, population-based study of 553 cases and 438 controls from British Columbia reported a 2.7-fold higher risk of NHL with HPgV viraemia after accounting for other risk factors (95% confidence interval [CI] 1.22–6.69) (Krajden, et al 2010), and a case-control study nested within the prospective Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial cohort (658 cases and 1316 controls) confirmed an association (Odds ratio [OR]=3.43, 95% CI 1.35–8.71) using pre-diagnosis, banked samples (Chang, et al 2014).
To further address the association of HPgV and risk of developing lymphoma, we conducted the largest study to date of over 2000 lymphoma cases and 1500 controls. We also correlated HPgV status with circulating cytokine levels in a subset of cases and controls (Charbonneau, et al 2012). Finally, because the virus has also been associated with prolonged survival in the setting of co-infection with HIV (Heringlake, et al 1998, Lefrere, et al 1999, Nunnari, et al 2003, Schwarze-Zander, et al 2010, Tillmann, et al 2001, Toyoda, et al 1998, Vahidnia, et al 2012, Williams, et al 2004, Xiang, et al 2001, Yeo, et al 2000, Zhang, et al 2006), we assessed the association of HPgV with event-free (EFS) and overall (OS) survival for the major lymphoma subtypes.
Methods
Study populations
All subjects provided written, informed consent and all studies were approved by the Human Subjects Institutional Review Boards of the Mayo Clinic and the University of Iowa.
Molecular Epidemiology Resource (MER) Cohort Study
The MER Cohort Study of the Iowa/Mayo Lymphoma Specialized Program of Research Excellence (SPORE) has been previously described (Cerhan, et al 2017). Briefly, from 1 September 2002 to 30 June 2015, we prospectively offered enrolment to all consecutive patients with lymphoma who were within 9 months of their initial diagnosis at presentation, aged 18 years and older, and had no prior history of lymphoma, leukaemia or HIV/acquired immunodeficiency syndrome (AIDs); 6972 participated (5256 at Mayo and 1716 at Iowa). At enrolment, participants completed a medical history questionnaire, provided a blood specimen (for DNA, plasma and serum), and consented for access to their medical records. Serum and plasma were stored at −70°C. Clinical and treatment data were abstracted, and pathology was centrally reviewed by a haematopathologist to confirm the diagnosis and to provide a lymphoma subtype according to the World Health Organization Classification of Neoplastic Diseases of the Haematopoietic and Lymphoid Tissues (Jaffe, et al 2001, Swerdlow, et al 2008). All participants were contacted every 6 months for the first 3 years after diagnosis and then annually thereafter to ascertain outcomes; disease progression/relapse, retreatment, transformation, new cancers and cause of death were validated against medical records.
Case-Control Study
The Mayo Clinic Case-Control study of Lymphoma has been previously described (Cerhan, et al 2011). Briefly, cases were Mayo patients enrolled into the MER from 1 September 2002 to 31 August 2014 who were residents of Minnesota, Iowa and Wisconsin at the time of diagnosis (n=3214). All of these cases are a subset of the full MER cohort described in the previous section; the MER cases from Iowa did not have risk factor data or a matched control group and were thus not eligible. We recruited controls from Mayo Clinic Rochester patients with prescheduled general medical examinations in the Department of Medicine. A total 2489 controls with no history of lymphoma, leukaemia, or HIV/AIDS were frequency matched to the case distribution by 5-year age group, sex, and geographic location of residence. Participants completed a series of self-administered risk-factor questionnaires that included demographic data, history of hepatitis, blood transfusion, autoimmune disease, atopy, family history of lymphoma, farming, obesity, smoking, alcohol use and sun exposure.
Analysis Datasets
The current analyses were based 2948 MER cases with an available baseline plasma sample when we extracted banked samples to run the HPgV assay in 2012. Of these 2948 cases, 2094 were also in the case-control study, as were 1572 controls. Of the 2948 cases, 1296 (61.9%) had a plasma sample collected prior to lymphoma treatment and 798 (38.1%) were collected after initiation of treatment.
Laboratory methods
HPgV RNA extraction, detection and quantification
HPgV replication was assessed by measuring HPgV RNA in plasma by real-time reverse transcription polymerase chain reaction (RT-PCR) as described below. To enhance sensitivity compared to previous studies, two variations from reported studies were used (Chang, et al 2014, Rydze, et al 2012, Souza, et al 2006). First, a larger volume was used for both plasma RNA extraction and in the reaction, and secondly the number of cycles of real-time PCR was increased to 45. Briefly, RNA was extracted from 560 μl of plasma using QIamp Viral RNA Mini Kit (Qiagen, Hilden, Germany), and each PCR reaction contained an 26 μl aliquot of RNA (representing 250 μl of plasma), 30 μl TaqMan Master Mix (2X) (ABI, Thermo Fisher Scientific, Waltham, MA, USA), 1.5 μl superscript III RT platinum Taq mix, 0.72 μl each of forward and reverse primers (50 μM, Integrated DNA Technologies, Coralville, IA, USA), 0.12 μl Taqman probe (100 μM, ABI) and 0.3 μl of Rnasin (40U/μl). Primers included the forward primer: 5′ GGC GAC CGG CCA AAA 3′ (96–110), antisense primer: 5′ CTT AAG ACC CAC CTA TAG TGG CTA CC (163- 188), and probe: 5′ FAM-TGA CCG GGA TTT ACG ACC TAC CAA CCC T-TAMRA (131–158) (George, et al 2003). Quantitative one step Real Time PCR was performed using an ABI 7500 system. The running conditions were 50°C for 20 min, 95°C for 2 min, 45 cycles at 95°C for 15s and 58°C for 1 min. A HPgV RNA quantitation standard used was prepared as described.(Rydze, et al 2012) HPgV genome sequences from nt 1 to 850 (from GenBank Accession Number AF121950; https://www.ncbi.nlm.nih.gov/nuccore/af121950) were cloned into pCR2.1 plas mid (Invitrogen, Inc., Camarillo, CA) downstream of the T7 polymerase promoter. The plasmid was linearized with Kpn1 and run off transcripts were generated (Riboprobe, Promega, Madison, WI, USA). RNA was quantified by A260/280, divided into 50 μl aliquots (concentration of 1 × 1010 genome copies/ml) and stored at −80°C. For each real-time PCR experiment, a fresh aliquot was used in serial 10-fold dilutions, starting at a 1:100 dilution in RNAse-free water. Linear regression analyses of standard curves were routinely excellent (r2> 0.97), and variation between cycle threshold (CT) values between experiments were within 2.1 CT.
Samples with CT values greater than 35 were repeated and further tested by nested RT-PCR as described (Souza, et al 2006). Briefly, 26 μl RNA was amplified in a nested RT-PCR to amplify HPgV RNA using primers from either the 5′ntr and E2 coding regions as described(Souza, et al 2006): 5′NTR Outer sense: 5′ – AAG CCC CAG AAA CCG ACG CC - 3′; 5′NTR Outer antisense: 5′ – TGA AGG GCG ACG TGG ACC GT - 3′; 5′NTR Inner sense: 5′ - CGG CCA AAA GGT GGT GGA TG - 3′; 5′NTR Inner antisense: 5′ – GTA ACG GGC TCG GTT TAA CG - 3′; E2 Outer sense: 5′ – TGT GGG GTT CCG TDT CTT GGT T -3′; E2 Outer antisense: 5′ - RAA CGT HCC RCT VGG AGG CT - 3′; E2 Inner sense: 5′ - TGG NTC WGC CAG CTG YAC CAT AG - 3′; E2 Inner antisense: 5′ – DTC YCG GAT CTT GGT CAT GG - 3′. The sequence is based on the full-length Iowa HPgV isolate (GenBank Accession Number AF121950) (Xiang, et al 2000). Nested RT-PCR reaction conditions were described previously, and PCR products were identified by agarose gel electrophoresis and ethidium bromide staining (Souza et al 2006). The 5′NTR and E2 amplicons were 204 nt and 304 nts in length (Souza, et al 2006). Negative and positive controls were included with each sample undergoing PCR testing. If neither of the nested RT reactions were positive, the sample was classified as negative. If the real-time and either nested RT-PCR were positive, the sample was classified as positive.
Serum cytokine assay
In a previously published case-control study from the MER (Charbonneau, et al 2012), we used a multiplexed assay to measure 30 cytokine concentrations in stored pre-treatment serum from 234 follicular lymphoma (FL) and 188 diffuse large B cell lymphoma (DLBCL) cases and 400 controls using a multiplex enzyme-linked immunosorbent assay (InvitrogenInc.). The cytokines were analysed using the Luminex-100 system Version 1.7 (Luminex, Austin, TX). Data were acquired using STar Station software (Applied Cytometry, Sheffield, UK) and analysis was performed using the MasterPlex QT 1.0 system (MiraiBio, Alameda, CA). Samples were randomly assigned to plates, stratified by case (DLBCL and FL) and control status, so that each plate was mixture of cases and controls. There were 136 FL, 102 DLBCL and 379 controls with cytokine and HPgV data for analysis.
Data analysis
Case-control analyses
We used unconditional logistic regression to calculate the ORs and 95% CI to estimate the association of HPgV viraemia positivity with lymphoma risk, first adjusting for age and sex and then further adjusting for additional lymphoma risk factors. In addition to overall lymphoma risk, we also evaluated associations with the following lymphoma subtypes: HL, CLL/SLL, DLBCL, FL, mantle cell lymphoma (MCL), marginal zone lymphoma (MZL), T cell lymphoma (TCL) and other NHL. In secondary analyses, we also evaluated the association of viraemic load with lymphoma risk, using the tertile cutpoints of viraemic positive controls for the analysis.
Survival analyses
We used Cox proportional hazards regression models to assess the association of HPgV viraemia status with EFS and OS. For all subtypes except CLL/SLL, EFS was defined as the time from diagnosis to disease progression, retreatment, or death due to any cause; for CLL/SLL we used time to first treatment instead of EFS. OS was defined as the time from diagnosis to death due to any cause. Patients without an event or death were censored at time of last known follow-up. Lymphoma subtype models were adjusted for the following disease-specific prognostic indices: International Prognostic Index (IPI) for DLBCL, TCL, MZL and other NHL; MCL IPI (MIPI) for MCL; FL IPI (FLIPI) for FL; International Prognostic Score (IPS) for HL; and Rai Stage for CLL/SLL). For analysis with HPgV viral load, we used tertile cutpoints among the HPgV-positive cases.
Correlative analysis with cytokines
We correlated cytokine levels with HPgV viraemia status using a Kruskal–Wallis rank test. We also evaluated the association of viraemic load, using the tertile cutpoints of viraemic positive controls for the analysis, with HPgV viraemia status in FL and DLBCL cases using a spearman correlation.
Results
HPgV and lymphoma risk: case-control analysis
The demographic and other selected characteristics by case and control status are reported in Table I. The mean age of cases was 61.1 years and 58.2% were male, while the mean age of controls was 59.6 years and 51.1% were male; these factors were adjusted for in all models. Cases were more likely to have a family history of NHL and a higher body mass index (BMI) at age 18 years, and were less likely to have a history of blood transfusion or ever drink alcohol.
Table I.
Selected baseline characteristics of study participants in the case-control analysis
| Cases (N=2094) | Controls (N=1572) | P-value | |
|---|---|---|---|
|
| |||
| N (%) | N (%) | ||
| Age, mean ± SD, years | 59.6 ± 14.1 | 61.1 ± 13.7 | 0.0015 |
| Gender | <0.0001 | ||
| Female | 875 (41.8) | 768 (48.9) | |
| Male | 1219 (58.2) | 804 (51.1) | |
| Highest educational level | 0.0613 | ||
| Less than high school graduate | 86 (5.1) | 45 (3.3) | |
| High school graduate/GED | 359 (21.2) | 333 (24.1) | |
| Some college/vocational school | 500 (29.5) | 4.6 (29.3) | |
| College graduate | 337 (19.9) | 265 (19.1) | |
| Graduate school | 414 (24.4) | 335 (24.2) | |
| Missing | 398 | 188 | |
| Family History of NHL | 0.0001 | ||
| No | 1442 (88.5) | 1235 (92.9) | |
| Yes | 188 (11.5) | 95 (7.1) | |
| Missing | 464 | 242 | |
| Ever diagnosed with hepatitis | 0.57 | ||
| No | 1650 (96.8) | 1346 (97.2) | |
| Yes | 54 (3.2) | 39 (2.8) | |
| Missing | 390 | 187 | |
| Blood transfusion | 0.034 | ||
| Never | 1465 (86.4) | 1145 (83.7) | |
| Ever | 230 (13.6) | 223 (16.3) | |
| Missing | 399 | 204 | |
| Body Mass Index (kg/m2) at age 18 years | 0.0001 | ||
| <18.5 | 147 (8.8) | 148 (10.9) | |
| 18.5–24.9 | 1205 (72.0) | 1036 (76.0) | |
| 25.0–29.9 | 267 (16.0) | 150 (11.0) | |
| 30.0–34.9 | 54 (3.2) | 30 (2.2) | |
| Missing | 421 | 208 | |
| Ever drink alcohol | 0.032 | ||
| Never | 204 (12.1) | 133 (9.7) | |
| Ever | 1476 (87.9) | 1239 (90.3) | |
| Missing | 414 | 200 | |
GED: General Equivalency Development; NHL: Non-Hodgkin lymphoma; SD: standard deviation
HPgV viraemia was detected in 78 controls (5.0%) and 211 cases (10.1%) (P<0.0001). There was no major difference in the prevalence by age group or sex among both cases and controls (data not shown). Among controls, HPgV viraemia was not associated with family history of NHL, history of hepatitis, blood transfusion, BMI at age 18 years, or ever use of alcohol (Supplemental Table I).
After adjustment for age and sex, there was a strong positive association of HPgV viraemia with risk of lymphoma overall (OR=2.14; 95% CI 1.63–2.80), and there was evidence of a trend with increasing viral load (P-trend<0.001) (Table II). These associations remained after further adjustment for education, family history of NHL, history of hepatitis, blood transfusion, BMI at age 18 years and ever use of alcohol. The risk associations by lymphoma subtype are shown in Table III: there were positive associations for all subtypes except CLL/SLL (OR=0.90; 95% CI 0.53–1.53) and HL (OR=1.47; 95% CI 0.76–2.85), with the strongest association observed for FL (OR=3.20; 95% CI 2.27–4.51). For lymphoma cases, the HPgV RNA positivity was detected in 117 (9.0%) plasma samples collected before the beginning of the treatment, and in 94 (11.8%) samples collected after initiation of therapy (P=0.26). After excluding cases without pre-treatment data, there were no notable changes in the risk associations (data not shown).
Table II.
Association of HPgV viraemia status with risk of lymphoma
| HPgV status | Cases N (%) |
Controls N (%) |
Age- and Sex Adjusted Model | Full Model* | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| OR | 95% CI | OR* | 95% CI | |||
| Negative | 1883 (89.9) | 1494 (95) | 1.00 | reference | 1.00 | reference |
| Positive | 211 (10.1) | 78 (5.0) | 2.14 | 1.63–2.80 | 2.37 | 1.73–3.24 |
| By viral load | ||||||
| 200–1111 | 52 (2.5) | 27 (1.7) | 1.58 | 0.99–2.54 | 1.89 | 1.09–3.31 |
| 1112–5399 | 58 (2.7) | 27 (1.7) | 1.74 | 1.10–2.77 | 1.64 | 0.97–2.79 |
| >5399 | 101 (4.8) | 26 (1.6) | 2.97 | 1.92–4.60 | 3.80 | 2.23–6.49 |
| P-trend | <0.001 | <0.001 | ||||
Adjusted for age, sex, education, family history of NHL, history of hepatitis, blood transfusion, BMI at age 18 years, and alcohol use.
BMI: body mass index; CI: confidence interval; HPgV: human pegivirus; NHL: Non-Hodgkin lymphoma; OR: odds ratio
Table III.
Association of HPgV viraemia status with risk of major lymphoma subtypes
| Age and Sex Adjusted Model | Full Model* | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Group | Total N | HPgV Positive N (%) |
OR | 95% CI | OR | 95% CI |
| Controls | 1572 | 78 (5.0) | 1.00 | Reference | 1.00 | Reference |
| Follicular | 483 | 70 (14.5) | 3.20 | 2.27–4.51 | 3.57 | 2.40–5.31 |
| DLBCL | 388 | 38 (9.8) | 2.14 | 1.64–2.81 | 2.36 | 1.46–3.81 |
| CLL/SLL | 400 | 18 (4.5) | 0.90 | 0.53–1.53 | 1.10 | 0.58–2.06 |
| Mantle cell | 115 | 13 (11.3) | 2.62 | 1.39–4.95 | 3.35 | 1.61–7.00 |
| Marginal zone | 184 | 17 (9.2) | 1.98 | 1.14–3.43 | 1.93 | 1.03–3.63 |
| Other B-NHL | 242 | 30 (12.4) | 2.71 | 1.72–4.27 | 3.51 | 2.09–5.90 |
| T-cell | 112 | 12 (10.7) | 2.25 | 1.18–4.28 | 3.36 | 1.61–7.02 |
| Hodgkin lymphoma | 170 | 13 (7.6) | 1.47 | 0.76–2.85 | 0.91 | 0.36–2.35 |
Adjusted for age, sex, education, family history of NHL, hepatitis, blood transfusion, BMI at age 18 years, and alcohol use.
B-NHL: B cell Non-Hodgkin lymphoma; CI: confidence interval; CLL/SLL: chronic lymphocytic leukaemia/small cell lymphoma; HPgV: human pegivirus; OR: odds ratio.
HPgV viraemia and cytokine levels
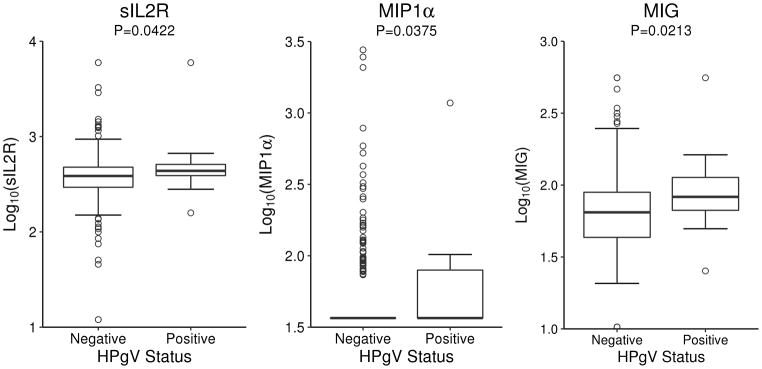
To better understand potential mechanisms by which HPgV might influence lymphoma risk, we correlated circulating serum cytokines in 379 controls from a previous study (Charbonneau, et al 2012) with the newly determined HPgV viraemia status (Supplemental Table II). As shown in Fig. 1, serum levels of soluble IL2R, macrophage inflammatory protein-1 alpha (MIP1α) and monokine induced by gamma interferon (MIG, also termed CXCL9) were significantly higher in HPgV viraemia positive controls compared to HPgV negative controls. Pre-treatment serum cytokines were available for 136 FL and 102 DLBCL cases, but there was no association of any cytokines with HPgV viraemic status for either group (data not shown).
Fig. 1.
Association of sIL2R, MIP1, and MIG levels by HPgV viraemic status.
HPgV: human pegivirus; MIG: monokine induced by gamma interferon; MIP1α: macrophage inflammatory protein-1 alpha; sIL2R: soluble interleukin 2 receptor
HPgV viraemia and lymphoma outcome
HPgV viraemia was not associated with EFS (Hazard ratio [HR]=1.00; 95% CI 0.85–1.18) or OS (HR = 0.97; 95% CI 0.79–1.20) in lymphoma overall or with each of the major subtypes (Table IV). For lymphoma overall, there was no association of EFS or OS with viraemic load (data not shown).
Table IV.
Association of HPgV viraemia status with event-free and overall survival for major lymphoma subtypes
| Event-Free Survival | Overall Survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| Subtype/GBV status | N | N events | % | HR* | 95% CI | N events | (%) | HR* | 95% CI |
| Follicular | |||||||||
| GBV− | 518 | 297 | 57 | 1.00 | reference | 98 | 19 | 1.00 | reference |
| GBV+ | 72 | 45 | 63 | 1.08 | 0.79–1.48 | 19 | 26 | 1.41 | 0.85–2.34 |
| DLBCL | |||||||||
| GBV− | 617 | 292 | 47 | 1.00 | reference | 231 | 37 | 1.00 | reference |
| GBV+ | 66 | 35 | 53 | 1.24 | 0.87–1.77 | 26 | 39 | 1.15 | 0.76–1.72 |
| CLL/SLL | |||||||||
| GBV− | 464 | NA | NA | NA | NA | 144 | 31 | 1.00 | reference |
| GBV+ | 24 | NA | NA | NA | NA | 9 | 38 | 0.91 | 0.42–1.95 |
| MCL | |||||||||
| GBV− | 135 | 102 | 76 | 1.00 | reference | 78 | 58 | 1.00 | reference |
| GBV+ | 19 | 15 | 79 | 0.94 | 0.54–1.61 | 12 | 63 | 0.83 | 0.45–1.55 |
| MZL | |||||||||
| GBV− | 236 | 105 | 44 | 1.00 | reference | 39 | 17 | 1.00 | reference |
| GBV+ | 22 | 9 | 41 | 0.85 | 0.43–1.70 | 4 | 18 | 1.19 | 0.41–3.42 |
| Other B-NHL | |||||||||
| GBV− | 285 | 178 | 64 | 1.00 | reference | 130 | 46 | 1.00 | reference |
| GBV+ | 35 | 19 | 54 | 0.70 | 0.44–1.13 | 13 | 37 | 0.66 | 0.37–1.17 |
| TCL | |||||||||
| GBV− | 164 | 111 | 68 | 1.00 | reference | 85 | 52 | 1.00 | reference |
| GBV+ | 18 | 15 | 83 | 1.62 | 0.94–2.78 | 10 | 56 | 1.10 | 0.57–2.11 |
| Hodgkin lymphoma | |||||||||
| GBV− | 249 | 69 | 28 | 1.00 | reference | 43 | 17 | 1.00 | reference |
| GBV+ | 24 | 7 | 29 | 0.87 | 0.40–1.90 | 3 | 13 | 0.64 | 0.20–2.06 |
Adjusted for each disease-specific prognostic index: IPI (DLBCL, TCL, MZL, Other NHL), MIPI (MCL), FLIPI (FL), IPS (HL), Rai Stage (CLL/SLL)
B-NHL: B cell Non-Hodgkin lymphoma; CI: confidence interval; CLL/SLL: chronic lymphocytic leukaemia/small cell lymphoma; DLBCL: diffuse large B cell lymphoma; FL: follicular lymphoma; FLIPI: FL International Prognostic Index; GBV: GB virus; HPgV: human pegivirus; HR: hazard ratio; IPI: International Prognostic Index; IPS: International Prognostic Score; MCL: mantle cell lymphoma; MIPI: MCL International Prognostic Index; MZL: marginal zone lymphoma; TCL: T cell lymphoma
Discussion
In this clinic-based case-control study we found a positive association of HPgV infection with risk of lymphoma overall and each of the major lymphoma subtypes except CLL/SLL and HL. This study is the largest to date and was well-powered for the first robust assessment of associations with the major lymphoma subtypes of DLBCL, FL and CLL/SLL. These associations remained after adjustment for potential confounding factors. In contrast, we did not observe any associations of HPgV status with EFS or OS for any of the lymphoma subtypes.
There are limitations to our study. Samples were collected after the start of treatment in about one third of cases, although we and others observed that this did not alter these associations (Krajden, et al 2010). However, only pre-disease samples can definitively address this question, as discussed below. Our HPgV viraemia prevalence was much higher than in the two largest published studies (Chang, et al 2014, Krajden, et al 2010). This is most likely explained by the use of a more sensitive HPgV RNA detection method than in previous studies. Unfortunately, a reproducible antibody test is no longer commercially available, so prior infection cannot be determined in PCR negative subjects. Although we were able to adjust for a variety of potential confounding factors in the case-control study, there may residual confounding for confounding by other factors, and the possibility remains that patients destined to clinically develop lymphoma may have a pre-existing immune alteration that reduces their clearance of HPgV infection, leading to higher viraemia rates in new lymphoma diagnoses. Also, our case-control study was clinic and not population-based, which is a weaker design, although we have shown robust internal and external validity for a variety of results from this study (Cerhan, et al 2011, Morton, et al 2014). Finally, while we were able to adjust for disease-specific prognostic indices in our EFS and OS analyses, we were not able to adjust for treatment or other clinical factors.
Our results confirm and extend prior reports of a positive association of HPgV infection with overall lymphoma risk observed small case-control studies, the large population-based Canadian case-control study and the nested case-control PLCO study with banked, pre-disease samples. Compared to the Canadian study (Krajden, et al 2010), we observed a higher overall prevalence of HPgV infection (7.8% vs. 3.3%), and a somewhat different risk correlation according to the subtypes. In the Canadian study the association was strongest for DLBCL (OR=5.18, 95% CI 2.06–13.71; based on 12 exposed cases), and was not associated with FL (OR=1.24; 95% CI 0.27–4.49; based on 3 exposed cases), MZL (OR=1.02; 95% CI 0.05–6.03; based on 2 exposed cases) or all T-cell (OR=1.00; 95% CI 0.05–5.77; based on 1 exposed case), acknowledging the limited power for subtype associations in that study (Krajden, et al 2010). In the PLCO Cancer Screening Trial, statistically significant associations of HPgV viraemia with NHL overall (OR=3.43, 95% CI 1.35–8.71) and with DLBCL (OR=5.31, 95% CI 1.54–18.36; based on 4 exposed cases) were observed, with suggestive associations for FL (OR=4.25; 95% CI 0.89–21.3; based on 2 exposed cases) and CLL/SLL (OR=3.05; 95% CI 0.89–10.5; based on 4 exposed cases) (Chang, et al 2014). Importantly, the latter prospective study showed for the first time that HPgV infection precedes development of NHL by several years, and the risk appears strongest with the longest documented presence of viraemia. Of note, the low prevalence of the HPgV infection (0.9%) in the study was probably due to the differences in age and birth cohort differences, as well as the different methods applied to detect the viral RNA (Chang, et al 2014).
We found higher levels of IL-2R, MIP1α, and MIG in HPgV positive compared to HPgV negative controls. High levels of these cytokines have been shown in other infectious and autoimmune diseases, and they were associated with risk of DLBCL and FL as reported in a prior analysis from this study (Witkowska 2005). It is difficult, however, to establish whether elevated levels of these cytokines promote tumour growth through immunosuppression or other mechanisms, or whether it depends on the constitutive activation of the immune cells. Although our data only suggest potential pathways through which HPgV might influence the lymphomagenesis, further studies in larger numbers of patients are needed to fully examine the role of HPgV-mediated cytokine modulation and the risk of developing NHL.
Additional biological mechanisms that may contribute to a role for HPgV viraemia in lymphomagenesis include the fact that it is a lymphotropic virus that may cause persistent infection in both T and B lymphocytes (Chivero, et al 2014, George, et al 2006, Tucker, et al 2000). In cell lines derived from HIV-infected patients, HPgV reduces Fas-mediated apoptosis (Moenkemeyer, et al 2008) and virus impairs T-cell receptor and IL2R signalling in primary and transformed T cell lines (Bhattarai, et al 2012, Bhattarai, et al 2013) and in patients with HCV infection (Bhattarai, et al 2017), all of which may contribute to subclinical impairment of immune surveillance and thereby increase the risk of developing NHL (Pietersma, et al 2008). While the lack of association of HPgV with risk of CLL might temper this hypothesis, the associations of NHL subtypes with specific types of infections and/or immunosuppression (primary, transplantation, iatrogenic) have been quite heterogeneous (Cerhan, et al 2018). HPgV also influences cytokine and chemokine gene expression in lymphocytes in vitro, and in cells obtained from infected individuals, potentially influencing immune response (Lanteri, et al 2015, Stapleton, et al 2012, Stapleton, et al 2013, Xiang, et al 2004). Finally, persistent lymphocyte infection could lead to DNA mutation and potentially malignant transformation. The biological reason for the lack of an association of HPgV infection with lymphoma prognosis is not known, but would suggest that HPgV does not influence lymphoma aggressiveness or that other clinical or treatment factors dominate disease progression and survival. However, a limitation of this analysis is the relatively small number of HPgV viraemia positive patients, particularly for less common subtypes, which limits the power of the survival analyses.
If this association were ultimately found to be causal, then there are potential public health considerations given an HPgV viraemia prevalence of approximately 2% in healthy blood donors (Heuft, et al 1998, Linnen, et al 1996, Mohr and Stapleton 2009) and the number of blood products transfused in the US annually; more than 1,000 units of HPgV infected blood are predicted to be administered to individuals daily. Further, HPgV may be a biomarker that potentially can be treated or prevented, and screening of blood donors for this infection should be considered.
In conclusion, our clinic-based case-control study confirms that HPgV infection is a risk factor for lymphoma overall and extends results to each NHL subtype except HL and CLL. Further studies are needed to clarify the biological mechanisms leading to lymphoma development.
Supplementary Material
Acknowledgments
We thank Thomas Kaufman and Lars Larsen for technical assistance and James H McLinden for helpful discussions. Grant support: Department of Veterans Affairs Merit Review Grants CX00821 and BX000207 (JTS), and NIH P50 CA97274 and U01 CA 195568 (JRC and BKL).
Footnotes
Author’s contributions
AF, JX, DK, ALF, GSN, ML, SMA, AJN, YWA, TGC, TMH, JRC and JTS performed the research; JRC and JTS designed the research study; JTS contributed essential reagents or tools; AF, CA, MCL, MJM, SLS, JRC and JTS analysed the data; and AF, JRC and JTS wrote the paper.
Conflict of interest
The authors have no conflict of interest to disclose.
References
- Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Archives of Virology. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- Bailey AL, Lauck M, Mohns M, Peterson EJ, Beheler K, Brunner KG, Crosno K, Mejia A, Mutschler J, Gehrke M, Greene J, Ericsen AJ, Weiler A, Lehrer-Brey G, Friedrich TC, Sibley SD, Kallas EG, Capuano S, 3rd, Rogers J, Goldberg TL, Simmons HA, O’Connor DH. Durable sequence stability and bone marrow tropism in a macaque model of human pegivirus infection. Science Translational Medicine. 2015;7:305ra144. doi: 10.1126/scitranslmed.aab3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Muller AR, Platz KP, Hohne M, Bechstein WO, Hopf U, Wiedenmann B, Neuhaus P, Schreier E. Dynamics of GB virus C viremia early after orthotopic liver transplantation indicates extrahepatic tissues as the predominant site of GB virus C replication. Hepatology. 1999;29:245–249. doi: 10.1002/hep.510290121. [DOI] [PubMed] [Google Scholar]
- Bhattarai N, McLinden JH, Xiang J, Kaufman TM, Stapleton JT. GB virus C envelope protein E2 inhibits TCR-induced IL-2 production and alters IL-2-signaling pathways. Journal of Immunology. 2012;189:2211–2216. doi: 10.4049/jimmunol.1201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N, McLinden JH, Xiang J, Landay AL, Chivero ET, Stapleton JT. GB virus C particles inhibit T cell activation via envelope E2 protein-mediated inhibition of TCR signaling. Journal of Immunology. 2013;190:6351–6359. doi: 10.4049/jimmunol.1300589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N, McLinden JH, Xiang J, Mathahs MM, Schmidt WN, Kaufman TM, Stapleton JT. Hepatitis C virus infection inhibits a Src-kinase regulatory phosphatase and reduces T cell activation in vivo. PLoS Pathogens. 2017;13:e1006232. doi: 10.1371/journal.ppat.1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerhan JR, Fredericksen ZS, Wang AH, Habermann TM, Kay NE, Macon WR, Cunningham JM, Shanafelt TD, Ansell SM, Call TG, Witzig TE, Slager SL, Liebow M. Design and validity of a clinic-based case-control study on the molecular epidemiology of lymphoma. International Journal of Molecular Epidemiology and Genetics. 2011;2:95–113. [PMC free article] [PubMed] [Google Scholar]
- Cerhan JR, Link BK, Habermann TM, Maurer MJ, Feldman AL, Syrbu SI, Thompson CA, Farooq U, Novak AJ, Slager SL, Allmer C, Lunde JJ, Macon WR, Inwards DJ, Johnston PB, Micallef INM, Nowakowski GS, Ansell SM, Kay NE, Weiner GJ, Witzig TE. Cohort Profile: The Lymphoma Specialized Program of Research Excellence (SPORE) Molecular Epidemiology Resource (MER) Cohort Study. International Journal of Epidemiology. 2017;46:1753–1754i. doi: 10.1093/ije/dyx119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerhan JR, Vajdic CM, Spinelli JJ. The non-Hodgkin Lymphomas. In: Thun MJ, Linet MS, Cerhan JR, Haiman CA, Schottenfeld D, editors. Schottenfeld and Fraumeni Cancer Epidemiology and Prevention. Oxford University Press; New York: 2018. pp. 767–796. [Google Scholar]
- Chang CM, Stapleton JT, Klinzman D, McLinden JH, Purdue MP, Katki HA, Engels EA. GBV-C infection and risk of NHL among U.S. adults. Cancer Research. 2014;74:5553–5560. doi: 10.1158/0008-5472.CAN-14-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau B, Maurer MJ, Ansell SM, Slager SL, Fredericksen ZS, Ziesmer SC, Macon WR, Habermann TM, Witzig TE, Link BK, Cerhan JR, Novak AJ. Pretreatment circulating serum cytokines associated with follicular and diffuse large B-cell lymphoma: a clinic-based case-control study. Cytokine. 2012;60:882–889. doi: 10.1016/j.cyto.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivero ET, Bhattarai N, Rydze RT, Winters MA, Holodniy M, Stapleton JT. Human pegivirus RNA is found in multiple blood mononuclear cells in vivo and serum-derived viral RNA-containing particles are infectious in vitro. Journal of General Virology. 2014;95:1307–1319. doi: 10.1099/vir.0.063016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzo A, Persico E, de Marino F, di Giacomo Russo G, Notaro R, di Grazia C, Picardi M, Santoro L, Torella R, Rotoli B, Persico M. High prevalence of hepatitis G virus infection in Hodgkin’s disease and B-cell lymphoproliferative disorders: absence of correlation with hepatitis C virus infection. Haematologica. 2002;87:714–718. discussion 718. [PubMed] [Google Scholar]
- Engels EA, Chatterjee N, Cerhan JR, Davis S, Cozen W, Severson RK, Whitby D, Colt JS, Hartge P. Hepatitis C virus infection and non-Hodgkin lymphoma: results of the NCI-SEER multi-center case-control study. International Journal of Cancer. 2004;111:76–80. doi: 10.1002/ijc.20021. [DOI] [PubMed] [Google Scholar]
- Fan X, Xu Y, Solomon H, Ramrakhiani S, Neuschwander-Tetri BA, Di Bisceglie AM. Is hepatitis G/GB virus-C virus hepatotropic? Detection of hepatitis G/GB virus-C viral RNA in liver and serum. Journal of Medical Virology. 1999;58:160–164. [PubMed] [Google Scholar]
- George SL, Xiang J, Stapleton JT. Clinical isolates of GB virus type C vary in their ability to persist and replicate in peripheral blood mononuclear cell cultures. Virology. 2003;316:191–201. doi: 10.1016/s0042-6822(03)00585-3. [DOI] [PubMed] [Google Scholar]
- George SL, Varmaz D, Stapleton JT. GB virus C replicates in primary T and B lymphocytes. Journal of Infectious Diseases. 2006;193:451–454. doi: 10.1086/499435. [DOI] [PubMed] [Google Scholar]
- Giannoulis E, Economopoulos T, Mandraveli K, Giannoulis K, Nikolaides C, Zervou E, Papageorgiou E, Zoulas D, Tourkantonis A, Giannopoulos G, Fountzilas G. The prevalence of hepatitis C and hepatitis G virus infection in patients with B cell non-Hodgkin lymphomas in Greece: a Hellenic Cooperative Oncology Group Study. Acta Haematologica. 2004;112:189–193. doi: 10.1159/000081270. [DOI] [PubMed] [Google Scholar]
- Giordano TP, Henderson L, Landgren O, Chiao EY, Kramer JR, El-Serag H, Engels EA. Risk of non-Hodgkin lymphoma and lymphoproliferative precursor diseases in US veterans with hepatitis C virus. JAMA. 2007;297:2010–2017. doi: 10.1001/jama.297.18.2010. [DOI] [PubMed] [Google Scholar]
- Heringlake S, Ockenga J, Tillmann HL, Trautwein C, Meissner D, Stoll M, Hunt J, Jou C, Solomon N, Schmidt RE, Manns MP. GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? Journal of Infectious Diseases. 1998;177:1723–1726. doi: 10.1086/517431. [DOI] [PubMed] [Google Scholar]
- Heuft HG, Berg T, Schreier E, Kunkel U, Tacke M, Schwella N, Hopf U, Salama A, Huhn D. Epidemiological and clinical aspects of hepatitis G virus infection in blood donors and immunocompromised recipients of HGV-contaminated blood. Vox Sanguinis. 1998;74:161–167. [PubMed] [Google Scholar]
- Jablonska J, Zabek J, Pawelczyk A, Kubisa N, Fic M, Laskus T, Radkowski M. Hepatitis C virus (HCV) infection of peripheral blood mononuclear cells in patients with type II cryoglobulinemia. Human Immunology. 2013;74:1559–1562. doi: 10.1016/j.humimm.2013.08.273. [DOI] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Vardiman JW. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon, France: 2001. [Google Scholar]
- Kaya H, Polat MF, Erdem F, Gundogdu M. Prevalence of hepatitis C virus and hepatitis G virus in patients with non-Hodgkin’s lymphoma. Clinical and Laboratory Haematology. 2002;24:107–110. doi: 10.1046/j.1365-2257.2002.00427.x. [DOI] [PubMed] [Google Scholar]
- Kisiel E, Cortez KC, Pawelczyk A, Osko IB, Kubisa N, Laskus T, Radkowski M. Hepatitis G virus/GBV-C in serum, peripheral blood mononuclear cells and bone marrow in patients with hematological malignancies. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases. 2013;19:195–199. doi: 10.1016/j.meegid.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Krajden M, Yu A, Braybrook H, Lai AS, Mak A, Chow R, Cook D, Tellier R, Petric M, Gascoyne RD, Connors JM, Brooks-Wilson AR, Gallagher RP, Spinelli JJ. GBV-C/hepatitis G virus infection and non-Hodgkin lymphoma: a case control study. International Journal of Cancer. 2010;126:2885–2892. doi: 10.1002/ijc.25035. [DOI] [PubMed] [Google Scholar]
- Lanteri MC, Vahidnia F, Tan S, Stapleton JT, Norris PJ, Heitman J, Deng X, Keating SM, Brambilla D, Busch MP, Custer B. Downregulation of Cytokines and Chemokines by GB Virus C After Transmission Via Blood Transfusion in HIV-Positive Blood Recipients. Journal of Infectious Diseases. 2015;211:1585–1596. doi: 10.1093/infdis/jiu660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Detection of hepatitis G virus replication sites by using highly strand-specific Tth-based reverse transcriptase PCR. Journal of Virology. 1998;72:3072–3075. doi: 10.1128/jvi.72.4.3072-3075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrere JJ, Roudot-Thoraval F, Morand-Joubert L, Petit JC, Lerable J, Thauvin M, Mariotti M. Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. Journal of Infectious Diseases. 1999;179:783–789. doi: 10.1086/314671. [DOI] [PubMed] [Google Scholar]
- Linnen J, Wages J, Jr, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih JW, Young L, Piatak M, Jr, Hoover C, Fernandez J, Chen S, Zou JC, Morris T, Hyams KC, Ismay S, Lifson JD, Hess G, Foung SK, Thomas H, Bradley D, Margolis H, Kim JP. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Kusano A, Sugumar A, Nakamura S, Tajima K, Mueller NE. Effect of hepatitis C virus infection on the risk of non-Hodgkin’s lymphoma: a meta-analysis of epidemiological studies. Cancer Science. 2004;95:745–752. doi: 10.1111/j.1349-7006.2004.tb03256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton J, Iqbal A, Eskiturk A, Irving W, Davies J. Hepatitis G virus infection in lymphoma and in blood donors. Journal of Clinical Pathology. 1998;51:676–678. doi: 10.1136/jcp.51.9.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenkemeyer M, Schmidt RE, Wedemeyer H, Tillmann HL, Heiken H. GBV-C coinfection is negatively correlated to Fas expression and Fas-mediated apoptosis in HIV-1 infected patients. Journal of Medical Virology. 2008;80:1933–1940. doi: 10.1002/jmv.21305. [DOI] [PubMed] [Google Scholar]
- Mohr EL, Stapleton JT. GB virus type C interactions with HIV: the role of envelope glycoproteins. Journal of Viral Hepatitis. 2009;16:757–768. doi: 10.1111/j.1365-2893.2009.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LM, Sampson JN, Cerhan JR, Turner JJ, Vajdic CM, Wang SS, Smedby KE, de Sanjose S, Monnereau A, Benavente Y, Bracci PM, Chiu BC, Skibola CF, Zhang Y, Mbulaiteye SM, Spriggs M, Robinson D, Norman AD, Kane EV, Spinelli JJ, Kelly JL, La Vecchia C, Dal Maso L, Maynadie M, Kadin ME, Cocco P, Costantini AS, Clarke CA, Roman E, Miligi L, Colt JS, Berndt SI, Mannetje A, de Roos AJ, Kricker A, Nieters A, Franceschi S, Melbye M, Boffetta P, Clavel J, Linet MS, Weisenburger DD, Slager SL. Rationale and design of the International Lymphoma Epidemiology Consortium (InterLymph) non-Hodgkin Lymphoma Subtypes Project. Journal of the National Cancer Institute Monographs. 2014;2014:1–14. doi: 10.1093/jncimonographs/lgu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari G, Nigro L, Palermo F, Attanasio M, Berger A, Doerr HW, Pomerantz RJ, Cacopardo B. Slower progression of HIV-1 infection in persons with GB virus C co-infection correlates with an intact T-helper 1 cytokine profile. Annals of Internal Medicine. 2003;139:26–30. doi: 10.7326/0003-4819-139-1-200307010-00009. [DOI] [PubMed] [Google Scholar]
- Pietersma F, Piriou E, van Baarle D. Immune surveillance of EBV-infected B cells and the development of non-Hodgkin lymphomas in immunocompromised patients. Leukemia and Lymphoma. 2008;49:1028–1041. doi: 10.1080/10428190801911662. [DOI] [PubMed] [Google Scholar]
- Rydze RT, Bhattarai N, Stapleton JT. GB virus C infection is associated with a reduced rate of reactivation of latent HIV and protection against activation-induced T-cell death. Antiviral Therapy. 2012;17:1271–1279. doi: 10.3851/IMP2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze-Zander C, Neibecker M, Othman S, Tural C, Clotet B, Blackard JT, Kupfer B, Luechters G, Chung RT, Rockstroh JK, Spengler U. GB virus C coinfection in advanced HIV type-1 disease is associated with low CCR5 and CXCR4 surface expression on CD4(+) T-cells. Antiviral Therapy. 2010;15:745–752. doi: 10.3851/IMP1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT Ictv Report Consortium. ICTV Virus Taxonomy Profile: Flaviviridae. Journal of General Virology. 2017;98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK. Isolation of novel virus-like sequences associated with human hepatitis. Nature Medicine. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- Souza IE, Allen JB, Xiang J, Klinzman D, Diaz R, Zhang S, Chaloner K, Zdunek D, Hess G, Williams CF, Benning L, Stapleton JT. Effect of primer selection on estimates of GB virus C (GBV-C) prevalence and response to antiretroviral therapy for optimal testing for GBV-C viremia. Journal of Clinical Microbiology. 2006;44:3105–3113. doi: 10.1128/JCM.02663-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Chaloner K, Zhang J, Klinzman D, Souza IE, Xiang J, Landay A, Fahey J, Pollard R, Mitsuyasu R. GBV-C viremia is associated with reduced CD4 expansion in HIV-infected people receiving HAART and interleukin-2 therapy. AIDS. 2009;23:605–610. doi: 10.1097/QAD.0b013e32831f1b00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. Journal of General Virology. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Chaloner K, Martenson JA, Zhang J, Klinzman D, Xiang J, Sauter W, Desai SN, Landay A. GB virus C infection is associated with altered lymphocyte subset distribution and reduced T cell activation and proliferation in HIV-infected individuals. PloS One. 2012;7:e50563. doi: 10.1371/journal.pone.0050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JT, Martinson JA, Klinzman D, Xiang J, Desai SN, Landay A. GB virus C infection and B-cell, natural killer cell, and monocyte activation markers in HIV-infected individuals. AIDS. 2013;27:1829–1832. doi: 10.1097/QAD.0b013e328363089f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of the Haematopoietic and Lymphoid Tissues. In: Bosman FT, Jaffe ES, Lakhani SR, Ohgaki H, editors. World Health Organization Classification of Tumours. IARC Press; Lyon, France: 2008. [Google Scholar]
- Tacke M, Schmolke S, Schlueter V, Sauleda S, Esteban JI, Tanaka E, Kiyosawa K, Alter HJ, Schmitt U, Hess G, Ofenloch-Haehnle B, Engel AM. Humoral immune response to the E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G virus exposure among healthy blood donors. Hepatology. 1997;26:1626–1633. doi: 10.1002/hep.510260635. [DOI] [PubMed] [Google Scholar]
- Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA: A Cancer Journal for Clinicians. 2016;66:443–459. doi: 10.3322/caac.21357. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Nakatsuji Y, Shih JW, Alter HJ, Nelson KE, Astemborski JA, Lyles CM, Vlahov D. Persistence and clinical significance of hepatitis G virus infections in injecting drug users. Journal of Infectious Diseases. 1997;176:586–592. doi: 10.1086/514078. [DOI] [PubMed] [Google Scholar]
- Tillmann HL, Heringlake S, Trautwein C, Meissner D, Nashan B, Schlitt HJ, Kratochvil J, Hunt J, Qiu X, Lou SC, Pichlmayr R, Manns MP. Antibodies against the GB virus C envelope 2 protein before liver transplantation protect against GB virus C de novo infection. Hepatology. 1998;28:379–384. doi: 10.1002/hep.510280213. [DOI] [PubMed] [Google Scholar]
- Tillmann HL, Heiken H, Knapik-Botor A, Heringlake S, Ockenga J, Wilber JC, Goergen B, Detmer J, McMorrow M, Stoll M, Schmidt RE, Manns MP. Infection with GB virus C and reduced mortality among HIV-infected patients. New England Journal of Medicine. 2001;345:715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Fukuda Y, Hayakawa T, Takamatsu J, Saito H. Effect of GB virus C/hepatitis G virus coinfection on the course of HIV infection in hemophilia patients in Japan. Journal of Acquired Immune Deficiency Syndromes and Human Retrovirology. 1998;17:209–213. doi: 10.1097/00042560-199803010-00004. [DOI] [PubMed] [Google Scholar]
- Tucker TJ, Smuts HE, Eedes C, Knobel GD, Eickhaus P, Robson SC, Kirsch RE. Evidence that the GBV-C/hepatitis G virus is primarily a lymphotropic virus. Journal of Medical Virology. 2000;61:52–58. [PubMed] [Google Scholar]
- Vahidnia F, Petersen M, Stapleton JT, Rutherford GW, Busch M, Custer B. Acquisition of GB virus type C and lower mortality in patients with advanced HIV disease. Clinical Infectious Diseases. 2012;55:1012–1019. doi: 10.1093/cid/cis589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. Persistent GB virus C infection and survival in HIV-infected men. New England Journal of Medicine. 2004;350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- Witkowska AM. On the role of sIL-2R measurements in rheumatoid arthritis and cancers. Mediators of Inflammation. 2005;2005:121–130. doi: 10.1155/MI.2005.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Wunschmann S, Schmidt W, Shao J, Stapleton JT. Full-length GB virus C (Hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. Journal of Virology. 2000;74:9125–9133. doi: 10.1128/jvi.74.19.9125-9133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Wunschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, Stapleton JT. Effect of coinfection with GB virus C on survival among patients with HIV infection. New England Journal of Medicine. 2001;345:707–714. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- Xiang J, George SL, Wunschmann S, Chang Q, Klinzman D, Stapleton JT. Inhibition of HIV-1 replication by GB virus C infection through increases in RANTES, MIP-1alpha, MIP-1beta, and SDF-1. Lancet. 2004;363:2040–2046. doi: 10.1016/S0140-6736(04)16453-2. [DOI] [PubMed] [Google Scholar]
- Yeo AE, Matsumoto A, Hisada M, Shih JW, Alter HJ, Goedert JJ. Effect of hepatitis G virus infection on progression of HIV infection in patients with hemophilia. Multicenter Hemophilia Cohort Study. Annals of Internal Medicine. 2000;132:959–963. doi: 10.7326/0003-4819-132-12-200006200-00006. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chaloner K, Tillmann HL, Williams CF, Stapleton JT. Effect of early and late GB virus C viraemia on survival of HIV-infected individuals: a meta-analysis. HIV Medicine. 2006;7:173–180. doi: 10.1111/j.1468-1293.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- Zignego AL, Giannini C, Monti M, Gragnani L. Hepatitis C virus lymphotropism: lessons from a decade of studies. Digestive and Liver Disease: Official Journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2007;39(Suppl 1):S38–45. doi: 10.1016/s1590-8658(07)80009-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.