Abstract
The incidence of type 1 diabetes (T1D) is determined by both genetic and environmental factors. In recent years, the gut microbiota have been identified to be an important environmental factor that could modify diabetes susceptibility. We have previously shown that Myeloid differentiation primary response gene 88 (MyD88), a major adaptor protein downstream of most innate immune Toll-like receptor (TLR) signaling, is important for mediating diabetes susceptibility in the non-obese diabetic (NOD) mouse model of human T1D. Here we report the role of TIR-domain-containing adapter-inducing interferon-β (TRIF) in T1D development, as TRIF is an important adaptor protein downstream of TLR3 and TLR4 signaling. We found that TRIF-deficient (TRIF−/−) NOD mice were protected from development of diabetes, but only when housed with TRIF-deficient (TRIF−/−) NOD mice. When housed with TRIF-sufficient wild type (WT, i.e., TRIF+/+) NOD mice, the mice developed diabetes. We further investigated the gut microbiota as a potential cause for the altered diabetes development. Interestingly, TRIF−/−NOD mice had a different microbiota composition compared to WT NOD mice, only if they were housed with TRIF−/−NOD mice. However, the composition of gut microbiota in the TRIF−/−NOD mice was indistinguishable from WT NOD mice, if they were housed with WT NOD mice. The difference in the gut microbiota in TRIF−/−NOD mice, due to cohousing, accorded with the diabetes development in TRIF−/−NOD mice. Comparing the gut microbiota in TRIF−/− and WT NOD mice, we identified changes in percentage of Sutterella, Rikenella and Turicibacter species. Moreover, bacteria from WT NOD mice induced significantly stronger inflammatory immune responses in vitro compared to those from TRIF−/−NOD mice. Further immunological analysis revealed impaired function of dendritic cells and reduced T cell activation and proliferation in TRIF−/−NOD mice. Our data show that TRIF-deficiency protects NOD mice from diabetes development through alteration of the gut microbiota and reduced immune cell activation; however, that protection is over-ridden upon exposure to WT NOD bacteria. Therefore exposure to different microbiota can modify disease susceptibility determined by genetic factors related to innate immunity.
1. Introduction
Type 1 diabetes (T1D) is a T cell-mediated disease, which results in the progressive destruction of insulin-producing β cells in the pancreatic islets of Langerhans. Over the past few decades the incidence of T1D has risen and continues to do so [1, 2] at a rate too fast to be attributed solely to genetic changes. Increasing evidence suggests that environmental factors contribute to the rise of T1D, and gut microbiota are an important modifier of diabetes susceptibility [3–6]. The intestine has the largest surface area that is in direct contact with the environment, providing a site for many immune-microbe interactions. While some of the microbes are mutualistic and beneficial to the host, others can be detrimental. This balance (homeostasis) between host and microbes is critical for maintaining good health.
Toll-like receptors (TLRs) are pattern-recognition receptors, which play a crucial role in initiating the innate immune response to microbes to both fight infection and maintain homeostasis. There are many different TLRs, all of which bind to different pathogen associated molecular patterns. TLR4 binds lipopolysaccharide (a major cell wall component of gram-negative bacteria) [7] and this leads to the activation of two major adaptor molecules that mediate downstream TLR signaling – the myeloid differentiation primary response gene 88 (MyD88) and the TIR-domain-containing adapter-inducing interferon-β (TRIF). While most TLRs signal through MyD88, TLR3 that detects viruses and TLR4 can both signal through TRIF [8].
Recent studies suggest that the gut microbiome can be linked to various diseases including autoimmune diseases, cancer and inflammatory bowel disease [9]. We previously reported that T1D susceptibility was modified by the gut microbiota in MyD88-deficient non-obese diabetic (NOD) mice [3]. Further, studies have shown that different microbial-sensing TLRs have different effects on the development of T1D [10–15]. T1D development in TLR3-deficient NOD mice is similar to TLR3-sufficient NOD mice [11] but reduced in a virus-induced model system [16]. However, T1D development is increased in TLR4-deficient NOD mice [3, 12], although both TLR3 and TLR4 signal through TRIF. To investigate the role of TRIF in spontaneous autoimmune diabetes development and its effect on the gut microbiota, we generated TRIF-deficient (TRIF−/−) NOD mice. Our results showed that NOD mice are protected from diabetes development in the absence of TRIF. However, this protection from diabetes was not observed if TRIF-deficient NOD mice were housed with TRIF-sufficient mice. Furthermore, we demonstrated that the protection from disease was associated with alterations in the composition of gut microbiota. In addition to the important role that the gut microbiota play in T1D development in TRIF−/−NOD mice, TRIF deficiency also alters the function of dendritic cells (DCs) and T cells including regulatory T cells (Treg). Interestingly, the effect of TRIF deficiency on the immune cells can be overcome when introducing gut microbiota from TRIF-sufficient mice, illustrating the importance of both genes and environmental interactions in mediating diabetes susceptibility.
2. Materials and Methods
2.1. Mice
NOD/Caj mice were originally obtained from the Jackson Laboratory and have been maintained at Yale University for over 30 yrs. TRIF-deficient (TRIF−/−) NOD mice were generated by backcrossing TRIF−/−C57BL/6 mice (C57BL/6J-Ticam1Lps2/J purchased from the Jackson Laboratory) to our NOD/Caj mice, which are wild type (+/+) for the TRIF gene. The progeny of this breeding was designated as N1, which are all heterozygous, +/−, for the TRIF gene. N1mice were then bred again with our NOD/Caj mice to generate N2, which have progeny heterozygous for the TRIF knockout gene (+/−) or wild type (+/+). We selected N2 mice with TRIF+/− genotype to further backcross to our NOD/Caj mice to obtain N3. We repeated this back cross to our NOD/Caj mice for over 10 generations. We then intercrossed the N11 mice with the TRIF+/− genotype (N11 × N11) to generate the progeny with TRIF+/+, TRIF+/− and TRIF−/− genotypes, designated as TRIF+/+ NOD, TRIF+/− NOD and TRIF−/− NOD mice. To ensure the NOD genetic purity, two randomly selected TRIF−/− NOD mice, which were analyzed for SNPs throughout the mouse genome (DartMouse, Lebanon, NH). SNP genome analysis revealed that there was no evidence of non-NOD genomic regions except for the targeted TRIF locus. TRIF+/+NOD and TRIF−/−NOD mice were used in the study and for cohousing experiments, TRIF+/+ NOD and TRIF−/−NOD mice were littermates that were generated from TRIF+/− NOD breeding and cohoused in a 1:1 ratio. Recombinase-activating-gene deficient NOD (Rag−/−NOD) mice or severe-combined-immunodeficient NOD (NOD.scid) mice and BDC2.5 NOD mice were originally obtained from the Jackson Laboratory. All mice used in this study were kept under specific pathogen–free conditions, in a 12-hour dark/light cycle and housed in individually-ventilated filter cages with free access to water and autoclaved food at the Yale University animal facility. The use of the animals in this study was approved by the Yale University Institutional Animal Care and Use Committee.
2.2. Antibodies and Reagents
All the fluorochrome-conjugated mAbs used in this study were purchased from Biolegend or eBioscience unless otherwise stated. Supernatants from the hybridomas (2C11, GK1.5, TIB105, 10.2.16, 2.4G2) producing mAbs (to CD3, CD4, CD8, MHC-class II I-Ag7 and Fc receptor, respectively), used for cell purification or stimulation, were generously provided by the late Charles Janeway Jr. (Yale University). Magnetic beads conjugated with goat anti-mouse IgG, goat anti-mouse IgM or goat anti-rat IgG were purchased from QIAGEN. RPMI-1640 medium and heat-inactivated FCS were purchased from Invitrogen and Gemini, respectively.
2.3. Diabetes Assessment
Female wild-type (WT) NOD and TRIF−/−NOD mice were co-housed (littermates) or non-cohoused for observation of diabetes development until 30 weeks of age. Mice were tested for glycosuria weekly and diabetes was confirmed by blood glucose ≥250 mg/dl (13.9 mmol/L).
2.4. Adoptive transfer
Total splenocytes (10×106 cells/mouse) from diabetic female donor mice (WT NOD or TRIF−/−NOD mice) were injected (i.v.) into 4–5-week-old immune-deficient female NOD (Rag−/−NOD or NOD.scid mice) or irradiated (600 rads) 4–5-week-old female WT NOD or TRIF−/−NOD mice. The recipient mice were monitored for the development of diabetes until 120 days or 25 weeks post-injection.
2.5. Extraction of gut bacterial DNA
Fecal samples were collected fresh from 3 month-old female cohoused or non-cohoused WT NOD and TRIF−/−NOD mice. Bacterial DNA was extracted as previously described [17]. Briefly, the fecal samples (~15–25 mg/mouse) were re-suspended in 300 μl Tris-EDTA (TE) and incubated for one hour at 37°C in the presence of 7.5 μl SDS (0.5%) and 3 μl Proteinase K (20 mg/ml). One volume of phenol:chloroform:isoamyl alcohol (25:24:1), 200 μl of 20% SDS and 0.3 g of zirconium/silica beads (0.1 mm, Biospec Inc) were added and samples were mixed with a Mini-bead-beater for 2 mins. The sample was then mixed with 820 μl of phenol:chloroform:isoamyl alcohol (25:24:1), centrifuged and the aqueous layer collected into a new tube. The bacterial DNA was precipitated with 0.6 volume of isopropanol, washed with 70% ethanol, air-dried and resuspended in 100 μl of sterile water.
2.6. 16S rRNA sequencing, microbiota classification and functional composition prediction
16S rRNA sequencing was performed as described previously [18]. Briefly, the V4 region of the bacterial 16S ribosomal gene was amplified from each DNA sample with barcoded broadly conserved bacterial primers (forward, 5′-CATGCTGCCTCCCGTAGGAGT-3′; reverse, 5′-TCAGAGTTTGATCCTGGCTCAG-3′). The PCR products were purified (QIAGEN gel extraction kit) and quantified (Nanodrop spectrophotometer), and equimolar amounts of each sample were pooled and pyrosequenced on an Ion Torrent Personal Genome Machine sequencing system (Life Technologies). The results were analyzed using the Quantitative Insights Into Microbial Ecology (QIIME) software package (version 1.8) and highly accurate Operational taxonomic unit sequences from microbial amplicon reads (UPARSE) pipeline (version 7.0). After removing the primer sequences, the sequences were de-multiplexed, quality-filtered using the Quantitative Insights into Microbial Ecology (QIIME) software, and further quality and chimera-filtered using the UPARSE pipeline algorithm [19]. Operational taxonomic units were picked with 97% identity in UPARSE pipeline. In QIIME, the Greengenes reference database was used for taxonomy assignment, which was performed at various levels using representative sequences of each operational taxonomic unit. β-diversity was calculated to compare differences between microbial community profiles and the data is shown as Principal Coordinate Analysis (PCoA). The functional composition of the bacterial communities was predicted by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) using the 16S rRNA sequencing data as outlined by Langille et al [20]. Raw sequencing data of 16S rRNA genes from NOD and TRIF−/−NOD were deposited in the NCBI Sequence Read Archive (SRA) under BIOProject ID PRJNA394782 (SRR5849873). Sequence metadata and samples can be identified using data in Supplementary Table 1.
2.7. Fecal microbiota transfer
Fecal samples from 2–4 month-old female NOD donor mice (non-diabetic) were resuspended in sterile PBS. Female TRIF−/−NOD mice (4 week-old) were gavaged weekly with 200 μl of the suspension (~108 bacteria/mouse) until 30 weeks of age or diabetes onset. Recipient mice were screened weekly for diabetes development until 30 weeks of age.
2.8. In vitro bacterial co-culture
Large intestine and spleens were harvested from 2-month-old female WT NOD or TRIF−/−NOD. Large intestines were flushed using 1×PBS and pooled from 2 mice. The large intestinal flush was then centrifuged at low speed to remove dietary material (52×g, 5 min) prior to a further centrifugation to remove tissue cells (454×g, 5 min). Bacteria were isolated by high-speed centrifugation (1876×g, 5 min) and resuspended in 1×PBS. Bacterial concentration was measured with a spectrophotometer (Bio-rad) and 107 colony forming units (cfu)/ml were used to stimulate splenocytes (1×106) overnight (16 hours) cultured in RPMI complete media; phorbol 12-myristate 13-acetate (PMA, 50 ng/ml, Sigma), Ionomycin (500 ng/ml, Sigma) and Golgi-plug (eBioscience) were added in the final 4-hour culture.
2.9. Cell Surface Staining
Briefly, splenocytes and lymph node cells from 3-month-old female WT NOD and TRIF−/−NOD mice were harvested. Single cell suspensions were generated and red blood cells were lysed from the splenocytes. 1×106 cells were then incubated with a 2.4G2 Fc-blocking antibody prior to staining with antibodies to CD4 (GK1.5), CD8α (53–6.7), CD11b (M1/70), CD11c (N418), CD19 (6D5), B220 (RA3-6B2), CD80 (16-10A1), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3) and a viability dye (all from BioLegend). FoxP3 (FJK-16S, eBioscience) was stained after surface staining following the protocol from eBioscience. The samples were analyzed on a BD LSRII flow cytometer (BD Biosciences).
2.10. Intracellular cytokine (ICC) assay
ICC was performed according to the protocol from eBioscience. Briefly, splenocytes from 3-month-old female WT NOD and TRIF−/−NOD mice were stimulated with PMA (50 ng/ml, Sigma) and ionomycin (500 ng/ml, Sigma) in the presence of Golgi-plug (eBioscience) for 4 hours. The cells were then stained with antibodies to surface markers before fixation and permeabilization. Fc receptors were blocked with 2.4G2 Fc-blocking antibody before staining with fluorochrome-labeled antibodies to detect intracellular cytokines (IL-10 (JES5-16E3), IL-6 (MP5-20F3), TNFα (MP6-XT22) and TGFβ (TW7-16B4); all from BioLegend). The samples were analyzed on a BD LSRII flow cytometer (BD Biosciences).
2.11. Cell purification
Splenic B cells, CD11b+ and CD11c+ cells were purified from 2 to 3-month-old female WT NOD and TRIF−/−NOD mice using the EasySep™ Mouse B Cell Isolation Kit, the EasySep™ Mouse CD11b Positive Selection Kit and the EasySep™ Mouse CD11c Positive Selection Kit (STEMCELL Technologies) respectively, according to the manufacturer’s instructions. Total APC were enriched by depleting CD4+ and CD8+ T cells using hybridoma supernatants (anti-CD4, GK1.5 and anti-CD8, T1B105) and magnetic bead (conjugated with goat anti-rat IgG) separation as described previously [21]. BDC2.5 CD4+ T cells were purified from spleens of BDC2.5 NOD mice by depleting CD8+ T cells, MHC class II+ cells (anti-MHC II, 10.2.16) and B cells (anti-mouse IgM and IgG, QIAGEN), followed by goat anti-rat conjugated magnetic bead separation. Tregs (CD4+CD25+) were purified from 2-month-old female WT NOD and TRIF−/−NOD using EasySep™ Mouse CD4+CD25+ Regulatory T Cell Isolation Kit II from STEMCELL Technologies.
2.12. Generation of bone marrow-derived dendritic cells (BMDCs)
BMDCs were generated from BM cells of 2-month-old female WT NOD and TRIF−/−NOD mice. The BM cells were cultured in the presence of granulocyte-macrophage colony-stimulating factor (25 ng/ml) and IL-4 (25 ng/ml) in RPMI 1640 complete medium supplemented with 5% heat-inactivated FBS. Culture medium was replenished every 2 days. On day 5, adherent and loosely adherent cells were harvested. Approximately 70% of the cells were CD11c+ DCs by FACS analysis.
2.13. Cell proliferation assay
Either total splenocytes or purified splenic T cells from 2-month-old female WT NOD and TRIF−/−NOD mice were cultured in triplicate (105 cells/well) in the presence of different concentrations of anti-CD3 (2C11) and anti-CD28 (37.51, 1:300). 3H-thymidine was added during the last 18 hours of a 4-d culture and T cell proliferation was assessed by 3H-thymidine incorporation as counts per minute (CPM). For antigen-specific T cell responses, purified splenic BDC2.5 CD4+ T cells were cultured with irradiated antigen presenting cells – either total splenocytes or purified BMDCs, splenic CD11c+, CD11b+ or B cells in the absence or presence of different concentrations of the BDC2.5 mimotope peptide (RTRPLWVRME)[22]. 3H-thymidine incorporation was determined as described earlier and the data were presented as the stimulation index (SI, the mean cpm in the presence of antigen/the mean cpm in the absence of antigen).
2.14. Treg suppression assay
Purified BDC2.5 CD4+ T cells were cultured with purified Tregs from 2-month-old female WT NOD or TRIF−/−NOD mice (1:1) in the presence of irradiated total splenocytes from WT NOD (as antigen-presenting cells, APCs) and mimotope peptide. BDC CD4+ T cells cultured with the irradiated APCs without mimotope peptide were used as a control. Treg suppression was determined by 3H-thymidine incorporation and the data are presented as the SI.
2.15. Statistics
Statistical analysis was performed using GraphPad Prism software version 7.0. Diabetes incidence was compared using the Log-rank or Gehan-Breslow-Wilcoxon test. In vitro assays were analyzed by single or multiple Student’s t test, with Bonferroni correction or Analysis of variance (ANOVA). P<0.05 was considered significant. Analysis of similarities (ANOSIM) was used to analyze β-diversity of taxonomic families of gut microbiota.
3. Results
3.1. TRIF-deficiency protects NOD mice from type 1 diabetes development in a housing-dependent manner
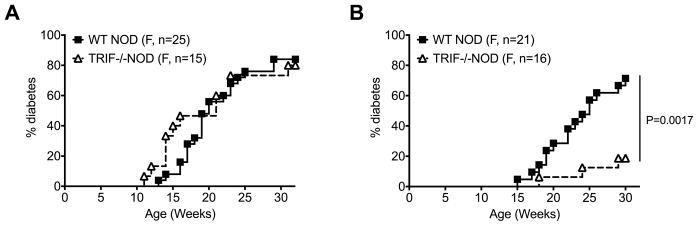
To understand if TRIF influences diabetes susceptibility, we generated TRIF-deficient (TRIF−/−) NOD mice by backcrossing TRIF−/−C57BL/6 mice to our NOD mice for over 10 generations. We first observed the natural history of spontaneous diabetes development in WT NOD (TRIF+/+) and TRIF−/−NOD littermates when both genotypes were housed together (cohoused) or when housed by genotype (e.g. TRIF−/−NOD mice housed only with other TRIF−/−NOD mice, designated as non-cohoused). We found that TRIF−/−NOD female mice developed a similar incidence of diabetes to WT NOD mice when they were cohoused (Fig. 1A). Interestingly, when TRIF−/−NOD mice were housed only with TRIF−/−NOD mice, diabetes development was significantly reduced compared to WT NOD mice (Fig. 1B). Together, the data suggested that the genotype of the cohoused mice influences the development of diabetes in TRIF−/−NOD mice, while this has no effect on diabetes development in WT NOD mice.
Fig. 1.
TRIF-deficiency protects NOD mice from diabetes development and is dependent on gut microbiota. The natural history of diabetes development was observed in female WT NOD and TRIF−/−NOD until 30–32 weeks. (A) Diabetes incidence of cohoused (mixed genotype) female WT NOD and TRIF−/−NOD littermates. (B) Diabetes incidence of non-cohoused (non-mixed genotype) female WT NOD and TRIF−/−NOD mice. Data were pooled from at least two independent experiments. Log-rank test for survival was used for analysis of diabetes incidence.
3.2. TRIF deficiency alters the gut microbiota composition
As TRIF is important in mediating downstream responses to pathogen associated molecular patterns (through TLR3 and partially through TLR4), we hypothesized that the gut microbiota may influence the diabetes development of TRIF−/−NOD mice. To assess gut microbiota composition, we performed 16S rRNA sequencing using fresh fecal samples from cohoused and non-cohoused TRIF−/−NOD mice and WT NOD mice.
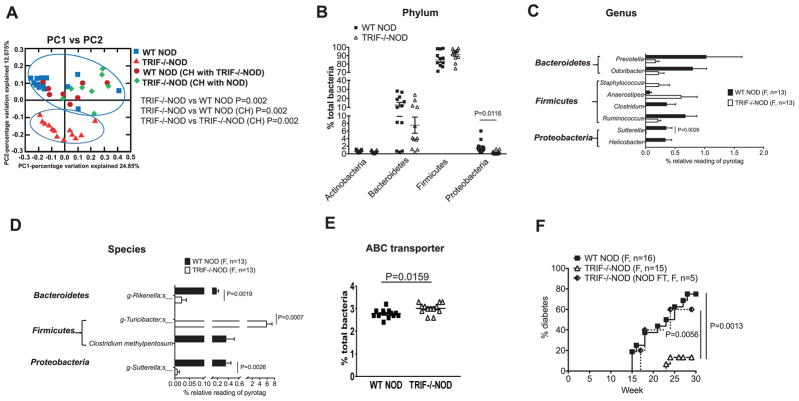
PCoA analysis of the fecal samples identified that the gut microbiota were very different between non-cohoused TRIF−/−NOD and WT NOD mice as shown by the separate clustering of the samples (Fig. 2A, red triangles and blue squares respectively). Interestingly, upon cohousing the mice, the gut microbiota were indistinguishable between TRIF-deficient and TRIF-sufficient mice (Fig. 2A, green diamonds and maroon dots). The effect of housing had little impact on the gut microbiota of WT NOD mice, supporting the data in Fig. 1, showing no differences in diabetes development. Therefore, our further analysis was focused on the mice that were non-cohoused (either TRIF−/−NOD or WT NOD mice), as these were significantly different to one another in both diabetes development and microbial composition.
Fig. 2.
TRIF deficiency alters the gut microbiota in NOD mice. Fecal samples from female WT NOD and TRIF−/−NOD mice were used for taxonomic analysis by 16S rRNA sequencing. (A) PCoA plot showing β-diversity. ANOSIM was used for statistical analysis. Significant taxonomic compositions of the gut microbiota of non-cohoused WT NOD and TRIF−/−NOD mice were shown by as phylum (B), genus (C) and species (D). (E) Predictive functional profiling illustrating increased expression of ABC transporter in TRIF−/−NOD mice. (F) Diabetes incidence from TRIF−/−NOD mice that were gavaged with a suspension of NOD feces (NOD FT) compared with incidence in WT NOD and TRIF−/−NOD mice. The functional composition of the metagenome of the bacteria communities was predicted using PICRUSt. Multiple t tests with Bonferroni correction were used for statistical analysis in (B–D) while a two-tailed Student’s t test was used for statistical analysis in (E) and a log-rank (mantel-cox) test was used in (F).
Investigation of the microbial composition revealed TRIF−/−NOD mice had a significant reduction of Proteobacteria in the gut microbiota, compared to WT NOD mice (Fig. 2B). Further analysis revealed a significant reduction of relative abundance in Sutterella (Proteobacteria) at the genus and species level and a reduction of Rikenella (Bacteroidetes) at the species level in TRIF−/−NOD compared to WT NOD mice (Fig. 2C and D). Furthermore, a significant expansion of Turicibacter species (Firmicutes) was found in TRIF−/−NOD mice compared to WT NOD (Fig. 2D). To assess if the altered microbial composition also altered the function of the bacteria, we analyzed the sequence results with PICRUSt software [20]. As shown in Fig. 2E, there was a significantly increased expression of ATP-binding cassette (ABC) transporter in the gut bacteria from TRIF−/−NOD mice. Together, our data illustrate that TRIF affects the composition and some functions of the gut microbiota in NOD mice.
To test if diabetes protection in TRIF−/−NOD mice was indeed due to the altered gut microbiota, we performed fecal microbiota transfer experiments by gavaging TRIF−/−NOD mice with fecal microbiota from WT NOD mice and assessed the mice for diabetes development. Interestingly, merely transferring the gut microbiota from WT NOD reversed the diabetes protection in TRIF−/−NOD mice, which developed a very similar incidence of diabetes to WT NOD mice (Fig. 2F). This result supports the role of the microbiota in mediating the disease protection seen in TRIF−/−NOD mice (Fig. 1B).
3.3. Gut microbiota alter the diabetogenic capability of immune cells
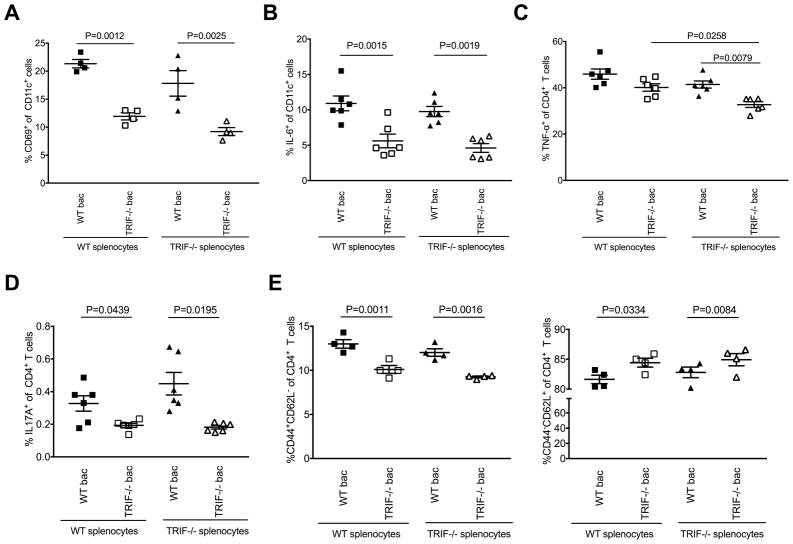
To further investigate the effect of changes in the gut microbiota by TRIF on host immune phenotype and function, and thus diabetes susceptibility, we cultured splenocytes from TRIF−/−NOD or WT NOD mice with heat-killed gut microbiota from either WT or TRIF−/−NOD mice. Interestingly, we found that, compared to WT NOD microbiota, TRIF−/−NOD microbiota induced attenuated CD69 expression on splenic CD11c+ DCs, CD11b+ macrophages and B cells from both WT NOD and TRIF−/−NOD mice (Fig. 3A, Supplemental Fig. 1A–B). Furthermore, TRIF−/−NOD microbiota stimulated significantly fewer pro-inflammatory IL-6-producing CD11c+ DCs (Fig. 3B), CD11b+ macrophages and B cells (Supplemental Fig. 1C and D) compared to WT microbiota. Proinflammatory cytokine (TNFα and IL-17)-expressing CD4+ T cells, regardless of the donor of the T cells, were also significantly lower when exposed to TRIF−/−NOD microbiota compared to WT NOD microbiota (Fig. 3C and D). We also found TRIF−/−NOD microbiota induced fewer effector CD4+ T cells than WT NOD microbiota (Fig. 3E). In line with the reduced percentage of effector CD4+ T cells, there was an increase in percentage of naïve CD4+ T cells when exposed to TRIF−/−NOD bacteria compared to WT NOD bacteria; however, this was only significant in CD4+ T cells from TRIF−/−NOD cells (Fig. 3E). A similar increase in naïve CD8+ T cells was also seen when the CD4+ T cells, from both TRIF-deficient and -sufficient hosts, were co-cultured with TRIF−/−NOD microbiota compared to WT NOD microbiota (Supplemental Fig. 1E). A lower number of TNFα-producing CD8+ T cells in response to TRIF−/−NOD microbiota were seen compared to the response to WT NOD microbiota although this was not statistically significant (Supplemental Fig. 1F). No differences were seen in the cultures without bacteria (Supplemental Fig. 2 and data not shown), suggesting that microbial stimulation is required to mediate these phenotypic and functional changes.
Fig. 3.
Direct effect of gut microbiota on immune cells. Splenocytes (106 cells/ml) of 2-month-old female WT NOD or TRIF−/−NOD mice were incubated overnight (16 hours) with gut bacteria (107 cfu/ml) isolated from large intestinal contents of WT NOD or TRIF−/−NOD mice, and the cells were stained with mAb for surface markers and ICC. For ICC staining, the splenocytes were also further stimulated with PMA and ionomycin in the presence of Golgi plug for the final 4 hours. (A) CD69-expressing CD11c+ cells. (B) IL-6 expressing CD11c+ cells. (C) TNF-α expressing CD4+ T cells. (D) IL-17A-expressing CD4+ T cells. (E) CD4+ T cell subsets (Left: CD44+CD62L−; right: CD44−CD62L+). Two-tailed Student’s t test was used for statistical analysis from (A–E).
3.4. Impaired function of APCs in TRIF−/−NOD mice
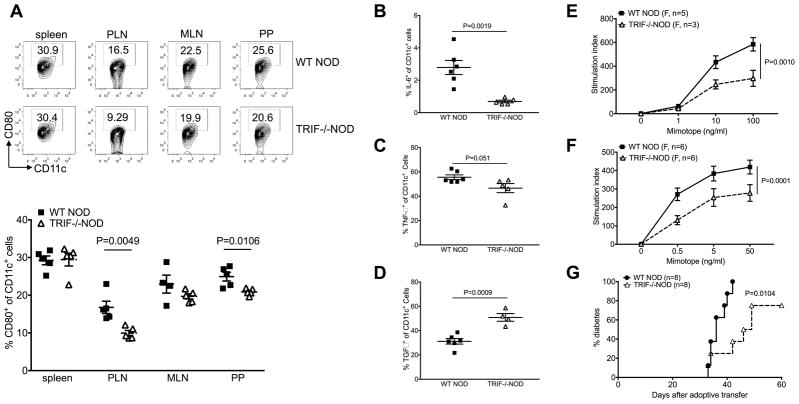
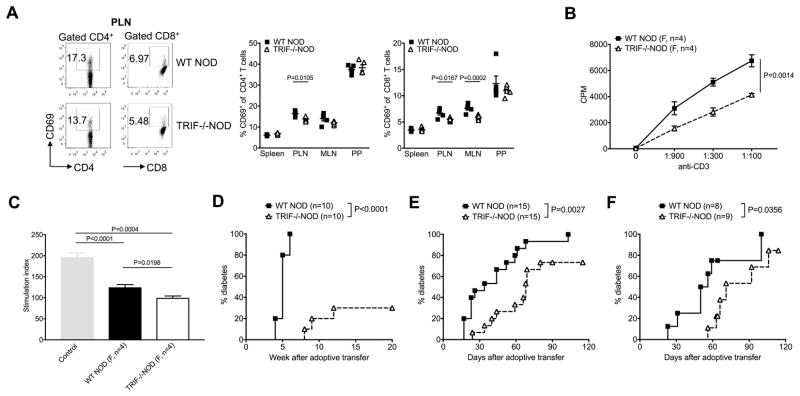
TRIF plays an important role in APC maturation [23–25] and we hypothesized that TRIF-deficiency would affect the function of APCs and thus, contribute to the diabetes-protected phenotype seen in TRIF−/−NOD mice. We examined the phenotype and function of APCs from TRIF−/−NOD mice and found that TRIF−/−NOD mice had reduced CD80 expression on CD11c+ DCs compared to WT NOD mice, within both the pancreatic draining lymph nodes (PLN) and the Peyer’s patches (PP, Fig. 4A). Additionally, we found that DCs from TRIF−/−NOD mice had reduced production of the proinflammatory cytokines, IL-6 and TNFα (Fig. 4B and C) while exhibiting enhanced anti-inflammatory TGFβ production (Fig. 4D) compared to TRIF-sufficient WT NOD mice.
Fig. 4.
Impaired APC function and attenuated activation of CD11c+ DCs in TRIF−/−NOD mice. (A) Percentage of CD80-expressing CD11c+ DCs. Immune cells from spleen, PLN, MLN and PP of 2-month-old female WT NOD and TRIF−/−NOD mice were stained with fluorochrome-conjugated anti-CD11c, anti-CD80, anti-CD11b, anti-B220 and anti-TCRβ antibodies. Representative FACS plots are shown on the left and the summary of CD80-expressing CD11c+ DCs is shown on the right. Data are shown as mean±SEM. The experiment was repeated more than three times. (B–D): Splenocytes from 2-month-old female WT NOD and TRIF−/−NOD mice were stimulated with PMA and ionomycin in the presence of Golgi plug followed by ICC staining. (B) IL-6; (C) TNF-α and (D) TGF-β. Data are shown as mean±SEM and pooled from two independent experiments. (E) Proliferation of BDC2.5 CD4+ T cells. Purified NOD BDC2.5 CD4+ T cells were cultured with irradiated T cell-depleted splenocytes from 2-month-old female WT NOD and TRIF−/−NOD mice in the presence of mimotope peptide. Proliferation was assessed using 3H-thymidine incorporation. n=3–5/mice/group/experiments from more than three experiments. (F) Proliferation of BDC2.5 CD4+ T cells. Purified NOD BDC2.5 CD4+ T cells were cultured with irradiated splenic CD11c+ DCs from 2-month-old female WT NOD and TRIF−/−NOD mice in the presence of mimotope peptide. Data were pooled from two independent experiments. (G) Diabetes incidence following adoptive transfer. Purified splenic T cells (7×106 cells/mouse) from diabetic WT NOD mice were i.v. injected into Rag−/−NOD or NOD.scid mice with T cell-depleted splenic APCs (7×106 cells/mouse) from either female WT NOD or TRIF−/−NOD mice. Data were pooled from two independent experiments. Log-rank test for survival was used for analysis of diabetes incidence. Two-way ANOVA was used for the comparison in (E) and (F). Two-tailed Student’s t test was used for statistical analysis in (B–D).
Next, we investigated the function of TRIF-deficient APCs in vitro and in vivo. We first tested the antigen-presenting capabilities of APCs from TRIF−/−NOD and TRIF+/+NOD mice by culturing diabetogenic BDC2.5 CD4+ T cells with the BDC2.5 mimotope peptide presented by mitomycin C-treated splenic APCs (T cell-depleted splenocytes). BDC2.5 CD4+ T cells were chosen as responder cells as they are potent diabetogenic T cells recognizing an islet autoantigen, which has recently been identified as a hybrid peptide of chromogranin A and insulin [26, 27]. We found that TRIF-deficient APCs had significantly impaired antigen-presenting function compared with TRIF-sufficient APCs (Fig. 4E). To identify the role of different APC subsets, we purified splenic CD11b+, CD11c+ and CD19+ cells and conducted the same antigen presentation assay. We found that TRIF-deficient CD11c+ DCs were responsible for the impaired antigen presentation to BDC2.5 T cells (Fig. 4F), as purified TRIF-deficient CD11b+ and CD19+ cells showed enhanced function (Supplemental Fig. 3A and B). The impaired antigen presentation function was also found in TRIF-deficient bone marrow-derived DCs (Supplemental Fig. 3C). We then tested APC function in vivo in an adoptive transfer system. We transferred purified T cells from diabetic WT NOD mice together with TRIF-deficient or -sufficient total APC into Rag−/−NOD mice. Consistent with the results from the experiments in vitro, we found that APCs from TRIF−/−NOD mice had an attenuated ability to facilitate diabetes development induced by diabetogenic T cells in the recipients compared to the APCs from TRIF-sufficient mice (Fig. 4G). Together, these data suggest that TRIF is important for APC function in mediating diabetes susceptibility.
3.5. Reduced T cell activation and function in TRIF−/−NOD mice
Having identified changes within the APC compartment, we asked if there were any TRIF-related effects on the T cells. Interestingly, TRIF−/−NOD mice exhibited reduced T cell activation, as assessed by CD69 expression, in both CD4+ and CD8+ T cell subsets specifically within the PLNs (Fig. 5A). Next, we tested whether TRIF expression influenced the T cell response to T cell receptor stimulation using anti-CD3 and anti-CD28. As shown in Fig. 5B, TRIF-deficient T cells showed impaired proliferation in response to pan-T cell stimulation compared with TRIF-sufficient T cells. As the T cell response to anti-CD3 stimulation requires the cross-linking of anti-CD3 molecules by APCs, to further probe whether the cause of impaired T cell proliferation was T cell intrinsic or secondary due to the impaired APC function, we cultured WT NOD T cells with either WT NOD APCs or TRIF−/−NOD APCs and vice versa in the presence of anti-CD3. Interestingly, we found that TRIF-deficiency affects the function of both T cells and APCs (Supplementary Fig. 4).
Fig. 5.
Reduced T cell activation and function in TRIF−/−NOD mice. (A) CD69-expressing CD4+ and CD8+ T cells in TRIF−/−NOD mice. FACS plots for PLNs are shown on the left and the summary of CD69-expressing CD4+ and CD8+ T cells are shown on the right. One of three independent experiments is presented, with data shown as mean±SEM. (B) BDC CD4+ T cell proliferation. Purified splenic CD4+ T cells from BDC2.5 mice were cocultured with total splenocytes from either female WT NOD or TRIF−/−NOD mice on stimulation by anti-CD3/anti-CD28. This experiment was repeated more than three times. (C) Treg suppression assay. Purified Treg (CD4+CD25+ cells) from 2-month-old female WT NOD or TRIF−/−NOD mice were cultured with BDC2.5 CD4+ T cells (1:1) in the presence of irradiated WT NOD splenocytes in the presence of BDC2.5 mimotope peptide. Data are shown as SI. (D) Diabetes incidence following adoptive transfer. Total splenocytes (8×106 cells/mouse) from female diabetic WT NOD or TRIF−/−NOD mice were injected i.v. into immune-deficient NOD mice (Rag−/−NOD or NOD.scid mice) for observation of diabetes incidence. Data were pooled from two independent experiments. Total splenocytes (8×106 cells/mouse) from female diabetic WT NOD or TRIF−/−NOD mice were injected i.v. into sub-lethally irradiated 4–5 week-old female WT NOD (E) or TRIF−/−NOD (F) mice followed by observation for development of diabetes. Data were pooled from 2–3 independent experiments. Two-tailed Student’s t test was used for statistical analysis in (A) and (C) Two-way ANOVA was used for the comparison in (B). Log-rank test for survival was used for analysis of diabetes incidence in (D–F).
Next we investigated if Treg cells would be affected by the absence of TRIF expression. We found no difference in the Treg frequency or absolute number between WT NOD and TRIF−/−NOD mice (Supplemental Fig. 5A and B). We also tested the function of the Treg cells in an antigen-specific Treg suppression assay, in which pathogenic BDC2.5 CD4+ T cells responder cells were cultured with purified splenic Tregs from WT or TRIF−/−NOD mice in the presence of antigenic peptide. It is interesting that TRIF-deficient Tregs showed stronger suppression of pathogenic responder cells than WT Tregs (Fig. 5C).
Our results indicated that TRIF-deficiency affected the function of both APCs and T cells including Tregs. To confirm that the changes seen in vitro contributed to diabetes protection in vivo, we performed a series of adoptive transfer experiments. Firstly, we adoptively transferred total splenocytes from WT or TRIF−/−NOD mice into Rag-deficient hosts. We found TRIF−/− splenocytes were significantly less able to induce diabetes compared to TRIF-sufficient counterparts (Fig. 5D). Secondly, we transferred total splenocytes from WT NOD or TRIF−/−NOD mice into irradiated WT NOD (Fig. 5E) and TRIF−/−NOD (Fig. 5F) recipients to ensure the protection was related to the immune system and not the non-hematopoietic cells. In both cases we found TRIF-deficient splenocytes had significantly weaker ability to induce diabetes development in all the recipients (Fig. 5E and F). Our data provide strong evidence that the TRIF-deficiency in the immune cells was responsible for the diabetes-protected phenotype in TRIF−/−NOD mice.
4. Discussion
TLRs and their adaptor molecule MyD88 are known to influence T1D development mediated by gut microbiota [3, 10, 13, 15]. In this study, we investigated the role of TRIF, another key adaptor molecule, in T1D development in NOD mice. We found that TRIF deficiency resulted in a significant reduced diabetes development. Similar to MyD88-deficient NOD mice, in the absence of TRIF, diabetes protection is mediated through changes in the gut microbiome. However, unlike MyD88-deficient NOD mice, co-housing TRIF-deficient NOD mice with TRIF-sufficient NOD mice was able to reverse diabetes protection in TRIF-deficient mice. Our results clearly demonstrate that the gut microbiota alter diabetogencity in these mice. However, TRIF deficiency not only alters the composition of gut microbiota but also alters the function of innate and adaptive immune cells. In the absence of TRIF, dendritic cells exhibited reduced costimulation, reduced inflammatory cytokine expression and impaired antigen-presenting function to diabetogenic CD4+ T cells. In the absence of TRIF, T cells were not able to mount an optimal response to anti-CD3 stimulation whereas Treg cells showed enhanced suppressive function in the absence of TRIF. Furthermore, diabetes protection could be maintained when transferring WT NOD T cells from a diabetic donor together with TRIF-deficient APCs or transferring TRIF-deficient splenocytes to TRIF-sufficient or -deficient hosts. Our results revealed the inter-relationship between genetic factors and the gut microbiota, as the genetic deletion of TRIF changed the composition of gut microbiota leading to diabetes protection. However, this effect on gut microbiota and diabetes protection could be overridden by introducing gut microbiota from TRIF-sufficient NOD mice. Our results also demonstrated that TRIF plays a direct role in immune cell functions that are most likely to be independent of gut microbiota.
Burrows and colleagues recently reported that TRIF-deficient NOD mice and TRIF-sufficient NOD mice developed a similar incidence of diabetes when co-housed [15], with which our results are consistent. However, the authors did not investigate the effect of non-cohousing the TRIF-deficient or TRIF-sufficient NOD mice on diabetes development. Instead, they studied the effect of TRIF on MyD88-mediated diabetes protection. It is interesting that the authors found that approximately 20% of TRIF and MyD88 double-deficient NOD mice developed diabetes at an old age (>25 weeks) whereas MyD88-deficient TRIF-sufficient NOD mice were diabetes free [15]. Although our study design and approaches were different to those of Burrows and colleagues, the conclusion is complementary. Both studies have shown that deficiency of innate immune molecules, such as TRIF, modulates T1D development and this effect is mediated by gut microbiota. In addition to the effect the innate immune system to change gut microbiota, we also demonstrated that the TRIF may play a direct role in the functions of innate and adaptive immune cells.
Several studies in human T1D have also shown alterations in gut microbiota [28–30]. The changes include a reduction of diversity in the composition of gut microbiota but an increased relative abundance of some gut bacterial species in patients with T1D compared to healthy individuals [28, 31–34]. Recent studies also indicate that gut bacterial products can modify T1D development in both humans [30] and NOD mice [18, 35]. In our current study, we found a significantly reduced relative abundance of Sutterella (Proteobacteria) in TRIF-deficient mice that are protected from T1D development. Sutterella reside within the human intestine and are reported to be associated with Crohn’s disease [36, 37] and other health issues [38]. Interestingly, Turicibacter was significantly increased in diabetes-protected TRIF-deficient NOD mice. Turicibacter has been placed in the class of Erysipelotrichia and been reported to be associated with the TNBS-induced colitis mouse model [39] and obesity in humans [40]. Thus, it is clear that the composition of gut microbiota plays an important role in health and disease.
Given the microbial differences seen between TRIF−/−NOD and WT NOD mice, we further investigated their functional profile using PICRUSt [20]. We found that gut microbiota from TRIF−/−NOD mice have an increased relative abundance of bacteria with ABC transporters compared to WT NOD mice. ABC transporters are a large superfamily, conserved between bacteria and humans. They are vital for the survival and function of prokaryotic or eukaryotic cells. It is not clear how ABC transporter-expressing gut bacteria affect the immune cells and T1D protection in TRIF−/−NOD mice. However, stimulating the immune cells with gut microbiota from TRIF−/−NOD mice, but not from WT NOD mice, reduced CD69 expression on the three major types of antigen presenting cells – dendritic cells, macrophages and B cells. Interestingly, CD69 expression on dendritic cells is important in regulating the migration of dendritic cells [41]. As DCs sample microbial antigens from the gut lumen [42], it is conceivable that the microbial products including metabolites in TRIF−/−NOD mice suppress CD69 expression on DCs, which in turn limits DC migration to the islets to initiate autoinflammation. It is also possible that through a similar mechanism, gut microbiota from TRIF−/−NOD mice induce lower levels of proinflammatory cytokines (IL-6, TNFα and IL-17) by DC and CD4+ T cells regardless of the TRIF expression. It is noteworthy that a significantly lower frequency of IL-6-expressing DCs was induced in both the in vitro bacterial co-culture assays and in ex vivo ICC assays, emphasizing the direct influence of gut microbiota from TRIF−/−NOD mice. Increasing evidence from human studies indicates an alteration of gut microbiota in patients with T1D compared to healthy control subjects [28–30], but less is known about the immune cell response to the different microbiota. Our results indicate that the microbiota from diabetes-protected hosts suppress the proinflammatory cytokine milieu that is important in promoting autoimmunity. Our results also highlight the importance of dendritic cell-microbiota interactions in shaping the development of autoimmunity.
Studies in both mice and humans have shown that TGFβ induces Treg differentiation and/or conversion in the periphery [43]. We observed a higher frequency of TGFβ-producing DCs when TRIF was deficient; TRIF-deficient Tregs also exhibited stronger immune suppressive function, both of which likely contributed to disease protection in this model. Interestingly, T1D patients had reduced TGFβ in the circulation [44].
It is known that the development and maturation of the immune system is dependent on the commensal bacteria [45]. Our results provide further evidence that commensal gut bacteria can shape the diabetogenicity of immune cells. By changing the constituent mice housed together, we can alter the composition of microbiota and are able to reverse the diabetes-protective effect seen in non-cohoused TRIF-deficient NOD mice. Interestingly, the altered gut microbiota in TRIF-deficient NOD mice did not confer diabetes protection on TRIF-sufficient NOD mice, which suggests a requirement for TRIF deficiency in the host immune cells, in addition to the altered microbiota for the diabetes-protected phenotype to manifest.
In conclusion, our study provides evidence that innate immunity modifies the composition of the gut microbiota, which in turn regulates autoimmune diabetes development. In the absence of TRIF, NOD mice harbor altered gut microbiota, leading to a reduced proinflammatory phenotype and function of immune cells that are associated with diabetes protection. However, this protective effect, which is mediated by TRIF-deficiency, can be reversed by introducing gut microbiota from TRIF-sufficient NOD mice. Our study also provides further evidence of the interaction between the genes and the gut microbiota, which is important for mediating immune tolerance (or lack of tolerance). Lastly, our results also demonstrate that innate immunity can affect T cell function directly. Further studies are required not only to identify the specific bacterial strain(s) that promote immune tolerance but also the molecular mechanism by which non-pathogenic gut bacteria regulate the function of immune cells. The more we understand mechanistically, the greater the potential for therapeutic applications which will help in designing new therapies for T1D to reduce disease severity or even prevent the disease by modulation of the gut microbiota.
Supplementary Material
Highlights.
TRIF-deficient NOD mice are protected from T1D development
Altered gut microbiota contributes to protection from T1D in TRIF-deficient NOD mice
Gut bacteria from TRIF-deficient NOD mice induce less inflammatory immune responses
TRIF-deficient DCs are functionally altered and reduce activation of T cells
TRIF deficiency-induced immune effects can be overridden by altering microbiota
Acknowledgments
7. Funding
This work was supported by a fellowship from the German Research Foundation (GU 1122/3-1) to EG, a scholarship from the China Scholarship Council (2010637116) to CC, a Fulbright-Diabetes UK Research scholarship and a JDRF postdoctoral fellowship to JAP (3-PDF-2016-197-A-N) and research grants from NIH (DK092882, DK100500, and P30 DK945735) and ADA (1-14-BS-222) to LW.
We thank Karl Hager (Lab Medicine, Yale) for the assistance with 16S rRNA sequencing, Xiaojun Zhang for care of the mice used in the study and all the lab members for their technical help and critical scientific comments during the study.
Footnotes
5. Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
6. Author Contributions
EG, CC, NT, JAP and LW designed the experiments; EG, CC, NT, JAP, JP and MMS carried out the experiments; EG, CC, NT, JAP, JP, FSW and LW analyzed data, ZZ, FSW and LW supervised the study; EG, NT, JAP, FSW and LW wrote the manuscript. The project was conceived by LW, who assumes responsibility for the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harjutsalo V, Sjoberg L, Tuomilehto J. Time trends in the incidence of type 1 diabetes in Finnish children: a cohort study. Lancet. 2008;371:1777–82. doi: 10.1016/S0140-6736(08)60765-5. [DOI] [PubMed] [Google Scholar]
- 2.Patterson CC, Gyurus E, Rosenbauer J, Cinek O, Neu A, Schober E, et al. Trends in childhood type 1 diabetes incidence in Europe during 1989–2008: evidence of non-uniformity over time in rates of increase. Diabetologia. 2012;55:2142–7. doi: 10.1007/s00125-012-2571-8. [DOI] [PubMed] [Google Scholar]
- 3.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulden E, Wong FS, Wen L. The gut microbiota and Type 1 Diabetes. Clin Immunol. 2015;159:143–53. doi: 10.1016/j.clim.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12:154–67. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 9.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 10.Kim HS, Han MS, Chung KW, Kim S, Kim E, Kim MJ, et al. Toll-like receptor 2 senses beta-cell death and contributes to the initiation of autoimmune diabetes. Immunity. 2007;27:321–33. doi: 10.1016/j.immuni.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Wong FS, Hu C, Zhang L, Du W, Alexopoulou L, Flavell RA, et al. The role of Toll-like receptors 3 and 9 in the development of autoimmune diabetes in NOD mice. Ann N Y Acad Sci. 2008;1150:146–8. doi: 10.1196/annals.1447.039. [DOI] [PubMed] [Google Scholar]
- 12.Gulden E, Ihira M, Ohashi A, Reinbeck AL, Freudenberg MA, Kolb H, et al. Toll-like receptor 4 deficiency accelerates the development of insulin-deficient diabetes in non-obese diabetic mice. PLoS One. 2013;8:e75385. doi: 10.1371/journal.pone.0075385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tai N, Wong FS, Wen L. TLR9 deficiency promotes CD73 expression in T cells and diabetes protection in nonobese diabetic mice. J Immunol. 2013;191:2926–37. doi: 10.4049/jimmunol.1300547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Lee AS, Shameli A, Geng X, Finegood D, Santamaria P, et al. TLR9 blockade inhibits activation of diabetogenic CD8+ T cells and delays autoimmune diabetes. J Immunol. 2010;184:5645–53. doi: 10.4049/jimmunol.0901814. [DOI] [PubMed] [Google Scholar]
- 15.Burrows MP, Volchkov P, Kobayashi KS, Chervonsky AV. Microbiota regulates type 1 diabetes through Toll-like receptors. Proc Natl Acad Sci U S A. 2015;112:9973–7. doi: 10.1073/pnas.1508740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCall KD, Thuma JR, Courreges MC, Benencia F, James CB, Malgor R, et al. Toll-like receptor 3 is critical for coxsackievirus B4-induced type 1 diabetes in female NOD mice. Endocrinology. 2015;156:453–61. doi: 10.1210/en.2013-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng J, Narasimhan S, Marchesi JR, Benson A, Wong FS, Wen L. Long term effect of gut microbiota transfer on diabetes development. J Autoimmun. 2014;53:85–94. doi: 10.1016/j.jaut.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tai N, Peng J, Liu F, Gulden E, Hu Y, Zhang X, et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med. 2016;213:2129–46. doi: 10.1084/jem.20160526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 20.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu C, Ding H, Li Y, Pearson JA, Zhang X, Flavell RA, et al. NLRP3 deficiency protects from type 1 diabetes through the regulation of chemotaxis into the pancreatic islets. Proc Natl Acad Sci U S A. 2015;112:11318–23. doi: 10.1073/pnas.1513509112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judkowski V, Pinilla C, Schroder K, Tucker L, Sarvetnick N, Wilson DB. Identification of MHC class II-restricted peptide ligands, including a glutamic acid decarboxylase 65 sequence, that stimulate diabetogenic T cells from transgenic BDC2. 5 nonobese diabetic mice. J Immunol. 2001;166:908–17. doi: 10.4049/jimmunol.166.2.908. [DOI] [PubMed] [Google Scholar]
- 23.Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, et al. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat Immunol. 2003;4:1223–9. doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- 24.Shen H, Tesar BM, Walker WE, Goldstein DR. Dual signaling of MyD88 and TRIF is critical for maximal TLR4-induced dendritic cell maturation. J Immunol. 2008;181:1849–58. doi: 10.4049/jimmunol.181.3.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W, Jain A, Gao Y, Dozmorov IM, Mandraju R, Wakeland EK, et al. Differential outcome of TRIF-mediated signaling in TLR4 and TLR3 induced DC maturation. Proc Natl Acad Sci U S A. 2015;112:13994–9. doi: 10.1073/pnas.1510760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haskins K, Portas M, Bradley B, Wegmann D, Lafferty K. T-lymphocyte clone specific for pancreatic islet antigen. Diabetes. 1988;37:1444–8. doi: 10.2337/diab.37.10.1444. [DOI] [PubMed] [Google Scholar]
- 27.Delong T, Wiles TA, Baker RL, Bradley B, Barbour G, Reisdorph R, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–4. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–73. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–53. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–77. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 33.Endesfelder D, zu Castell W, Ardissone A, Davis-Richardson AG, Achenbach P, Hagen M, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes. 2014;63:2006–14. doi: 10.2337/db13-1676. [DOI] [PubMed] [Google Scholar]
- 34.Pinto E, Anselmo M, Calha M, Bottrill A, Duarte I, Andrew PW, et al. The intestinal proteome of diabetic and control children is enriched with different microbial and host proteins. Microbiology. 2017;163:161–74. doi: 10.1099/mic.0.000412. [DOI] [PubMed] [Google Scholar]
- 35.Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18:552–62. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 36.Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y, et al. Molecular inventory of faecal microflora in patients with Crohn’s disease. FEMS Microbiol Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–41. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiippala K, Kainulainen V, Kalliomaki M, Arkkila P, Satokari R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front Microbiol. 2016;7:1706. doi: 10.3389/fmicb.2016.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones-Hall YL, Kozik A, Nakatsu C. Ablation of tumor necrosis factor is associated with decreased inflammation and alterations of the microbiota in a mouse model of inflammatory bowel disease. PLoS One. 2015;10:e0119441. doi: 10.1371/journal.pone.0119441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greiner T, Backhed F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab. 2011;22:117–23. doi: 10.1016/j.tem.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Lamana A, Martin P, de la Fuente H, Martinez-Muñoz L, Cruz-Adalia A, Ramirez-Huesca M, et al. CD69 modulates sphingosine-1-phosphate-induced migration of skin dendritic cells. J Invest Dermatol. 2011;131:1503–12. doi: 10.1038/jid.2011.54. [DOI] [PubMed] [Google Scholar]
- 42.Farache J, Koren I, Milo I, Gurevich I, Kim KW, Zigmond E, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38:581–95. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanjabi S, Oh SA, Li MO. Regulation of the Immune Response by TGF-beta: From Conception to Autoimmunity and Infection. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiao YC, Shen J, Hong XZ, Liang L, Bo CS, Sui Y, et al. Changes of regulatory T cells, transforming growth factor-beta and interleukin-10 in patients with type 1 diabetes mellitus: A systematic review and meta-analysis. Clin Immunol. 2016;170:61–9. doi: 10.1016/j.clim.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.