Abstract
Transfer RNA (tRNA) from all domains of life contains multiple modified nucleosides, the functions of which remain incompletely understood. Genetic interactions between tRNA modification genes in Saccharomyces cerevisiae suggest that different tRNA modifications collaborate to maintain translational efficiency. Here we characterize such collaborative functions in the ochre suppressor tRNA SUP4. We quantified ochre read-through efficiency in mutants lacking either of the 7 known modifications in the extended anticodon stem loop (G26-C48). Absence of U34, U35, A37, U47 and C48 modifications partially impaired SUP4 function. We systematically combined modification defects and scored additive or synergistic negative effects on SUP4 performance. Our data reveal different degrees of functional redundancy between specific modifications, the strongest of which was demonstrated for those occurring at positions U34 and A37. SUP4 activity in the absence of critical modifications, however, can be rescued in a gene dosage dependent fashion by TEF1 which encodes elongation factor eEF1A required for tRNA delivery to the ribosome. Strikingly, the rescue ability of higher-than-normal eEF1A levels extends to tRNA modification defects in natural non-suppressor tRNAs suggesting that elevated eEF1A abundance can partially compensate for functional defects induced by loss of tRNA modifications.
Introduction
Transfer RNA (tRNA) is known for the presence of extensive post-transcriptional modifications. Some of the non-standard ribonucleosides are thought to be important for tRNA stability and folding or to improve codon-anticodon recognition1–3. The latter has been attributed to the wobble uridine modification 5-methoxy-carbonyl-methyl-2-thiouridine (mcm5s2U)4–7. In yeast, this modification is naturally present in tRNALysUUU, tRNAGlnUUG and tRNAGluUUC and is formed by two separate pathways which mediate mcm5 side chain addition at position 5 of the uracil base and exchange of the oxygen for sulfur at position 2 (wobble uridine thiolation)8. Genes required for addition of the mcm5 side chain include ELP1-ELP6 (Elongator), TRM9-TRM112 (wobble methyltransferase) and KTI11-KTI14 or SIT4, other potentially regulatory loci4,9–17. The thiolation of tRNALysUUU, tRNAGlnUUG and tRNAGluUUC depends on the ubiquitin like modifier Urm1 and several other sulfur transfer proteins11,18–21.
Several lines of evidence support a role of mcm5s2U in improvement of tRNA binding to the ribosomal A-site; in the absence of the modification, reduced A-site binding evokes downstream effects, including ribosomal slow down and associated protein folding defects as well as increased frameshift errors5,22–24. In vitro studies with bacterial ribosomes suggest that absence of s2U already increases tRNA rejection both before and after GTP hydrolysis by EF-Tu and impedes ribosomal translocation7. In all cases analysed, the combined absence of mcm5 and s2U led to more severe effects and phenotypes compared to loss of either part alone, suggesting that mcm5 and s2U are to some extent functionally redundant in maintaining tRNA decoding efficiency22,24–26.
The genes required for mcm5 and s2U formation form an extensive network of negative genetic interactions with loci required for other tRNA modifications, such as DEG1 (pseudouridine, ψ38/39) and TCD1 (cyclic N6-threonylcarbamoyadenosine, ct6A37)27–29. In vivo studies support the conclusion that malfunction of tRNALysUUU in the combined absence of mcm5/s2U and ct6A and malfunction of tRNAGlnUUG in the combined absence of mcm5/s2U and ψ38 is the underlying cause of the observed negative genetic interactions30. Hence, modifications at distinct positions of the anticodon loop might cooperatively promote tRNA function, an idea that is further supported by in vitro studies indicating an anticodon-prestructuring role shared between U34 and A37 modifications1,2,6,26,31. Functional redundancy between different modifications may explain why the majority of single tRNA modification defects has only mild or no negative effects on growth, whereas combined modification mutants show more severe phenotypes in several cases. However, so far, no study is available to the best of our knowledge that quantified the vivo function of a defined tRNA in the combined absence of different modifications.
We utilized in here the non-sense ochre suppressor tRNA SUP4 to measure its translational efficiency and UAA read-through in the presence and absence of modifications. SUP4 is a variant of tRNATyrGψU with a G to U exchange at the wobble base that renders the anticodon cognate for ochre stop codons32. The U34 position of SUP4 is known to be modified in an Elongator dependent manner to mcm5U and previously, loss of mcm5 was shown to severely (but not entirely) diminish nonsense suppression4,10,33. Apart from mcm5U also i6A37 (N6-isopentenyl-adenosine) is known to be required for efficient UAA read-through by SUP434. Further, ψ35 formation was shown to require the presence of an intron in the tRNATyr gene and nonsense suppression by the tyrosine inserting ochre suppressor SUP6 depends on the presence of the intron in the SUP6 gene35. Thus, it appears likely that ψ35 is important for efficient nonsense suppression by tyrosine inserting suppressor tRNAs (SUP4 and SUP6) as well.
We quantified in vivo UAA read-through efficiency of SUP4 and relative importance of all modifications present in the extended anticodon stem and loop (ASL) including the variable loop (VL) (G26 to C48). Systematic combination of modification defects and subsequent measurement of suppressor activity provided support for functional collaboration between distinct modifications in the entire extended ASL. The strongest functional impairment of SUP4 was observed in the combined absence of mcm5U and i6A37 reinforcing the notion that U34 and different A37 modifications in general might fulfil overlapping roles in maintaining the anticodon open loop configuration2,26. Genetic interaction networks of the studied modification genes further suggested a role of eEF1A gene dosage in changing the dependency of tRNA on the presence of modifications, which was directly demonstrated for SUP4 tRNA.
Results
Comparison of relative contributions of single tRNA modifications for SUP4 functioning
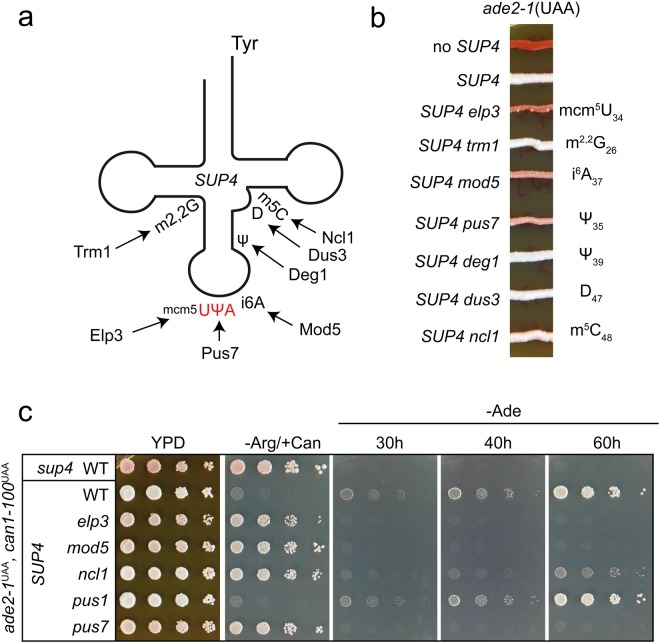
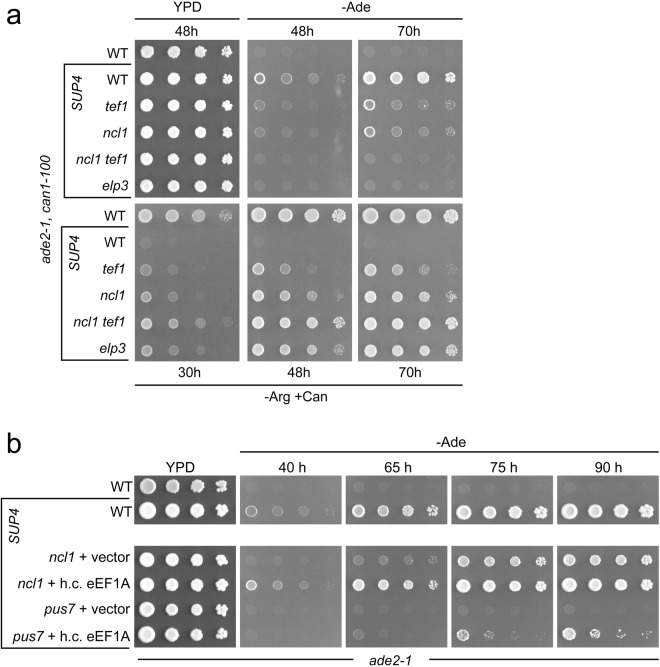
To systematically investigate the role of individual modifications in SUP4, we targeted all known modifications of the extended ASL spanning from position 26 to 48. In tRNATyrGψA, this region is reported to harbour modified nucleosides N2,N2-dimethylguanosine (m2,2G) at position 26, pseudouridine (ψ) at positions 35 and 39, N6-isopentenyladenosine (i6A) at position 37, dihydrouridine (D) at position 47 and 5-methylcytosine (m5C) at position 48 (Fig. 1A)36. In addition, the G:U exchange at position 34 renders SUP4 an Elongator substrate carrying mcm5U10. These modifications are introduced by well characterized modifiers that can be inactivated by single gene deletions10,37–41. We used existing elp3 (mcm5U) and deg1 (ψ39) mutants in a W303-1B derived SUP4 strain10,42 and additionally deleted TRM1 (m2,2G), PUS7 (ψ35), MOD5 (i6A), DUS3 (D47) and NCL1 (m5C) to assemble a set of mutants individually defective in each of the 7 modifications of the extended ASL of SUP4. We also deleted PUS1 since earlier evidence indicated an ability of recombinant Pus1 to modify U35 in pre-tRNATyrGUA/UUA in vitro43. The parental strain utilized carries ade2-1 and can1-100 alleles containing premature UAA stop codons which can be suppressed by SUP410. Nonsense suppression of ade2-1 enables growth on −Ade media and suppresses red pigmentation normally associated with loss-of-function ade2 alleles. UAA stop codon read-through of can1-100 by functional SUP4 results in sensitivity to the arginine analogue canavanine. As expected, the presence of SUP4 entirely suppressed red pigmentation in the wild type and loss of ELP3 (mcm5U), MOD5 (i6A) and PUS7 (ψ35) partially restored pigment formation, albeit to a lesser extent compared to the parental strain lacking SUP4 tRNA altogether (Fig. 1B). When all single mutants were analysed for can1-100 and ade2-1 suppression using serial dilution spot assays on appropriate media, loss of suppression to comparable degrees of both ade2-1 and can1-100 was observed for elp3, mod5 and pus7 (Fig. 1C). The ncl1 mutant defective in m5C formation41 entirely lost can1-100 suppression and was partially defective in ade2-1 suppression, observable due to its delayed growth on −Ade media. Absence of DEG1, TRM1, PUS1 and DUS3 did not detectably affect suppression of the two reporter alleles (Figs 1C, S1).
Figure 1.
Relevance of individual tRNA modifications for ochre suppression by SUP4. (a) Schematic representation of SUP4 tRNA. Modifications are indicated according to36 together with proteins essential for their formation. (b) Red pigmentation of yeast strains of the indicated genetic background as an indicator for ade2-1 suppression. Strains were streaked on YPD, grown for 4 days, followed by incubation at 4 °C for an additional 4 days prior to documentation. (c) Plate assays of ade2-1 and can1-100 suppression by SUP4. Strains of the indicated genetic backgrounds were spotted on YPD, −Ade or −Arg/+Can media and photographed after 40 h of incubation at 30 °C (YPD and −Arg/+Can) or at the indicated time points (−Ade).
The change of pigmentation from white to pink suggested residual non-sense suppression in elp3, mod5 and pus7 mutants since the complete absence of SUP4 resulted in much stronger red pigmentation (Fig. 1B). SUP4 nonsense suppression was previously quantified using lacZ reporters with a premature UAA codon and indeed significant residual nonsense suppression in elp3 mutants was observed, suggesting that canavanine resistance and inability of modification mutant strains to grow on −Ade media does not necessarily indicate a complete loss of SUP4 function4,42.
SUP4 dependent nonsense suppression in the absence of nonsense mediated decay
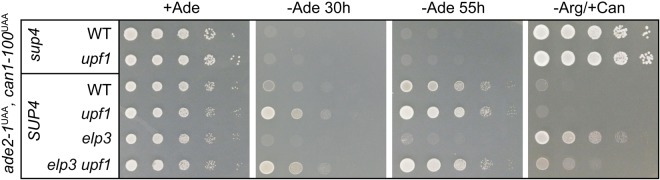
Since ade2-1 and can1-100 nonsense mRNAs are drastically stabilized in the absence of nonsense mediated decay (NMD)44,45, we investigated SUP4 dependent nonsense suppression in the absence of a functional NMD pathway. Because nonsense suppressor tRNA is in competition with translation termination factors46, which also interact with Upf1 to initiate NMD of transcripts with premature termination codons47,48, it appeared possible that SUP4 may affect nonsense suppression indirectly by inhibiting NMD. To test the role of NMD in SUP4 readouts used in this study, we deleted the UPF1 gene essential for decay of can1-100 and ade2-1 mRNAs44,45 in wild type and SUP4 strains and scored phenotypic suppression of both reporter genes. Deletion of UPF1 did not enable growth on −Ade media or suppress canavanine resistance in the absence of SUP4 (Fig. 2). However, growth on −Ade media is detectably improved in a SUP4 upf1 strain as compared to the SUP4 UPF1 parent (Fig. 2), indicating that SUP4 dependent generation of functional Ade2 protein from the nonsense transcript is improved by stabilization of the mRNA. Also, the above described defect in SUP4 dependent suppression of ade2-1 and can1-100 in the absence of a functional ELP3 gene can be bypassed by inactivation of NMD. The latter can be concluded due to the reversion of adenine auxotrophy and canavanine resistance in SUP4 elp3 by deletion of UPF1 (Fig. 2). Together these results indicate that SUP4 functions independently of NMD in ade2-1 and can1-100 suppression and that absence of NMD improves the absolute SUP4 dependent nonsense readthrough.
Figure 2.
SUP4 dependent ochre suppression in absence of nonsense mediated decay. Plate assays of ade2-1 and can1-100 suppression by SUP4. Strains of the indicated genetic backgrounds were spotted on +Ade, −Ade or −Arg/+Can media and photographed after 40 h of incubation at 30 °C (+Ade and −Arg/+Can) or at the indicated time points (−Ade).
Quantification of SUP4 dependent readthrough using a dual luciferase reporter
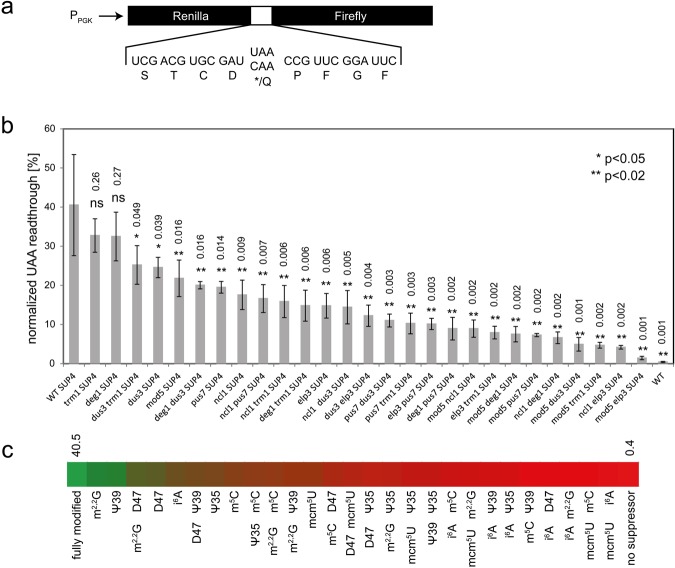
Since we demonstrated SUP4 to function independently of NMD but found ade2-1 and can1-100 suppression to be influenced by levels of the respective mRNAs, we next employed a nonsense readthrough reporter insensitive to changes in mRNA levels to quantify UAA readthrough in absence of tRNA modifications. For this purpose, we used dual luciferase constructs consisting of an upstream (renilla) and a downstream (firefly) luciferase gene, separated by a linker region containing either a sense or an in-frame nonsense (UAA) codon49. Since relative readthrough levels are determined based on the two reporter activities that are encoded on a single mRNA and utilizing a shared AUG start codon, they are insensitive to changes of reporter mRNA levels. Both constructs were introduced into the set of mutants and wild type control strains (carrying or lacking SUP4) and normalized UAA read-through levels were determined (Fig. 3). In the absence of modification defects, the presence of SUP4 correlated with a 94-fold increase in UAA read-through levels (from 0.4 to ~40%), indicating the suitability of this assay system to quantify SUP4 function. Comparable to results obtained with lacZ based reporters, loss of ELP3 resulted in a significant drop of read-through efficiency to ~15%, equivalent to 37% of wild type level. The normalized read-through level of the mutant was significantly different (p < 0.02) from both the SUP4 strain without modification defect and the control strain lacking SUP4, revealing a substantial residual read-through activity of SUP4 in the absence of mcm5U. As expected from results obtained with can1-100 and ade2-1 alleles (Fig. 1), loss of PUS7 (ψ35), MOD5 (i6A) and NCL1 (m5C) also significantly reduced SUP4 mediated UAA read-through in the dual luciferase reporter, without abolishing it entirely (17–20% residual read-through level, p < 0.02; Fig. 3). In contrast and consistent with results obtained for ade2-1 and can1-100 suppression, deg1 (ψ39) and trm1 (m2,2G) mutations did not significantly lower SUP4 activity. However, a dus3 (D47) mutation, which did not result in detectable loss of ade2-1 and can1-100 suppression mildly lowered read-through in the dual luciferase construct to ~25% (p < 0.05) (Fig. 3).
Figure 3.
Quantification of SUP4 mediated ochre suppression by dual luciferase assays in single and double tRNA modification mutants. (a) Schematic representation of dual luciferase control (pDB688) and ochre stop (pDB723) constructs. (b) Normalized UAA readthrough was calculated for indicated strains by dividing the renilla/firefly activity ratios of the ochre stop construct (pDB723) by the one obtained with the control construct (pDB688) multiplied by 100. All strains are sorted according to readthrough efficiency. Values significantly different from wild type (WT) SUP4 as per two-tailed t-test are indicated (*p < 0.05; **p < 0.02). Actual p-values are indicated. (c) Heat bar representing SUP4 translational efficiency (from 40.5% UAA readthrough to 0.4%) in the absence of indicated modifications.
Combined SUP4 modification loss uncovers functional redundancy
Since individual loss of m5C, mcm5U, i6A, ψ35 and D47 detectably lowered SUP4 activity without abolishing it, whereas m2,2G and ψ39 did not appear to be important (Fig. 3), we decided to investigate the consequence of combined modification loss on SUP4 function and performance. We attempted to generate all possible combinations of modification defects within the extended ASL to quantify the combined impact on SUP4 activity. 20 out of 21 theoretically possible mutant combinations could be generated, all of which grew without severe defects at 30 °C (Fig. S2A). Solely the combination elp3 deg1 was not obtained due to synthetic lethality in the W303-1B derived SUP4 strain (Fig. S2B). We previously generated this mutant combination in a distinct strain background (S288C derived) and observed most severe growth defects and cytological abnormalities30. Thus, we continued the SUP4 functional profiling with the remaining set of 20 viable double mutant combinations and quantified UAA read-through using the dual luciferase assay (Fig. 3). Double and single mutants were sorted according to their remaining SUP4 activity. Most of the modification defects that reduced SUP4 function in single mutants led to stronger reduction in the combination mutants, indicative of independent contributions of the modifications to in vivo function of the tRNA.
Residual read-through levels (UAA read-through %) in double mutants were normalized to the wild type SUP4 strain and compared to levels of the single mutants to categorize the extent of genetic interaction between the modification genes of interest. Using the normalized read-through levels, we calculated the value expected for the respective double mutants (relative activity ∆A * relative activity ∆B) and subtracted it from the experimentally determined value for the double mutant (∆A ∆B), providing a measure of genetic interaction. When the resulting values were sorted, strong synergistic effects could be distinguished from more additive ones (Fig. S3). Strikingly, strong synergistic effects were observed in combinations involving the deg1 and trm1 mutations, which combine ψ39 or m2,2G deficiencies with other modification defects. This result indicates that m2,2G and ψ39, which are dispensable for SUP4 function in the presence of all other modifications become functionally important in the absence of other modifications. Also, the combination of ψ39 and m2,2G deficiency led to a substantial reduction in SUP4 function, despite the absence of detectable effects in the respective single mutants. Whereas most combinations of modification defects still allowed residual SUP4 function (Fig. 3), a near complete loss of SUP4 activity was observed for the mod5 elp3 double mutant simultaneously lacking mcm5U and i6A (Fig. 3).
Genetic interaction of SUP4 relevant modification genes with TEF1/TEF2
The above data suggested a functional interaction between distinct tRNA modifications in maintaining translational efficiency of SUP4. Since tRNA function in translation is strictly dependent on the formation of a ternary complex between eEF1A/GTP and aminoacyl-tRNA, we investigated existing synthetic genetic array datasets27 for genetic interactions between eEF1A (encoded by the paralogs TEF1 and TEF2) and SUP4-relevant tRNA modification genes. TEF1 and TEF2 display negative genetic interactions with 307 and 635 genes, respectively27. Among the strongest negative interactors, we identified several tRNA modification genes involved in mcm5/mcm5s2U formation (Table S2). Gene ontology (GO) analysis also revealed the GO term “tRNA modification” to be significantly enriched in tef1/tef2 negative genetic interactors (p < 2 * 10−5) and identified several of the modification genes shown in here to be relevant for SUP4 function (Elongator and interactor genes, PUS7, MOD5; Table S2). These results suggest that reduced eEF1A abundance enforces the requirement for modifications to maintain tRNA function and prompted us to investigate the functional impact of reducing eEF1A abundance on SUP4 function. Of note, it was already demonstrated that halving the eEF1A gene copy number lowers the efficiency of a tyrosine inserting amber suppressor (SUP7-a)50. While this work in progress, it was further demonstrated that yeast phenotypes caused by loss of the tRNA methyltransferase Trm7 can be aggravated or suppressed, depending on whether eEF1A abundance is lowered or increased51.
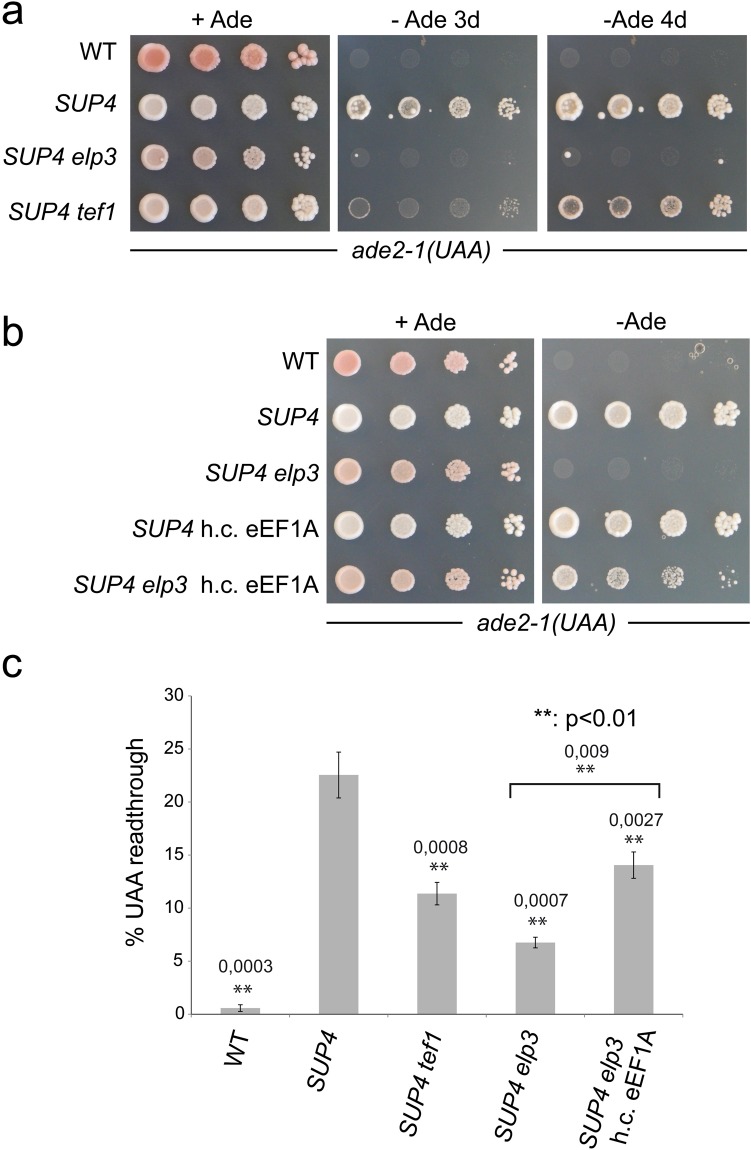
To test whether eEF1A copy number reduction also impairs the ochre suppressor SUP4, we deleted the TEF1 gene in the SUP4 strain used in this study and scored ade2-1 read-through efficiency (Fig. 4). Indeed, TEF1 deletion clearly impaired ade2-1 suppression by SUP4. Reduced nonsense suppression in the tef1 mutant strain was also confirmed using lacZ based reporter constructs52 carrying a premature UAA codon (Fig. 4). In both ade2-1 and lacZ-UAA, tef1 mutation lowered nonsense suppression significantly (p = 0.0008), confirming that eEF1A copy number is critical for SUP4 mediated nonsense read-through. To test whether the partial SUP4 defect caused by half the eEF1A copy number is additive with the negative effect caused by SUP4 hypomodification, we used the SUP4 ncl1 mutant that was partially defective in ade2-1 nonsense suppression (Fig. 1) to generate a SUP4 ncl1 tef1 double mutant. In this mutant both, ade2-1 and can1-100 alleles are less efficiently suppressed by SUP4 compared to either single mutant (Fig. 5). Thus, high eEF1A levels and a complete tRNA modification set independently maintain UAA read-through capacity of SUP4.
Figure 4.
Effect of TEF1 (eEF1A) deletion or overexpression on SUP4 mediated ochre suppression efficiency. (a) Plate assays of ade2-1 suppression in wild type (no SUP4), wild type SUP4 (SUP4) and SUP4 elp3 as well as SUP4 tef1 mutants. −Ade plates were photographed after 3 and 4 days; +Ade plate was photographed after 3 days. (b) Plate assay of ade2-1 suppression of indicated strains. h.c. eEF1A refers to multi-copy TEF1 expression construct (pTEF1). −Ade plate was photographed after 3 days, −Ade plate was photographed after 4 days. (c) Quantification of UAA readthrough in the indicated strains using a lacZ-UAA and a lacZ control construct42. Values significantly different from wild type (WT) SUP4 as per two-tailed t-test are indicated above bars (**p < 0.01). The same test revealed significant difference between SUP4 elp3 and SUP4 elp3 h.c. eEF1A (indicated by bracket). Actual p-values are indicated.
Figure 5.
Additive SUP4 defect in ncl1 tef1 mutants and SUP4 rescue efficiency of other modification mutants by overexpression of TEF1. (a) Plate assays for ade2-1 (−Ade) and can1-100 (−Arg/+Can) readthrough in ncl1 tef1 mutants compared to respective single mutants. (b) Plate assay for ade2-1 readthrough in indicated tRNA modification mutants carrying either vector (YEplac181) or overexpression construct for TEF1 (h.c. eEFA, pTEF1). Plates were photographed at indicated time points. YPD plate in (B) was photographed after 48 h.
Functional rescue of hypomodified SUP4 by elevated eEF1A levels
Since the above results suggest that eEF1A abundance and tRNA modifications independently promote translational efficiency of the SUP4 tRNA, we investigated whether SUP4 defects associated with modification loss can be compensated by elevated eEF1A levels. We used a multicopy construct containing the TEF1 gene (pTEF1)53 and introduced it into the SUP4 elp3 strain defective in ade2-1 ochre suppression. As shown in Fig. 4B, there is a clear restoration of growth on −Ade media of the SUP4 elp3 strain, dependent on the TEF1 expression construct. Also, read-through in the lacZ-UAA reporter is partially restored in elp3 cells overexpressing eEF1A compared to the elp3 strain without extra pTEF1. Thus, the functional defect in SUP4 tRNA induced by loss of the mcm5U modification can be partially compensated by elevated eEF1A levels. To test whether this effect is unique to the mcm5U defective elp3 strain or more general and extends to other modification genes, deletion of which also impairs SUP4 nonsense suppressor efficiency, we analysed rescue by elevated eEF1A levels of ade2-1 suppression in pus7 and ncl1 mutants. As shown in Fig. 5B, overexpression of TEF1 indeed partially restored nonsense suppression in these mutant backgrounds, consistent with an ability of elevated eEF1A levels to compensate functional defects induced by various tRNA modification defects.
Rescue of modification defects in non-suppressor tRNA by elevated eEF1A levels
Combined loss of mcm5/s2U and ψ38 was shown to induce severe growth defects that correlate with translational inefficiency of tRNAGlnUUG30,54. Since these defects were suppressed by elevated copy numbers of hypomodified tRNAGlnUUG and this tRNA represents the single tRNA in yeast, which carries mcm5s2U and ψ38, simultaneous absence of these modifications apparently results in a severe loss of function of particularly this tRNA species. TEF1/TEF2 are negative genetic interactors of several tRNA modification genes (Table S2) and elevated levels of eEF1A were shown in this study to rescue hypomodified SUP4 (Figs 4B, 5B). Therefore, we investigated whether higher than normal levels of eEF1A can also rescue functional defects in natural, non-suppressor tRNAs.
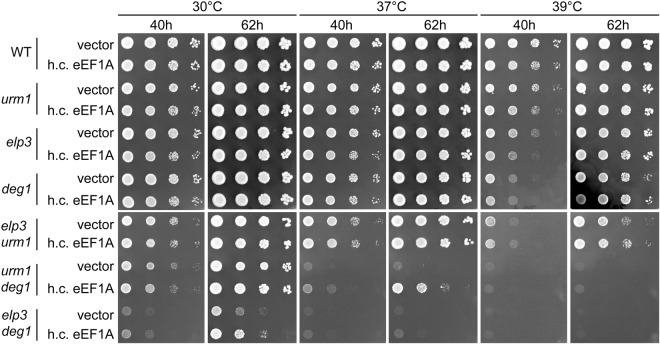
To do so we tested whether growth defects in deg1 urm1 (ψ38/s2U) or deg1 elp3 (ψ38/mcm5U) under non-stressed or temperature stress (TS) conditions are ameliorated upon introduction of the TEF1 multicopy construct (pTEF1) that rescued hypomodified SUP4 (Fig. 5B). As shown in Fig. 6, there is indeed a clear improvement of growth of the deg1 urm1 and deg1 elp3 mutants, but not of the respective single mutants upon overexpression of eEF1A. Other than deg1 elp3, which is already severely impaired in growth under non-stressed conditions, the deg1 urm1 mutant shows a severe growth defect only under conditions of mild heat stress (37 °C; Fig. 6). Growth of deg1 elp3 under non-stressed conditions was partially rescued by pTEF1, whereas in urm1 deg1, pTEF1 was clearly beneficial under mild heat stress condition (Fig. 6). Rescue of the TS phenotype in urm1 deg1 was only partial at 37 °C and undetectable at further elevated temperatures, suggesting that elevated eEF1A levels can improve functionality of hypomodified tRNA, but not to the level of fully modified tRNA. A sharp difference of growth responses between 37° and 39 °C was observed for several yeast strains defective in tRMA modification (Fig. S2)30,54. A partial phenotypic rescue at elevated temperature was also detected in a distinct double mutant (tcd1 urm1) lacking ct6A and s2U, in which tRNALysUUU displays reduced functionality30 (Fig. S4), suggesting that functional defects in tRNAGlnUUG and tRNALysUUU in absence of critical modifications can be partly suppressed by elevating eEF1A abundance.
Figure 6.
Heat sensitivity and growth defects in single and double tRNA modification mutants in the presence and absence of TEF1 overexpression. h.c. eEF1A refers to overexpression of TEF1 (pTEF1). Vector refers to YEplac181. Cells were taken from selective YNB medium and dilutions prepared in sterile water. After spotting of cell dilutions on YPD plates, these were incubated at 30 °C, 37 °C or 39 °C as indicated. Plates were photographed at the indicated time points.
Discussion
In this study, we quantified the functional relevance of all modifications within the extended ASL (including the VL) of a single tRNA species (SUP4) in vivo. Previous studies indicated a relevant contribution of ASL modifications mcm5U, ψ35 and i6A10,34,35. Indeed, all reporters used to measure SUP4 efficiency identified loss of nonsense suppression in elp3, pus7 and mod5 mutants lacking these modifications individually. This result confirms previous studies of elp3 and mod5 mutants10,34 and provides first direct evidence for a crucial role of ψ35 in SUP4 function. While it was shown that absence of an intron in the SUP6 tRNATyr derived ochre suppressor gene abolished both ψ35 formation and nonsense suppression35, a direct demonstration that the modification itself is important for suppressor tRNA function was lacking and is provided in the current study (Figs 1; 3). Since ψ can stabilize base pairs in RNA duplexes55, functional roles of the U to ψ isomerization at position 35 in SUP4 could be to improve codon binding and competition against release factors. Consistent with Pus7 as the sole in vivo modifier for U35 in pre-tRNATyr38, we detected no functional relevance of the related Pus1 synthase in supporting nonsense suppression of ade2-1 or can1-100 by SUP4.
Classical reporters used to detect defects in nonsense suppression rely on the reversion of phenotypes linked to nonsense codons in genes causing auxotrophies or drug resistances. We find that elp3, mod5 and pus7 mutations confer adenine auxotrophy to a SUP4 ade2-1 can1-100 strain and cause canavanine resistance, at first sight consistent with full loss of SUP4 function. However, red pigmentation associated with ADE2 gene defects is incompletely restored (Fig. 1) suggesting residual SUP4 translational activity even in the absence of these modifications, which was already demonstrated for an elp3 mutant using lacZ based reporter constructs4,42.
Although formally possible, we demonstrated that SUP4 dependent suppression of ade2-1 and can1-100 does not involve an inhibition of NMD. First, inhibition of NMD by deletion of UPF1 in the absence of SUP4 suppressor tRNA did not revert ade2-1 or can1-100 phenotypes under the conditions tested (Fig. 2). Second, SUP4 clearly affects ade2-1 and can1-100 phenotypes in the absence of NMD, ruling out the option that SUP4 functions mainly via NMD inhibition. However, the restoration of SUP4 dependent ade2-1 and can1-100 suppression in the absence of ELP3 by inactivation of NMD (Fig. 2) also indicates that changes in reporter mRNA levels can affect total SUP4 dependent nonsense read-through.
To quantify the effect of modification defects on SUP4 function independent of potential mRNA fluctuations, we utilized dual luciferase reporter constructs49 and measured residual SUP4 function in modification mutants. These constructs utilize an internal normalization of luciferase activity generated after nonsense read-through to an upstream reporter expressed independently of nonsense suppression and are therefore insensitive to changes of reporter mRNA levels. As expected, mod5, pus7 and elp3 mutations significantly reduced SUP4 dependent nonsense suppression in this reporter without entirely abolishing it. In addition, ncl1 and dus3 mutations (lack of m5C or D47) reduced SUP4 functions significantly. The ncl1 effect correlated with a full loss of can1-100 suppression and a partial defect in ade2-1 suppression, while the dus3 mutation did not affect these reporters and lowered dual luciferase based SUP4 read-out less drastically compared to ncl1, elp3, mod5 or pus7 mutations.
In tRNATyr, the Ncl1 dependent m5C is present at position 48, which forms a non-canonical basepair (Levitt basepair) with the nucleoside in position 15 during tRNA folding2,56. However, multiple tRNAs naturally lack this modification and form the Levitt pair without m5C modification2,36. Therefore, the contribution of the m5C modification to tRNA stability and function remains obscure. To our knowledge, this study provides first in vivo evidence for a clear positive role of m5C48 modification in maintaining function of a defined tRNA. While absence of Ncl1 is tolerated without severe phenotypes in yeast (Fig. S2), mutation of the human NCL1 orthologue NSUN2 is linked to intellectual disability, like several other mutations affecting tRNA modifiers26,57. Thus, m5C likely fulfils important functions in yeast and human tRNAs.
In contrast to anticodon loop modifications and m5C, neither m2,2G nor ψ39 influenced SUP4 function significantly in any of the reporters used. Hence, there is a differential importance of individual modifications for SUP4 function. While all the anticodon loop modifications are important on their own, stem and VL modifications can be either dispensable (m2,2G, ψ39) or important (m5C, D47). The dispensability of ψ39 for SUP4 function was also observed using a lacZ based reporter42 and provides an example for an extreme position effect of modification importance: Pus7 dependent ψ within the anticodon loop (position 35) is crucial, whereas Deg1 dependent introduction of the same modification in the anticodon stem (position 39) is much less important. Since ψ is thought to contribute rigidity and reduce flexibility of the sugar phosphate backbone58, its presence in the loop might be of higher functional relevance compared to the stem location due to a potential contribution to stabilizing the open loop configuration. Indeed, in the sup70–65 amber suppressor derived from tRNAGlnCUG, in which a Deg1 dependent ψ is in position 38 (loop), this modification is crucial for stop codon read-through42,59. However, not only ψ35 but also ψ39 appears to be conserved within tRNATyr as a large majority of sequenced eukaryotic tRNATyr species contain this modification or one of its derivatives (1-methylpseudouridine, 2′-O-methylpseudouridine)36. Thus, despite the absence of effects on SUP4 upon its removal alone, a subtler contribution of ψ39 to SUP4 function appears likely.
One possibility to explain absence of effects in single mutant backgrounds is functional redundancy with other modifications. In such case, effects would only be observable in combined absence with modifications that contribute to the same functional aspect. Indeed, our double mutant approach revealed a reduction of SUP4 function upon removal of ψ39 when the tRNA was already missing any of the extended ASL modifications, except for mcm5U which could not be tested due to synthetic lethality (Fig. 3). Due to its position in the anticodon stem, ψ39 might improve RNA duplex stability which may become critical only after additional modification defects that destabilize tRNA by other means. A similar trend was observed for Trm1 dependent m2,2G, which was dispensable alone but absence of which clearly reduced SUP4 function when this tRNA was already lacking any of the anticodon loop modifications. m2,2G contributes to tRNA folding by preventing base pairing within the internal loop but this contribution is not essential, since several tRNAs lack this modification and trm1 mutation is tolerated without severe phenotypes in yeast2,36 (Fig. S2). Again, upon introduction of additional modification defects that may weaken structural stability of the tRNA via a distinct mechanism, more drastic functional defects might be induced. Such effect was most pronounced for the combination of i6A and m2,2G defects, which had a strong synergistic negative effect on SUP4 function (Figs 3, S3). In general, absence of i6A amplified the negative effects induced by most additional modification defects, and combination of i6A and mcm5U deficiencies nearly entirely eliminated residual SUP4 function (Fig. 3). A37 is at risk of pairing with the conserved U33, which results in destabilization of the anticodon loop and which is why A37 is often modified by bulky side chains (i6A, ct6A, yW) occupying the Watson-Crick interface1,6,36. Absence of i6A37 modification therefore likely has a destabilizing effect on the anticodon loop which may sensitize tRNA to loss of other modifications (such as m2,2G) that would further contribute to tRNA structure and stability. Since mcm5s2U was previously identified to contribute to tRNALysUUU anticodon loop prestructuring along with the A37 modification 2-methylthio-N6-threonylcarbamoyladenosine6, it appears very likely that a similar functional collaboration exists between the mcm5U and i6A modifications in the SUP4 tRNA.
Our study further indicates that eEF1A abundance constitutes an independent factor which modulates the UAA read-through capacity of the SUP4 suppressor tRNA. In the presence of all modifications, SUP4 efficiency is clearly decreased upon reducing eEF1A gene dosage from its natural two copies (TEF1/TEF2) to one (tef1/TEF2) (Figs 4A,C, 5A). This negative effect is additive with a partial loss of SUP4 function caused by the absence of the m5C modification (Fig. 5A). While m5C deficient SUP4 is still able to mediate partial suppression of ade2-1, this property is lost upon additional deletion of one of the two eEF1A encoding genes. This result indicates that tRNA modifications which are of minor functional relevance under standard conditions, can become rather critical under conditions of reduced eEF1A availability. One prediction of such contribution of eEF1A abundance to tRNA efficiency would be that functional impairments by loss of critical modifications could potentially be compensated by elevating eEF1A levels. Indeed, our results support this notion, since ade2-1 nonsense suppression defects of SUP4 tRNA lacking either mcm5U, ψ35 or m5C can be significantly rescued by overexpression of eEF1A (Figs 4B, 5B). In addition, the TS phenotype caused by dual modification loss in two natural non-suppressor tRNA species, tRNAGlnUUG or tRNALysUUU, can be partially suppressed by overexpression of eEF1A (Figs 6, S4). Interestingly, a similar TS phenotype caused by a defect of tRNAPhe in trm7 mutants was recently shown to be partially suppressible by an additional copy of TEF1/251. Thus, the positive effect of increasing eEF1A abundance on tRNA function in vivo extends from nonsense suppressor tRNA to at least three different natural tRNAs lacking critical modifications. Together our results indicate that tRNA modifications and eEF1A abundance constitute factors that appear to independently control the translational efficiency of tRNAs. If both are impaired, additive negative effects can be demonstrated, whereas elevated levels of eEF1A might generally compensate for defects caused under conditions of inappropriate tRNA modifications.
Methods
General methods
Yeast strains used in this study are listed in Table S1 and were cultivated in yeast peptone dextrose (YPD) or yeast nitrogen base minimal medium lacking specific nutrients to select for gene deletions or plasmids60. For targeted gene deletion, PCR products containing auxotrophic marker genes (KlLEU2 or SpHIS5) with flanking regions of 50 nucleotides homology to target genes were generated according to61 and transformed into yeast strains using the lithium acetate method62. Correct target gene replacement was verified using PCR and involved marker gene specific oligonucleotides in combination with primers binding the target locus outside of the region covered by the gene deletion PCR product. A plasmid shuffle assay to check for viability of an elp3 deg1 double mutant involved pFF8 (CEN-ELP3-URA3)63 and subsequent counterselection on 5-FOA containing medium as described earlier30.
Phenotypic growth and nonsense suppression assays
Yeast strains were grown on YPD or leucine free YNB solid media to select for the presence of plasmids YEp181 or pTEF1 (multi copy TEF1)53 for 2 to 3 days. Subsequently, cells were recovered from the plates and resuspended in sterile water and dilutions prepared with OD600nm values of 0.15, 0.015, 0.0015 and 0.00015. These were spotted on either solid YPD medium or YNB medium lacking adenine or lacking arginine and containing canavanine sulfate (60 µg/ml). Subsequently, plates were incubated at indicated temperatures for 30–90 h and photographed at indicated time points.
Beta galactosidase assay
Yeast strains transformed with pUKC815 (wild type lacZ) or pUKC817 (UAA ochre insertion in lacZ)52 were grown in uracil free YNB medium to OD600 nm of 2–3. Cells were washed once with Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 50 mM 2-mercaptoethanol, pH 7) and subsequently resuspended using the same buffer. Cell density at 600 nm was measured and 500 µL aliquots transferred to new tubes. To each, two drops of 0.01% sodium dodecyl sulphate (SDS) solution and chloroform were added, followed by vigorous mixing and incubation at 37 °C for 5 min. Reactions were started by addition of 100 µL of 4 mg·mL−1 ortho-nitrophenyl-β-galactoside dissolved in Z-buffer and stopped by the addition of 250 µL 1 M Na2CO3. Activity units were calculated using OD420 nm absorbance of samples by employing Miller’s formula64. Relative read-through efficiency (%) was determined using the ratio of the beta galactosidase activities measured with the pUKC817 and pUKC815 constructs. For each strain, at least three independent cultures were measured with both constructs. Significant differences of SUP4 activity in the wild type and mutant strains or between the elp3 mutant with and without eEF1A overexpression were determined using two-tailed t-test as described before for yeast beta galactosidase reporter assays23.
Dual luciferase assay
Dual luciferase assays were performed using the dual luciferase reporter (DLR) assay kit (Promega). Yeast strains of the W303-1B background with or without SUP4 were transformed separately with two different dual luciferase reporter plasmids49, the reporter construct containing a UAA nonsense codon (pDB723) or the control construct (pDB688) containing a sense codon at the same position between the renilla and firefly luciferase genes. Strains were grown in uracil free YNB medium and assayed for luminescence with a Promega Glomax luminometer as described49. The relative read-through level (read-through %) was calculated using the firefly-renilla luciferase activity (nonsense) ratio divided by the firefly-renilla luciferase activity (sense) ratio multiplied by 100. At least four independent transformants for each strain and construct were measured. Significant differences of SUP4 activity in the absence and presence of modification defects were determined by two-tailed t-test as described before for dual luciferase assays in yeast23.
Electronic supplementary material
Acknowledgements
We thank D. Bedwell, D. Amberg and M.F. Tuite for providing plasmids. We gratefully acknowledge funding support by the Deutsche Forschungsgemeinschaft (DFG) to R.S. (SCHA750/15-2) and their Priority Programme SPP1784 ‘Chemical Biology of Native Nucleic Acid Modifications’ to R.S. (SCHA750/20-2) and R.K. (KL2937/1-2). In addition, part of this work was carried out in the framework of the European Union Cost Action EPITRAN CA16120.
Author Contributions
R.K. performed experiments; R.K. and R.S. analysed the data; R.K. and R.S. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31158-2.
References
- 1.Agris, P. F. et al. Celebrating wobble decoding. Half a century and still much is new. RNA biology 1–17 (2017). [DOI] [PMC free article] [PubMed]
- 2.Väre, V. Y. P., Eruysal, E. R., Narendran, A., Sarachan, K. L. & Agris, P. F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules7 (2017). [DOI] [PMC free article] [PubMed]
- 3.Grosjean H, Westhof E. An integrated, structure- and energy-based view of the genetic code. Nucleic acids research. 2016;44:8020–8040. doi: 10.1093/nar/gkw608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson MJO, Esberg A, Huang B, Björk GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Molecular and cellular biology. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rezgui VAN, et al. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12289–12294. doi: 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vendeix FAP, et al. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. Journal of molecular biology. 2012;416:467–485. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranjan N, Rodnina MV. Thio-Modification of tRNA at the Wobble Position as Regulator of the Kinetics of Decoding and Translocation on the Ribosome. Journal of the American Chemical Society. 2017;139:5857–5864. doi: 10.1021/jacs.7b00727. [DOI] [PubMed] [Google Scholar]
- 8.Schaffrath R, Leidel SA. Wobble uridine modifications–a reason to live, a reason to die?! RNA biology. 2017;14:1209–1222. doi: 10.1080/15476286.2017.1295204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Molecular and cellular biology. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang B, Johansson MJO, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA (New York, N.Y.) 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA (New York, N.Y.) 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fichtner L, et al. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Molecular microbiology. 2002;43:783–791. doi: 10.1046/j.1365-2958.2002.02794.x. [DOI] [PubMed] [Google Scholar]
- 13.Bär C, Zabel R, Liu S, Stark MJR, Schaffrath R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes. Kti11/Dph3. Molecular microbiology. 2008;69:1221–1233. doi: 10.1111/j.1365-2958.2008.06350.x. [DOI] [PubMed] [Google Scholar]
- 14.Zabel R, Bär C, Mehlgarten C, Schaffrath R. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Molecular microbiology. 2008;69:175–187. doi: 10.1111/j.1365-2958.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- 15.Mehlgarten C, Jablonowski D, Breunig KD, Stark MJR, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Molecular microbiology. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Fattah W, et al. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS genetics. 2015;11:e1004931. doi: 10.1371/journal.pgen.1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehlgarten, C. et al. Use of a Yeast tRNase Killer Toxin to Diagnose Kti12 Motifs Required for tRNA Modification by Elongator. Toxins9 (2017). [DOI] [PMC free article] [PubMed]
- 18.Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. The Journal of biological chemistry. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 19.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic acids research. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leidel S, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 21.Jüdes A, Bruch A, Klassen R, Helm M, Schaffrath R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microbial cell (Graz, Austria) 2016;3:554–564. doi: 10.15698/mic2016.11.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tükenmez H, Xu H, Esberg A, Byström AS. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic acids research. 2015;43:9489–9499. doi: 10.1093/nar/gkv832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klassen R, Bruch A, Schaffrath R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA biology. 2017;14:1252–1259. doi: 10.1080/15476286.2016.1267098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klassen R, et al. Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PloS one. 2015;10:e0119261. doi: 10.1371/journal.pone.0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokołowski, M., Klassen, R., Bruch, A., Schaffrath, R. & Glatt, S. Cooperativity between different tRNA modifications and their modification pathways. Biochimica et biophysica acta1861, 409–418 (2017). [DOI] [PubMed]
- 27.Costanzo, M. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science (New York, N.Y.)353 (2016). [DOI] [PMC free article] [PubMed]
- 28.Lecointe F, et al. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. The Journal of biological chemistry. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 29.Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nature chemical biology. 2013;9:105–111. doi: 10.1038/nchembio.1137. [DOI] [PubMed] [Google Scholar]
- 30.Klassen R, et al. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic acids research. 2016;44:10946–10959. doi: 10.1093/nar/gkw705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agris PF. Bringing order to translation. The contributions of transfer RNA anticodon-domain modifications. EMBO Reports. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman HM, Olson MV, Hall BD. Nucleotide sequence of a mutant eukaryotic gene. The yeast tyrosine-inserting ochre suppressor SUP4-o. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator’s key role for zymocin-induced cell death in yeast. Molecular microbiology. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- 34.Laten HM. Antisuppression of class I suppressors in an isopentenylated-transfer RNA deficient mutant of Saccharomyces cerevisiae. Current genetics. 1984;8:29–32. doi: 10.1007/BF00405428. [DOI] [PubMed] [Google Scholar]
- 35.Johnson PF, Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983;302:681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- 36.Boccaletto P, et al. MODOMICS. A database of RNA modification pathways. 2017 update. Nucleic acids research. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. The Journal of biological chemistry. 1986;261:9703–9709. [PubMed] [Google Scholar]
- 38.Behm-Ansmant I, et al. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA (New York, N.Y.) 2003;9:1371–1382. doi: 10.1261/rna.5520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dihanich ME, et al. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Molecular and cellular biology. 1987;7:177–184. doi: 10.1128/MCB.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing F, Hiley SL, Hughes TR, Phizicky EM. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. The Journal of biological chemistry. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 41.Motorin Y, Grosjean H. Multisite-specific tRNA. M5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA (New York, N.Y.) 1999;5:1105–1118. doi: 10.1017/S1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klassen, R. & Schaffrath, R. Role of Pseudouridine Formation by Deg1 for Functionality of Two Glutamine Isoacceptor tRNAs. Biomolecules7 (2017). [DOI] [PMC free article] [PubMed]
- 43.Motorin Y, et al. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA (New York, N.Y.) 1998;4:856–869. doi: 10.1017/S1355838298980396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Molecular and cellular biology. 2000;20:4591–4603. doi: 10.1128/MCB.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luke B, et al. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic acids research. 2007;35:7688–7697. doi: 10.1093/nar/gkm912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kervestin S, Jacobson A. NMD. A multifaceted response to premature translational termination. Nature reviews. Molecular cell biology. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi T, Funakoshi Y, Hoshino S-I, Katada T. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. The Journal of biological chemistry. 2004;279:45693–45700. doi: 10.1074/jbc.M405163200. [DOI] [PubMed] [Google Scholar]
- 48.Czaplinski K, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & development. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keeling KM, et al. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA (New York, N.Y.) 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song JM, et al. Elongation factor EF-1 alpha gene dosage alters translational fidelity in Saccharomyces cerevisiae. Molecular and cellular biology. 1989;9:4571–4575. doi: 10.1128/MCB.9.10.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han L, Guy MP, Kon Y, Phizicky EM. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS genetics. 2018;14:e1007288. doi: 10.1371/journal.pgen.1007288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stansfield I, Akhmaloka, Tuite MF. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Current genetics. 1995;27:417–426. doi: 10.1007/BF00311210. [DOI] [PubMed] [Google Scholar]
- 53.Haarer B, et al. Actin dosage lethality screening in yeast mediated by selective ploidy ablation reveals links to urmylation/wobble codon recognition and chromosome stability. G3 (Bethesda, Md.) 2013;3:553–561. doi: 10.1534/g3.113.005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han L, Kon Y, Phizicky EM. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA (New York, N.Y.) 2015;21:188–201. doi: 10.1261/rna.048173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kierzek E, et al. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic acids research. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliva R, Tramontano A, Cavallo L. Mg2+ binding and archaeosine modification stabilize the G15 C48 Levitt base pair in tRNAs. RNA (New York, N.Y.) 2007;13:1427–1436. doi: 10.1261/rna.574407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abbasi-Moheb L, et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. American journal of human genetics. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spenkuch F, Motorin Y, Helm M. Pseudouridine. Still mysterious, but never a fake (uridine)! RNA biology. 2014;11:1540–1554. doi: 10.4161/15476286.2014.992278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kemp AJ, et al. A yeast tRNA mutant that causes pseudohyphal growth exhibits reduced rates of CAG codon translation. Molecular microbiology. 2013;87:284–300. doi: 10.1111/mmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman F. Getting started with yeast. Methods in enzymology. 2002;350:3–41. doi: 10.1016/S0076-6879(02)50954-X. [DOI] [PubMed] [Google Scholar]
- 61.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic acids research. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature protocols. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 63.Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. The EMBO journal. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller, J. H. Experiments in Molecular Genetics (Cold Spring Harbor Laboratory Press, New York, NY, USA, 1972).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.