Abstract
Background
Autophagy is a conserved, self-degradation system that is critical for maintaining cellular homeostasis during stress conditions. Dysregulated autophagy has implications in health and disease. Specifically, in cancer, autophagy plays a dichotomous role by inhibiting tumor initiation but supporting tumor progression. Early results of clinical trials repurposing hydroxychloroquine for cancer suggest autophagy inhibition could be a promising approach for advanced cancers.
Methods
Literature Review
Results
Here we review fundamental advances in the biology of autophagy, approaches to targeting autophagy, the preclinical rationale and clinical experience with HCQ in cancer clinical trials, the potential role of autophagy in tumor immunity, and recent developments in next generation autophagy inhibitors that have clinical potential.
Conclusions
Autophagy is a promising target for drug development in cancer.
Keywords: Autophagy, HCQ, chemotherapy, immunotherapy
Autophagy in health and disease
Autophagy is a tightly orchestrated process that sequesters misfolded proteins, damaged or aged organelles, and mutated proteins in double membrane vesicles called autophagosomes that ultimately fuse to lysosomes leading to the degradation of the sequestered components1. In 2016 the Nobel Prize in Medicine was awarded to Dr. Yoshinori Ohsumi for his groundbreaking work in yeast unraveling the regulation of autophagy2. The recycling capacity of autophagy is conserved from yeast to man, and regulates cellular homeostasis in both physiological and pathophysiological contexts. Dysregulated autophagy has been implicated in a number of diseases including neurodegenerative diseases3, cardiomyopathy4, infectious disease5, type II diabetes6, fatty liver7, and cancer8. Another way autophagy is categorized is into nonselective and selective autophagy9. On the one hand, nonselective autophagy occurs when cells degrade their cytoplasm in a bulk manner. On the other hand, the study of how selective autophagy targets specific organelles or proteins has engendered terms such as mitophagy, pexophagy, and xenophagy10. Recently autophagy has been identified as a target for therapeutic intervention in a number of diseases including cancer. In order for effective development of therapeutics that target autophagy, a thorough understanding of molecular components of autophagy, with a special focus on druggable targets, will be reviewed here.
A mechanistic understanding of autophagy
The process of autophagy is broken down into four critical steps: initiation, nucleation, maturation, and degradation10. During the initiation step, the Unc-51-like kinase 1(ULK1)-Autophagy related gene 13 (ATG13)-family interacting protein 200kD (FIP200) kinase complex gets activated by coordinated inputs from the mechanistic target of rapamycin complex 1 (mTORC1), and AMP-activated protein Kinase (AMPK). In the nucleation step of autophagy, The ULK1 complex phosphorylates and activates the Beclin-1-VPS34 complex. This complex includes Beclin-1, VPS34 (a class III phosphatidylinositol 3-kinase (PI3K)), and other proteins such as VPS15, ATG14L, and autophagy and beclin 1 regulator 1 (AMBRA-1), depending on the subcellular localization of the complex1. Both the initiation and nucleation proteins promote the formation of the autophagic vesicle membrane. This membrane can be derived from mitochondria, plasma membrane, or the endoplasmic reticulum11–13. During the maturation step, two unique protein conjugation events are necessary for autophagosome formation: 1) ATG7 and ATG10 conjugate ATG5 to ATG12. 2) ATG7 and ATG3 conjugate LC3 (ATG8) to the lipid phosphatidylethanolamine (PE)14. The ATG5-ATG12 conjugate forms a complex with ATG16L1, and the ATG5-ATG12-ATG16L1 complex gets anchored onto phosphoinositol 3-phosphate generated by VPS34 on emerging autophagosomal membranes through WIPI-2b15.. Meanwhile, the cleavage of LC3 by ATG4 leads to the soluble form (LC3-1), which is then conjugated to (PE) on the surface of the emerging autophagosome by ATG3 and ATG7, and guided by the ATG5-ATG12-ATG16L1 complex 16. The lipidated form of LC3 is referred to as LC3-II. LC3-II migrates faster than LC3-I on gel electrophoresis allowing, the ratio of lipidated to free LC3 to utilized to reflect the number of autophagosomes forming at any given time.
The ultimate effect of this complex system is to place an ubiquitin-like protein (LC3-I) on membrane to label it as an autophagic membrane and to allow for interaction with cargo receptors that bring autophagic cargo to the autophagosome. SQSTM1/P62 and NBR1 are cargo receptors that recruit cargo destined for autophagic degradation to LC3-II on forming autophagasomes17. Once the isolation membrane is enclosed, it is called the autophagosome18. After autophagosome formation and cargo sequestration, the cargo-bound autophagosomes are transported on microtubules to the perinuclear region where lysosomes are present19. The multiprotein HOPS complex along with syntaxin 17 helps tether the autophagosomes to the lysosome20 Emerging evidence indicates there are adaptor proteins such as EPG5 that recognize autophagy proteins to increase the specificity of autophagosome-lysosome fusion events21. Lastly, in the degradation phase, autophagic cargo are degraded by lysosomal hydrolases18. This degraded material is then recycled through nutrient transporters and used to fuel growth of the cell22.
The dichotomous role of autophagy in cancer
Autophagy plays a dichotomous role in cancer by suppressing benign tumor growth but promoting advanced cancer growth. In the past decade, numerous research groups have established autophagy as a potential therapeutic target in cancer. However, there is some debate about whether to inhibit or induce autophagy. The rationale for inducing autophagy is based on studies which show that mice with loss of one allele of the autophagy gene Beclin 1 developed spontaneous tumors23–25. Initially this was thought to translate into patients with breast, ovarian, and prostate cancers, which are known to harbor monoallelic loss of Beclin 123, 24, 26. However, it was shown that the human homolog of the mouse Beclin1/Atg6 gene, BECN1 gene resides adjacent to BRCA1 on the same chromosome and other tumor suppressor genes. Tumorigenesis in human tumors may therefore be driven by neighboring genes lost rather than BECN1.
Many preclinical studies have demonstrated a variety of targeted therapies and DNA damaging agents that can induce autophagy, but in the majority of these studies the autophagy induced by the anticancer agent is cytoprotective, rather than cytotoxic27. Even if the goal was to induce autophagy to the point of activating cell death (what some refer to as autophagic cell death), to date there are almost no specific autophagy inducers. Most agents that induce autophagy inhibit other important process in the cell, such as mTOR signaling, or activate other stress responses such as the unfolded protein response28. Only Tat-Beclin1, a fusion peptide with an unclear mechanism of action, has been reported as a specific autophagy inducer29. These inducers could be useful in preventing the development of benign lesions such as polyps, but further work is needed to identify the best targets and chemical agents that can specifically induce autophagy.
There is mounting evidence that autophagy inhibition could be an effective approach in advanced cancer18. Genetically engineered mouse models (GEMMs) of lung cancer, pancreatic cancer and melanoma driven by either mutant RAS or BRAF in which autophagy genes were deleted have demonstrated that autophagy suppresses the growth of benign tumors, but accelerates the growth of advanced cancers30–34. This was also found in a mouse model of breast cancer35, 36.
Mouse models of tumorigenesis should not be used to understand the utility of inhibiting autophagy in the therapeutic context because autophagy genes are usually deleted in utero at the same time that oncogenes and tumor suppressor genes are altered. In patients, autophagy inhibitors will be deployed after the cancer is already formed in the adult, and likely in combination with other agents. Therefore modulating autophagy in this context may produce different results than modulating autophagy at the origin of tumorigenesis. Accumulating evidence supports that autophagy promotes resistance during cancer therapy in established tumors. This was first demonstrated in a therapeutic mouse model of lymphoma, where autophagy inhibition augmented the efficacy of chemotherapy37. Recently a complex GEMM model which allows sudden genetic suppression of autophagy by conditionally deleting ATG7 throughout the adult animal harboring a growing tumor was reported38, 39. This model is the closest model of autophagy inhibition in a cancer therapeutic context to the human clinic. In this model complete loss of autophagy in the mouse was well tolerated for months, during which time dramatic tumor shrinkage was observed. After a few months of complete genetic suppression of autophagy throughout the mouse, mice began to develop fatal neurodegeneration. Despite this fatal toxicity, collectively, these findings strongly support the use of autophagy inhibitors for cancer in clinic, and there may be a therapeutic window for potent extra-central nervous system (CNS) autophagy inhibition. Chronic autophagy inhibition, especially with agents that cross the blood brain barrier must be evaluated cautiously to balance between potency and toxicity, as autophagy plays an important role in normal cell and organismal homeostasis40.
Autophagy inhibitors for laboratory research
There are a number of tool compounds that can be used to study autophagy in the laboratory. Examples include inhibitors which block the activity Beclin-vps34 complex (3 methyladenine41–43, LY29400244, 45, and Wortmannin46, the Spautin47, 48); potent and specific VPS34 inhibitors (SAR40549–51; PIK-III52); the ULK1 inhibitor (SBI-020696553); ATG4B inhibitors (UAMC-252654; autophagin-155, NSC18505856 ); vacuolar-type H+-ATPase inhibitors (bafilomycin57, salinomycin58); lysosomal inhibitors (ROC32559, 60, VATG-02761, Mefloquine61, Verteporfin62, 63). 3-methyladenine may impact cancer cell metabolism independent of autophagy by serving as an ROS scavenger at high concentrations typically used. PI3K complex inhibitors (LY294002, Wortmannin) have activity against both class I and class III PI3K so interpretation of effects on autophagy may be difficult especially at the high doses often utilized. Spautin targets deubiquitinases that regulate the degradation of other client proteins besides BECLIN. Vps34 inhibitors target endocytic trafficking in addition to autophagy as vps34 activity is required for many of these autophagy independent trafficking events. SBI-020695 is also a potent FAK1 inhibitor. The potency of ATG4 inhibitors described in the literature thus far have been low, raising the possibility that these inhibitors also inhibit the protease activity of other cysteine proteases. There is very little in vivo evidence of efficacy published for any of the upstream autophagy inhibitors. In contrast, lysosomal inhibitors have had the most convincing in vivo activity. However the lack of a molecular target for these agents makes it even more difficult to determine their autophagy-dependent and autophagy independent effects. In summary while numerous compounds are available, concerns about off-target effects, and suitability for in vivo systems underscores the need to develop more potent, specific and translatable inhibitors of autophagy.
Autophagy inhibition in clinical trials
Despite a growing number of tool compounds that can be used to study autophagy in the laboratory, to date, no specific inhibitor that targets an autophagy protein has entered clinical trials. Hydroxychloroquine (HCQ) is the clinically available drug that could function as an autophagy inhibitor. HCQ is thought to inhibit autophagy by acting as a weak base that when trapped inside acidic cellular compartments, such as lysosomes, increases the pH of those compartments64. However numerous drugs are weak bases, and they do not function as autophagy inhibitors. Recent work completed with potent lysosomal inhibitors suggests there may actually be a molecular target for chloroquine (CQ) and HCQ (See below). Preclinical studies in tumor cell lines and animal models have shown that HCQ increases tumor cell death alone or through enhancing tumor killing in combination with targeted agents or cytotoxic chemotherapy. One study showed that with the addition of HCQ with the mTOR inhibitor temsirolimus (TEM), there was an increase in cytotoxicity against renal cell carcinoma cell lines in vitro65. Additionally, the combination of HCQ and tamoxifen (TMX) was more effective than either monotherapy in estrogen receptor-positive breast cancer cells66. HCQ treatment has been effective against pancreatic ductal adenocarcinomas tumors as well61. These are examples of preclinical papers where chloroquine was used as an anticancer agent. A pubmed search of chloroquine and cancer results in > 9000 entries, so there are too many examples to discuss here. 67.
As outlined below numerous groups have launched clinical trials with either chloroquine (CQ) or HCQ. The rationale for choosing HCQ in the majority of the studies comes from the predicted ocular toxicity when chloroquine is administered. Dose escalation with chloroquine from the standard 150 mg/day is likely to cause significant toxicity. In contrast dose escalation with HCQ has been accomplished successfully in rheumatoid arthritis68. The first phase I trials involving HCQ that included autophagy markers, combined HCQ with vorinostat (VOR), temsirolimus (TEM), temozolomide (TMZ), doxorubicin (DOX), or bortezomib in patients with refractory solid tumors, melanoma, glioblastoma multiforme (GBM) and relapsed/refractory myeloma (Table 1) 46, 61, 63, 69–75. In these trials across over 200 patients enrolled on these studies there was a < 10% grade 3–4 non hematological adverse event rate, which is surprising since each of the drugs that were combined with HCQ has significant toxicity (e.g. fatigue (VOR), mouth sores and hyperglycemia (TEM), myelosuppression(TMZ), neuropathy (BOR)) that could become dose limiting, and the population was a very sick phase I population or GBM patients. Multiple patients with melanoma, colorectal carcinoma, myeloma, and renal cell carcinoma demonstrated partial response or prolonged stable disease on these HCQ combinations; however, overall response rates were not high (detailed below). None of the combinations tested were pursued in phase II studies except VOR and HCQ (see below). These studies were important however, because they incorporated pharmacokinetic-pharmacodynamic assays that demonstrated for the first time that the highest doses of HCQ allowed by the FDA (600 mg p.o. b.i.d.) were able to produce a modest but reproducible degree of autophagy inhibition in patient tumors or surrogate tissues (peripheral blood mononuclear cells)46, 61, 63, 69–75. This allowed the launch of numerous phase II trials with more effective chemotherapy or targeted therapy backbones. Promising preliminary results for some of these trials have been presented in abstract form at meetings, including trials in colon cancer (presented by O’Hara et al ASCO 2015), pancreatic cancer (presented by Miller-Ocuin et al ASCO annual meeting 2017), and melanoma (presented by Gangadhar et al. Society of Melanoma Research 2017). Below we have provided more details about the published HCQ studies (Table 1).
Table 1.
Published Clinical trials involving HCQ
| Trial | Disease | DLT/MTD | N | Comments | Reference |
|---|---|---|---|---|---|
| Randomized double blind phase II study carmustine, radiation and chloroquine | Glioma | None | 30 | Median survival 24 v 11 months favoring chloroquine but not significant | 78 |
| Randomized Phase II trial of chloroquine and whole brain radiation | No-n-small cell lung cancer; breast cancer | None | 73 | Chloroquine significantly improved CNS progression-free survival | 80 |
| Phase I erlotinib + HCQ | Prior EGFR inhibitor-treated non small cell lung cancer | None | 35 | Well tolerated, erlotinib + HCQ: 5% response rate | 81 |
| Phase II HCQ | Pancreas ca | Grade 3 lymhopenia, transaminitis | 20 | 2/19 evaluable with stable disease | 71 |
| Phase I/II temozolomide/radiation + HCQ | Glioma | Myelo-suppression at 800 mg 600 mg MTD | 92 | MTD was lower than other tirals. No survival benefit in a9 center trial | 76 |
| Phase I bortezomib + HCQ | Myeloma | None | 30 | Responses seen in bortezomib naïve patients | 73 |
| Phase I temozolomide + HCQ | Solid tumors, Melanoma expansion | None | 40 | PK-PD established dose dependent autophagy inhibition | 75 |
| Phase I temsirolimus + HCQ | Solid Tumors, melanoma expansion | None | 35 | Serial PET scans, tumor biopsies; 72% stable disease rate in BRAF WT melanoma | 74 |
| Phase I vorinostat + HCQ | Solid tumors, | Grade 3 fatigue at 800 mg 600 mg MTD | 24 | Tumor biopsies; Clinical activity | 77 |
| Phase IB vorinostat | Colon cancer | None | 19 | 5/19 patients stable disease | 82 |
| Pilot and phase I cyclophosphamide, dexamethasone, rapamycin+ HCQ | Myeloma | Grade 3 diarrhea, grade 4 thrombocytopenia HCQ 1200 mg daily; MTD HCQ 800 mg daily | 18 | 29% response rate at MTD | 83 |
| Phase I/II neoadjuvant gemcitabine + HCQ | Pancreatic cancer | None | 35 | 61% CA19-9 decrease 70% R0 resection; modulation of autophagy correlated with disease free survival | 84 |
Randomized phase II trial of temozolomide, radiation and chloroquine in malignant glioma
In a trial conducted in Mexico, 30 patients with resected glioma were randomized to standard temozolomide and radiation with placebo or chloroquine 150 mg/day76. No pharmacodynamic markers were assessed and adverse events were not recorded. A follow-up retrospective study in 41 additional patients treated with carmustine and CQ compared to 82 patients treated with carmustine alone reproduced the survival curves of the original randomized study77.
Randomized phase II trial of whole brain radiation with or without chloroquine
73 patients with non-small cell lung cancer or breast cancer with brain metastases appropriate for whole brain radiation were randomized to receive chloroquine 150 mg daily for 28 days or a matching placebo along with whole brain radiation. Patients treated with chloroquine had a 1 year brain metastases progression free survival rate of 83% versus 55% for the placebo treated arm. This benefit was not associated with a difference in response rate or overall survival.78,
Phase I trial of erlotinib and HCQ in EGFR inhibitor treated non-small cell lung cancer
A phase I dose escalation of either HCQ alone or HCQ and erlotinib was conducted in EGFR mutant non-small cell lung cancer patients who had previously benefited temporarily from EGFR inhibitor therapy. The HCQ alone arm was closed early due to lack of accrual. The combination of erlotinib and HCQ 1000 mg daily was well tolerated. One out of 19 patients had a partial response and 4 patients had stable disease79.
HCQ alone in refractory pancreatic cancer
The safety and activity of single agent HCQ in 20 patients with metastatic pancreatic cancer that did not respond to standard chemotherapy was assessed in a phase II study69. This trial included ten patients treated twice daily with 400 mg and the other ten patients with 600 mg of HCQ. Of note these patients were mostly Eastern Cooperative Oncology Group (ECOG) performance status 1–2 (restricted - minimal activity), reflecting a sick population of patients. Out of the 20 patients involved, only 2 patients did not experience progressive disease at 2 months69. Median progression-free-survival (PFS) was 46.5 days and overall survival was 69 days. One patient experienced grade 3–4 lymphopenia and one had grade 3–4 transaminitis69. Overall this study demonstrated no activity for single agent HCQ in ECOG PS 1–2 patients with treatment refractory pancreatic cancer.
Phase I trial of HCQ and temsirolimus in solid tumors with expansion in melanoma patients
This dose-escalation study included 27 patients with advanced solid malignancies followed by a cohort expansion at the top dose level in 12 patients with metastatic melanoma. 72. Overall the combination of HCQ and TEM was well tolerated with almost no Grade 3–4 toxicities. Common Grade 1 and 2 toxicities included anorexia, fatigue, and nausea. This study demonstrated that an effective and safe dose for this combination was 600 mg of HCQ twice daily in combination with 25 mg of TEM weekly. Fourteen of 21 patients in the dose escalation achieved stable disease. In melanoma patients including the dose escalation and disease specific cohort expansion 14/19 (74%) patients achieved stable disease. The median PFS in 13 melanoma patients treated with HCQ 600 mg twice daily and TEM on the disease specific cohort expansion was 3.5 months72.
Phase I trial of HCQ and bortezomib in multiple myeloma
The effects of HCQ and bortezomib were studied in 25 patients with relapsed and refractory multiple myeloma71. No dose limiting toxicities were observed. Lack of neurotoxicity exacerbation with HCQ was an important finding in this study. A recommended phase II HCQ dose was determined to be 600 mg twice daily in addition to standard doses of bortezomib. Partial response was observed in 3/22 patients, 3/22 had minor responses, and 10/22 had a period of stable disease. However, given the patients with response were bortezomib-naïve, this signal of activity was not deemed enough to warrant phase II study71.
HCQ and vorinostat in solid tumors with expansion in patients with colon cancer
Another phase I clinical trial studied the combination HCQ with VOR75. Twenty four evaluable patients with refractory solid tumors were treated on the dose escalation portion of the study which identified 600 mg daily HCQ and 400 mg daily VOR as the recommended phase II doses for this regimen. The dose limiting toxicities encountered in this study were fatigue and gastrointestinal disturbances75.. A phase I dose expansion study in 19 colorectal cancer patients found that the combination of VOR and HCQ produced a 2.9-month PFS in patients refractory to standard chemotherapy included regorafenib, and was tolerable80. Five of 19 patients experienced prolonged stable disease. This finding justified the launch of an ongoing randomized phase II study of VOR + HCQ versus regorafenib (NCT02316340).
Adjuvant HCQ and temozolomide in glioma
In a phase I/II clinical trial, HCQ in combination with radiation therapy and concurrent and adjuvant TMZ was tested in patients with newly diagnosed gliobastoma multiforme74. In this study patients treated with standard low dose continuous TMZ and HCQ 400 mg p.o. b.i.d. experienced Grade 3 and 4 neutropenia and thrombocytopenia74. A maximal tolerated dose (MTD) of 600mg/day HCQ was established and a phase II clinical trial was conducted through a national brain tumor consortium. PK-PD studies showed only modest treatment associated accumulation of autophagic vesicles, a surrogate for autophagy inhibition, at HCQ 600 mg/day. No significant improvement in overall survival of patients with malignant glioma was observed74. Concern was raised in regards to the dose limiting myelosuppression observed, that prevented adequate autophagy inhibition to produce efficacy.
A phase I study of dose intense temozolomide in patients with solid tumors
In this phase I clinical trial of HCQ and TMZ 40 patients with the vast majority being melanoma patients were enrolled73. For 7/14 days, these patients were treated orally with 200–1200 mg of HCQ daily with dose-intense oral TMZ 150 mg/m2. Unlike the low dose continuous TMZ study in glioma, the dose-intense TMZ and HCQ combination was well-tolerated with no recurrent dose-limiting toxicities. Grade 2 fatigue, anorexia, nausea, constipation, and diarrhea was occasionally observed. Clinical benefit was observed in 9/22 (41%) at the highest dose level of HCQ (600 mg bid) with refractory BRAF-wild type melanoma had a near complete response or prolonged stable disease73.
Phase I trial of HCQ, rapamycin, cyclophosphamide, and dexamethasone in multiple myeloma
A safety pilot and phase I study was conducted to determine the MTD of HCQ and safety of the 4-drug combinations81. Patients with relapsed or refractory multiple myeloma were eligible for this study if they received prior treatment with lenalidomide or bortezomib. All subjects received cyclophosphamide and dexamethasone. In the pilot study, 3 subjects were treated with 12 mg loading of rapamycin followed by 4 mg daily dose of rapamycin for 5 further days64. Also in the pilot study, 3 subjects received 800 mg of HCQ daily and had no dose limiting toxicities (DLT). Two DLTs including grade 3 diarrhea and grade 4 thrombocytopenia occurred at 1200 mg HCQ daily. Dose de-escalation to 800 mg HCQ daily resulted in one DLT (grade 4 thrombocytopenia). Therefore, 800 mg of HCQ was determined to be the MTD when combined with rapamycin, cyclophosphamide and dexamethasone64.
Phase I neoadjuvant study of HCQ and gemcitabine in pancreatic cancer
A phase I/II trial of 35 patients examined the neoadjuvant treatment with HCQ and gemcitabine in patients with borderline resectable pancreatic adenocarcinomas82. 1200 mg/day of HCQ was taken for 31 days up until the day of surgery combined with two doses of a fixed-dose 1500 mg/m2 gemcitabine69. DLT or grade 4/5 events did not occur during this trial. 19/35 patients had a decrease in surrogate biomarker response (CA 19-9) and 29/35 patients underwent surgical resection as scheduled69. This trial demonstrated that HCQ with gemcitabine is safe and tolerable, and showed encouraging activity in the neoadjuvant setting. A randomized phase II trial with gemcitabine, abraxane, with or without HCQ in resectable pancreatic cancer patients has been launched (NCT01978184), and a randomized phase II study of gemcitabine, abraxane, with or without HCQ in the metastatic setting has been launched (NCT01506973).
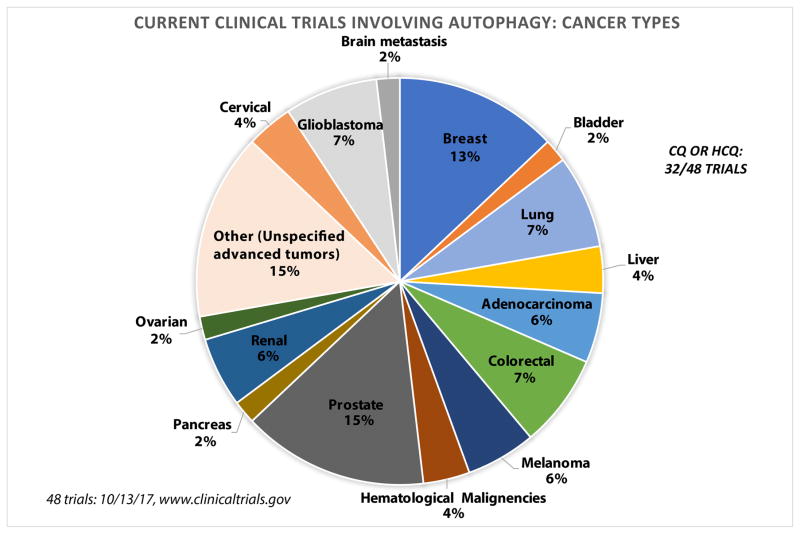
Overall, HCQ alone and in combination therapy is being used in clinical trials across a range of cancers (Figure 1). Examples of recently completed and actively recruiting clinical trials which may be reporting results soon are organized in Table 2. Even though HCQ provided the clinical and basic research communities insight into the use of autophagy inhibitors in clinic, the exact mode of action and potency remain to be issues with HCQ. Recent progress into a potential molecular target has been made with more potent autophagy inhibitors in the laboratory (see below). Another issue is that there are no validated biomarkers to identify tumors most likely to respond to HCQ studies. A recent paper identified patterns of expression at the protein level of aldehyde dehydrogenase1A1 (ALDH1A1) and helicase-like transcription factor (HLTF), to predict HCQ sensitivity and resistance83. This 2 gene signature can distinguish HCQ sensitive from HCQ resistant tumors in vitro. Analysis of The Cancer Genome Atlas (TCGA) demonstrated that the ALDH1A1/HLTF HCQ-S pattern is prevalent in a number of cancer83.
Figure 1.
Current clinical trials targeting autophagy in cancer. The pie chart shows the breakdown of which cancers have clinical trials targeting autophagy registered at www.clinicaltrials.gov.
Table 2.
Examples of ongoing or recently completed HCQ clinical trials
| Condition | Treatment | ClinicalTrials.gov Identifier | Phase | Recruiting |
|---|---|---|---|---|
| Colorectal Cancer | vorinostat + HCQ versus regorafenib | NCT02316340 | II | yes |
| Breast Cancer (Adjuvant ) | everolimus + HCQ | NCT03032406 | II | yes |
| Pancreatic Cancer | gemcitabine + nab-paclitaxel +/− HCQ | NCT01978184 | II | yes |
| Solid Tumor | HCQ | NCT03015324 | I | yes |
| Leukemia, Acute Myelogenous | mitoxantrone + etoposide + HCQ | NCT02631252 | I | yes |
| Metastatic Renal Cell Carcinoma | HCQ + IL-2 | NCT01550367 | I/II | yes |
| Advanced BRAF Mutant Melanoma | dabrafenib + trametinib + HCQ | NCT02257424 | I/II | yes |
| Hepatocellular Carcinoma | TACE + HCQ | NCT02013778 | I/II | yes |
| Prostate Cancer | HCQ | NCT00726596 | II | yes |
| Pancreatic Cancer | capecitabine + Proton or Photon Radiation Therapy + HCQ | NCT01494155 | II | no |
| Advanced Cancers | sirolimus + HCQ Vorinstat+ HCQ | NCT01266057 | I | no |
| Rectal, Colon Cancer; Metastasis; Adenocarcinoma | HCQ + oxaliplatin + leucovorin + 5-fluorouracil + bevacizumab | NCT01206530 | I/II | no |
| Metastatic Clear Cell Renal Cell Carcinoma | HCQ + RAD001 | NCT01510119 | I/II | no |
Autophagy in cancer immunity
The paradigm changing effect of immune checkpoint inhibitors and chimeric antigen receptor T cells has demonstrated the need to consider the immune system when targeting cancer pathways. Early studies demonstrated that loss of autophagy genes promotes tumor inflammation84, 85. In this context conflicting reports on the role of autophagy in tumor immunity continue to cloud the picture of whether or not anti-autophagy therapy will promote or suppress tumor immunity. Inhibiting autophagy may blunt cross-priming of tumor-specific CD8+T cells86. Additionally, autophagy has been shown to play an important role in effector and memory differentiation in T cell activation87. Autophagy has been shown to dictate the immunogenicity of cell death in tumors88. The absence of appropriate response to autophagy induction in tumor cells could potentially predict resistance to immune checkpoint blockers (ICB) therapies in human cancers89. Michaud et al demonstrated that autophagy was dispensable for chemotherapy-induced cell death but required for its immunogenicity since only autophagy competent cells attracted dendritic cells and T lymphocytes into the tumor. 90. These authors proposed that autophagy is essential for immunogenic release of ATP from dying cells, and increased extracellular ATP concentrations improve the efficacy of antineoplastic chemotherapies when autophagy is disabled90.
To the contrary, emerging data indicates that autophagy limits immune cell-mediated cytotoxicity, and therefore inhibiting autophagy would enhance antitumor immunity. During hypoxia, autophagy inhibits T cell mediated cytotoxicity in lung cancer cells91. Additionally, lysosomal exocytosis enhances anti-melanoma therapy91. It was found that autophagy inhibition also enhances cell death from natural killer (NK) cells92. Within the same study, deletion of Beclin1 caused influx of immune cells into the tumor microenvironment92. Emerging data also suggests autophagy deficiency in host cells or tumor cells promotes T cell mediated antitumor immunity36, 93. Therapeutic translation of the findings is ongoing. For instance studies designed to determine if combining an anti-PD-L1 or anti-PD-1 with autophagy inhibitor may produce a synergistic effect of cancer immunotherapy.
Novel lysosomal inhibitors
Lys05, a water-soluble dimeric chloroquine94, was reported as a novel lysosomal autophagy inhibitor that is 10-fold more potent than HCQ. In vivo studies of Lys05 alone95 and in combination with BRAF inhibition treatment96 demonstrated its single agent and in combination efficacy. The efficacy of Lys05 has also been demonstrated in a range of in vitro and in vivo tumor models97–100. Recently, our group developed an even more potent lysosomal inhibitor, DQ661, a derivative of quinacrine101. Leveraging its potency and increased localization to the lysosome, DQ661 was used to pull down its previously unknown molecular target palmitoyl-protein thioesterase 1 (PPT1). DQ661 has in vivo activity in models of melanoma, pancreatic and colorectal cancer 101. Current efforts are ongoing to develop Lys05 and or DQ661 derivatives into clinical drug candidates.
In conclusion, autophagy inhibition is gaining traction as a potentially new therapeutic approach in cancer. The field looks forward to clinical development of novel agents that target upstream components of autophagy (e.g. ULK1, ATG7, ATG4) or lysosomal agents. Efforts are underway to determine if PPT1 is the molecular target of dimeric chloroquines like Lys05, and even HCQ itself. This would change the paradigm of considering these agents as simply weak bases that accumulate and disrupt the lysosome, and instead they would be considered targeted therapies. More research is needed to understand the role of autophagy inhibition in the context of immunotherapy. Additionally, identifying tumor subsets that are susceptible to autophagy inhibition would enhance translational research outcomes. Substantial progress has been made both in the lab and the clinic to credential autophagy inhibition as a therapeutic strategy, but more work remains to determine how autophagy can be exploited to benefit clinical outcomes.
Summary box.
Overview of targeting autophagy in cancer
New fundamental mechanistic insights into autophagy uncover new druggable targets
Mouse models of tumorigenesis indicate autophagy suppresses benign tumors but promotes advanced cancer
More tool compounds are being developed to inhibit autophagy in cancer
Hydroxychloroquine is the first drug to be repurposed as an autophagy inhibitor in multiple cancer trials.
Phase I trials have demonstrated safety and target engagement biologically
Phase II trials may show promise in specific cancers
More potent and specific autophagy inhibitors and autophagy inducers are being developed.
Acknowledgments
Funding: This work was entirely supported by NIH grants R01CA169134.
Footnotes
Conflict of Interest Statement: None
Author Contributions: Conceptualization: AO, RO, RA; writing and editing: AO, MD, RO, RA; figure preparation: MD; overall supervision: RA
References
- 1.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Klionsky DJ. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci U S A. 2017;114:201–205. doi: 10.1073/pnas.1619876114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLendon PM, Ferguson BS, Osinska H, et al. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci U S A. 2014;111:E5178–5186. doi: 10.1073/pnas.1415589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M, Hong MJ, Sun H, et al. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat Med. 2014;20:503–510. doi: 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarparanta J, Garcia-Macia M, Singh R. Autophagy and Mitochondria in Obesity and Type 2 Diabetes. Curr Diabetes Rev. 2017;13:352–369. doi: 10.2174/1573399812666160217122530. [DOI] [PubMed] [Google Scholar]
- 7.Mao Y, Yu F, Wang J, Guo C, Fan X. Autophagy: a new target for nonalcoholic fatty liver disease therapy. Hepat Med. 2016;8:27–37. doi: 10.2147/HMER.S98120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amaravadi RK, Lippincott-Schwartz J, Yin XM, et al. Principles and Current Strategies for Targeting Autophagy for Cancer Treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9:424–425. doi: 10.4161/auto.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson MI, Dooley HC, Tooze SA. WIPI2b and Atg16L1: setting the stage for autophagosome formation. Biochem Soc Trans. 2014;42:1327–1334. doi: 10.1042/BST20140177. [DOI] [PubMed] [Google Scholar]
- 16.Kraya AA, Piao S, Xu X, et al. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy. 2015;11:60–74. doi: 10.4161/15548627.2014.984273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 18.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 19.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- 20.Takats S, Pircs K, Nagy P, et al. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Miao G, Xue X, et al. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol Cell. 2016;63:781–795. doi: 10.1016/j.molcel.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Kimmelman AC, White E. Autophagy and Tumor Metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Y, An Z, Zou Z, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife. 2015:4. doi: 10.7554/eLife.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Shoji-Kawata S, Sumpter RM, Jr, et al. Autosis is a Na+, K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 28.Kapuy O, Vinod PK, Banhegyi G. mTOR inhibition increases cell viability via autophagy induction during endoplasmic reticulum stress - An experimental and modeling study. FEBS Open Bio. 2014;4:704–713. doi: 10.1016/j.fob.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoji-Kawata S, Sumpter R, Leveno M, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strohecker AM, White E. Targeting mitochondrial metabolism by inhibiting autophagy in BRAF-driven cancers. Cancer Discov. 2014;4:766–772. doi: 10.1158/2159-8290.CD-14-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Wang X, Contino G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang A, Kimmelman AC. Inhibition of autophagy attenuates pancreatic cancer growth independent of TP53/TRP53 status. Autophagy. 2014;10:1683–1684. doi: 10.4161/auto.29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JY, Teng X, Laddha SV, et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo JY, White E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy. 2013;9:1636–1638. doi: 10.4161/auto.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Wang C, Yeo S, et al. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 2016;30:856–869. doi: 10.1101/gad.276428.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karsli-Uzunbas G, Guo JY, Price S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amaravadi R, Debnath J. Mouse models address key concerns regarding autophagy inhibition in cancer therapy. Cancer Discov. 2014;4:873–875. doi: 10.1158/2159-8290.CD-14-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancias JD, Kimmelman AC. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J Mol Biol. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo XL, Li D, Hu F, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–179. doi: 10.1016/j.canlet.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Yu H, Qin H, et al. Inhibition of autophagy enhances cisplatin cytotoxicity through endoplasmic reticulum stress in human cervical cancer cells. Cancer Lett. 2012;314:232–243. doi: 10.1016/j.canlet.2011.09.034. [DOI] [PubMed] [Google Scholar]
- 43.Lamoureux F, Thomas C, Crafter C, et al. Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor AZD5363. Clin Cancer Res. 2013;19:833–844. doi: 10.1158/1078-0432.CCR-12-3114. [DOI] [PubMed] [Google Scholar]
- 44.Borowa-Mazgaj B, Skwarska A, Augustin E, Radomińska-Pandya A, Mazerska Z. Antitumor DNA-Damaging C-1748 is a New Inhibitor of Autophagy that Triggers Apoptosis in Human Pancreatic Cancer Cell Lines. The FASEB Journal. 2016;30:1062–1065. [Google Scholar]
- 45.Avni D, Glucksam Y, Zor T. The phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 modulates cytokine expression in macrophages via p50 nuclear factor kappaB inhibition, in a PI3K-independent mechanism. Biochem Pharmacol. 2012;83:106–114. doi: 10.1016/j.bcp.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 46.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Xia H, Kim M, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao S, Li S, Qin Y, et al. Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int J Oncol. 2014;44:1661–1668. doi: 10.3892/ijo.2014.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasquier B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy. 2015;11:725–726. doi: 10.1080/15548627.2015.1033601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 51.Pasquier B, El-Ahmad Y, Filoche-Romme B, et al. Discovery of (2S)-8-[(3R)-3-methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: a novel potent and selective inhibitor of Vps34 for the treatment of solid tumors. J Med Chem. 2015;58:376–400. doi: 10.1021/jm5013352. [DOI] [PubMed] [Google Scholar]
- 52.Dowdle WE, Nyfeler B, Nagel J, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 53.Egan DF, Chun MG, Vamos M, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurdi A, Cleenewerck M, Vangestel C, et al. ATG4B inhibitors with a benzotropolone core structure block autophagy and augment efficiency of chemotherapy in mice. Biochem Pharmacol. 2017;138:150–162. doi: 10.1016/j.bcp.2017.06.119. [DOI] [PubMed] [Google Scholar]
- 55.Qiu Z, Kuhn B, Aebi J, et al. Discovery of Fluoromethylketone-Based Peptidomimetics as Covalent ATG4B (Autophagin-1) Inhibitors. ACS Med Chem Lett. 2016;7:802–806. doi: 10.1021/acsmedchemlett.6b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akin D, Wang SK, Habibzadegah-Tari P, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goulielmaki M, Koustas E, Moysidou E, et al. BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Oncotarget. 2016;7:9188–9221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endo S, Nakata K, Sagara A, et al. Autophagy inhibition enhances antiproliferative effect of salinomycin in pancreatic cancer cells. Pancreatology. 2017 doi: 10.1016/j.pan.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Carew JS, Espitia CM, Zhao W, et al. Disruption of Autophagic Degradation with ROC-325 Antagonizes Renal Cell Carcinoma Pathogenesis. Clin Cancer Res. 2017;23:2869–2879. doi: 10.1158/1078-0432.CCR-16-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carew JS, Nawrocki ST. Drain the lysosome: Development of the novel orally available autophagy inhibitor ROC-325. Autophagy. 2017;13:765–766. doi: 10.1080/15548627.2017.1280222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi TT, Yu XX, Yan LJ, Xiao HT. Research progress of hydroxychloroquine and autophagy inhibitors on cancer. Cancer Chemother Pharmacol. 2017;79:287–294. doi: 10.1007/s00280-016-3197-1. [DOI] [PubMed] [Google Scholar]
- 62.Donohue E, Thomas A, Maurer N, et al. The autophagy inhibitor verteporfin moderately enhances the antitumor activity of gemcitabine in a pancreatic ductal adenocarcinoma model. J Cancer. 2013;4:585–596. doi: 10.7150/jca.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donohue E, Tovey A, Vogl AW, et al. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem. 2011;286:7290–7300. doi: 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan Y, Gao Y, Chen L, et al. Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin Cancer Res. 2011;17:3248–3258. doi: 10.1158/1078-0432.CCR-10-0890. [DOI] [PubMed] [Google Scholar]
- 65.Lee HO, Mustafa A, Hudes GR, Kruger WD. Hydroxychloroquine Destabilizes Phospho-S6 in Human Renal Carcinoma Cells. PLoS One. 2015;10:e0131464. doi: 10.1371/journal.pone.0131464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cook KL, Warri A, Soto-Pantoja DR, et al. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin Cancer Res. 2014;20:3222–3232. doi: 10.1158/1078-0432.CCR-13-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene. 2016;35:1–11. doi: 10.1038/onc.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carmichael SJ, Charles B, Tett SE. Population pharmacokinetics of hydroxychloroquine in patients with rheumatoid arthritis. Ther Drug Monit. 2003;25:671–681. doi: 10.1097/00007691-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 69.Wolpin BM, Rubinson DA, Wang X, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10:1415–1425. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogl DT, Stadtmauer EA, Tan KS, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rangwala R, Chang YC, Hu J, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rangwala R, Leone R, Chang YC, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenfeld MR, Ye X, Supko JG, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahalingam D, Mita M, Sarantopoulos J, et al. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10:1403–1414. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 77.Briceno E, Calderon A, Sotelo J. Institutional experience with chloroquine as an adjuvant to the therapy for glioblastoma multiforme. Surg Neurol. 2007;67:388–391. doi: 10.1016/j.surneu.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 78.Rojas-Puentes LL, Gonzalez-Pinedo M, Crismatt A, et al. Phase II randomized, double-blind, placebo-controlled study of whole-brain irradiation with concomitant chloroquine for brain metastases. Radiat Oncol. 2013;8:209. doi: 10.1186/1748-717X-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goldberg SB, Supko JG, Neal JW, et al. A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. J Thorac Oncol. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel S, Hurez V, Nawrocki ST, et al. Vorinostat and hydroxychloroquine improve immunity and inhibit autophagy in metastatic colorectal cancer. Oncotarget. 2016;7:59087–59097. doi: 10.18632/oncotarget.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scott EC, Maziarz RT, Spurgeon SE, et al. Double autophagy stimulation using chemotherapy and mTOR inhibition combined with hydroxychloroquine for autophagy modulation in patients with relapsed or refractory multiple myeloma. Haematologica. 2017;102:e261–e265. doi: 10.3324/haematol.2016.162321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boone BA, Bahary N, Zureikat AH, et al. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol. 2015;22:4402–4410. doi: 10.1245/s10434-015-4566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piao S, Ojha R, Rebecca VW, et al. ALDH1A1 and HLTF modulate the activity of lysosomal autophagy inhibitors in cancer cells. Autophagy. 2017:1–16. doi: 10.1080/15548627.2017.1377377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo JY, Karsli-Uzunbas G, Mathew R, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16:991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- 87.Xu X, Araki K, Li S, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 2015;28:690–714. doi: 10.1016/j.ccell.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 89.Pitt JM, Vetizou M, Daillere R, et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity. 2016;44:1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 90.Michaud M, Martins I, Sukkurwala AQ, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 91.Rao S, Yang H, Penninger JM, Kroemer G. Autophagy in non-small cell lung carcinogenesis: A positive regulator of antitumor immunosurveillance. Autophagy. 2014;10:529–531. doi: 10.4161/auto.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mgrditchian T, Arakelian T, Paggetti J, et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc Natl Acad Sci U S A. 2017;114:E9271–e9279. doi: 10.1073/pnas.1703921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levy J, Cacheux W, Bara MA, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17:1062–1073. doi: 10.1038/ncb3206. [DOI] [PubMed] [Google Scholar]
- 94.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McAfee Q, Zhang Z, Samanta A, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma XH, Piao SF, Dey S, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DeVorkin L, Hattersley M, Kim P, et al. Autophagy Inhibition Enhances Sunitinib Efficacy in Clear Cell Ovarian Carcinoma. Mol Cancer Res. 2017;15:250–258. doi: 10.1158/1541-7786.MCR-16-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gade TPF, Tucker E, Nakazawa MS, et al. Ischemia Induces Quiescence and Autophagy Dependence in Hepatocellular Carcinoma. Radiology. 2017;283:702–710. doi: 10.1148/radiol.2017160728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frazier JP, Bertout JA, Kerwin WS, et al. Multidrug Analyses in Patients Distinguish Efficacious Cancer Agents Based on Both Tumor Cell Killing and Immunomodulation. Cancer Res. 2017;77:2869–2880. doi: 10.1158/0008-5472.CAN-17-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vijayaraghavan S, Karakas C, Doostan I, et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun. 2017;8:15916. doi: 10.1038/ncomms15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rebecca VW, Nicastri MC, McLaughlin N, et al. A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]